PheNetic: network-based interpretation of unstructured gene lists in E. coli†
Abstract
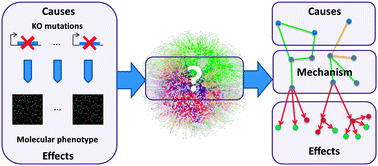
At the present time, omics experiments are commonly used in wet lab practice to identify leads involved in interesting phenotypes. These omics experiments often result in unstructured gene lists, the interpretation of which in terms of pathways or the mode of action is challenging. To aid in the interpretation of such gene lists, we developed PheNetic, a decision theoretic method that exploits publicly available information, captured in a comprehensive interaction network to obtain a mechanistic view of the listed genes. PheNetic selects from an interaction network the sub-networks highlighted by these gene lists. We applied PheNetic to an Escherichia coli interaction network to reanalyse a previously published KO compendium, assessing gene expression of 27 E. coli knock-out mutants under mild acidic conditions. Being able to unveil previously described mechanisms involved in acid resistance demonstrated both the performance of our method and the added value of our integrated E. coli network. PheNetic is available at http://bioi.biw.kuleuven.be/~driesdm/phenetic/.
- This article is part of the themed collection: Molecular BioSystems Network Biology

 Please wait while we load your content...
Please wait while we load your content...