Themed collection Molecular BioSystems Network Biology

Network Biology editorial 2013
The guest editors introduce the Network Biology themed issue.
Mol. BioSyst., 2013,9, 1557-1558
https://doi.org/10.1039/C3MB90018E
Social networks to biological networks: systems biology of Mycobacterium tuberculosis
Contextualizing relevant information to construct a network that represents a given biological process presents a fundamental challenge in the network science of biology.
Mol. BioSyst., 2013,9, 1584-1593
https://doi.org/10.1039/C3MB25546H
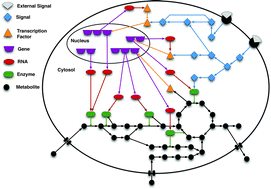
Bridging the layers: towards integration of signal transduction, regulation and metabolism into mathematical models
Mathematical modelling is increasingly becoming an indispensable tool for the study of cellular processes, allowing their analysis in a systematic and comprehensive manner.
Mol. BioSyst., 2013,9, 1576-1583
https://doi.org/10.1039/C3MB25489E
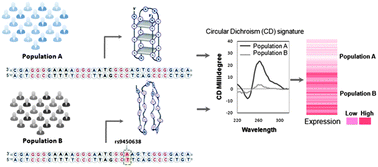
Emerging trends in G-quadruplex biology – role in epigenetic and evolutionary events
This review introduces quadruplex motifs in roles supporting epigenetic events, in determining evolutionary selection and as determinants of quantitative trait loci.
Mol. BioSyst., 2013,9, 1568-1575
https://doi.org/10.1039/C3MB25492E
Building synthetic gene circuits from combinatorial libraries: screening and selection strategies
An overview of how screening or selection of combinatorial libraries can be used to build synthetic gene circuits.
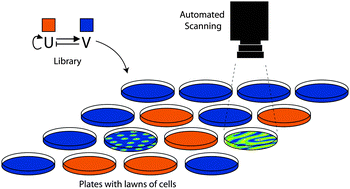
Mol. BioSyst., 2013,9, 1559-1567
https://doi.org/10.1039/C2MB25483B
A design automation framework for computational bioenergetics in biological networks
Investigating the bioenergetic activity of the mitochondrial and algal metabolisms using optimization methods.
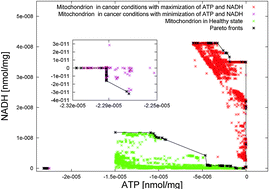
Mol. BioSyst., 2013,9, 2554-2564
https://doi.org/10.1039/C3MB25558A
Logical modelling of Drosophila signalling pathways
This manuscript presents predictive logical models for the nine main signalling pathways recurrently used in Drosophila development.
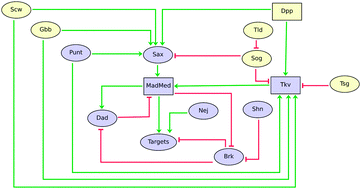
Mol. BioSyst., 2013,9, 2248-2258
https://doi.org/10.1039/C3MB70187E
Network properties of decoys and CASP predicted models: a comparison with native protein structures
The protein structure network [PSN] profile clearly distinguishes the native [black] from the non-native [red, pink] models.
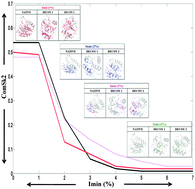
Mol. BioSyst., 2013,9, 1774-1788
https://doi.org/10.1039/C3MB70157C
Structural comparison of biological networks based on dominant vertices
A mathematical tool to compare topology and dynamics of six bacterial networks with equivalent homogeneous, scale-free and geometric random networks.
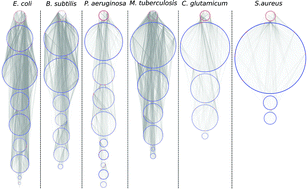
Mol. BioSyst., 2013,9, 1765-1773
https://doi.org/10.1039/C3MB70077A
Graphical modelling of molecular networks underlying sporadic inclusion body myositis
Here we present a novel statistical methodology that allows us to analyze gene expression data that have been collected from a number of different cases or conditions in a unified framework.
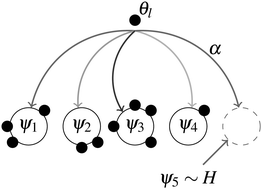
Mol. BioSyst., 2013,9, 1736-1742
https://doi.org/10.1039/C3MB25497F
Transcriptional feedback in the insulin signalling pathway modulates ageing in both Caenorhabditis elegans and Drosophila melanogaster
Using manually curated signalling models of the IIS/TOR pathways, transcriptional feedback is shown to modulate ageing in worms and flies.
Mol. BioSyst., 2013,9, 1756-1764
https://doi.org/10.1039/C3MB25485B
Large-scale metabolome analysis and quantitative integration with genomics and proteomics data in Mycoplasma pneumoniae
Increased availability of quantitative -omics data from the same biological source allows integrative studies, yielding functional conclusions on cell physiology.
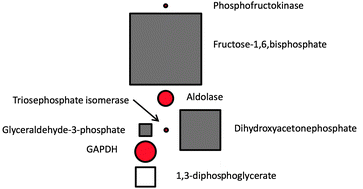
Mol. BioSyst., 2013,9, 1743-1755
https://doi.org/10.1039/C3MB70113A
Network function shapes network structure: the case of the Arabidopsis flower organ specification genetic network
Many biological networks have been reconstructed, leading to the question of how to interpret their structural features.
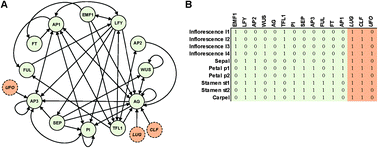
Mol. BioSyst., 2013,9, 1726-1735
https://doi.org/10.1039/C3MB25562J
Tethering preferences of domain families co-occurring in multi-domain proteins
Genomic data of several organisms have revealed that domain families in multi-domain proteins have diverse tethering preferences.
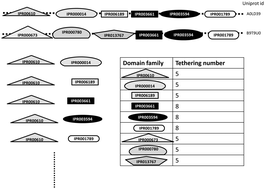
Mol. BioSyst., 2013,9, 1708-1725
https://doi.org/10.1039/C3MB25481J
Oligomerisation status and evolutionary conservation of interfaces of protein structural domain superfamilies
Protein-protein interactions can also be represented into networks.
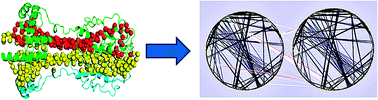
Mol. BioSyst., 2013,9, 1652-1661
https://doi.org/10.1039/C3MB25484D
Stress induces remodelling of yeast interaction and co-expression networks
Network approaches applied to microarray, RNA-seq and proteomics data reveal that stress induces modularization in co-expression and protein interaction networks.

Mol. BioSyst., 2013,9, 1697-1707
https://doi.org/10.1039/C3MB25548D
Finding the targets of a drug by integration of gene expression data with a protein interaction network
Finding the targets of a drug: a matter of looking into the neighborhood.
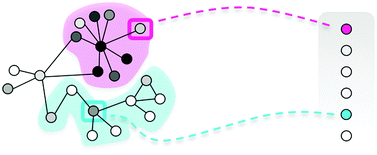
Mol. BioSyst., 2013,9, 1676-1685
https://doi.org/10.1039/C3MB25438K
A disease-drug -phenotype matrix inferred by walking on a functional domain network
The dcGOnet is a functional domain network that can be globally explored for linking together diseases, drugs and cross-species phenotypes.
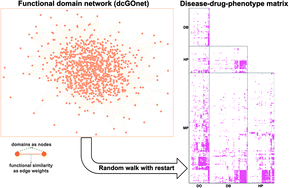
Mol. BioSyst., 2013,9, 1686-1696
https://doi.org/10.1039/C3MB25495J
Quantifying the similarity of monotonic trajectories in rough and smooth fitness landscapes
When selection is strong and mutations are rare, evolution can be thought of as an uphill trajectory in a rugged fitness landscape.
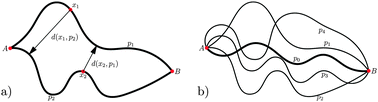
Mol. BioSyst., 2013,9, 1627-1631
https://doi.org/10.1039/C3MB25553K
DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle
The chromosomal DNA polymer constituting the cellular genetic material is primarily a device for coding information.
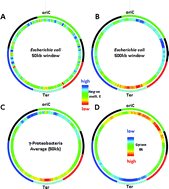
Mol. BioSyst., 2013,9, 1643-1651
https://doi.org/10.1039/C3MB25515H
A study of Caenorhabditis elegans DAF-2 mutants by metabolomics and differential correlation networks
Pearson and partial correlation analysis was used to build networks to explore the central role of daf-2 in regulating fatty acid and amino acid metabolism in Caenorhabditis elegans.
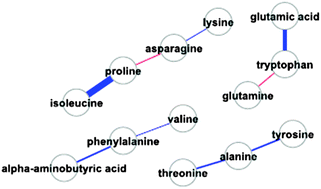
Mol. BioSyst., 2013,9, 1632-1642
https://doi.org/10.1039/C3MB25539E
MGclus: network clustering employing shared neighbors
MGclus is a new algorithm designed to detect network modules by accounting for shared network neighbors in large scale biological interaction networks.
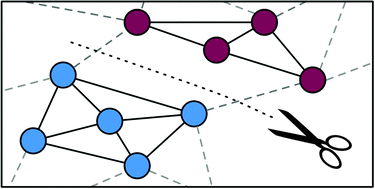
Mol. BioSyst., 2013,9, 1670-1675
https://doi.org/10.1039/C3MB25473A
Regulation of protein –protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes
A significant association was found between phosphorylation, disorder and binding for serine and threonine in human protein complexes.
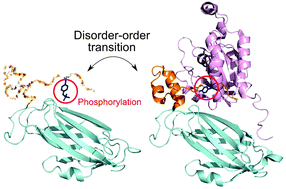
Mol. BioSyst., 2013,9, 1620-1626
https://doi.org/10.1039/C3MB25514J
iPoint: an integer programming based algorithm for inferring protein subnetworks
iPoint attains very compact and accurate solutions that outperform previous network inference algorithms.
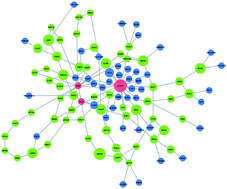
Mol. BioSyst., 2013,9, 1662-1669
https://doi.org/10.1039/C3MB25432A
Predicting cancer drug mechanisms of action using molecular network signatures
Molecular signatures are a powerful approach to characterize novel small molecules and derivatized small molecule libraries.
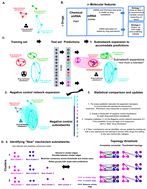
Mol. BioSyst., 2013,9, 1604-1619
https://doi.org/10.1039/C2MB25459J
PheNetic: network-based interpretation of unstructured gene lists in E. coli
PheNetic, a method to use interaction networks for the interpretation of in-house generated omics data.
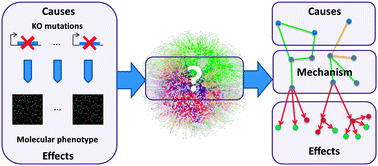
Mol. BioSyst., 2013,9, 1594-1603
https://doi.org/10.1039/C3MB25551D
About this collection
Molecular BioSystems brings together cutting-edge research addressing the many and varied aspects of Network Biology in this themed web collection. This collection of articles includes reviews and papers from a themed issue on Network Biology, guest edited by Edward Marcotte, Charles Boone, M. Madan Babu and Anne-Claude Gavin.