Structural comparison of biological networks based on dominant vertices†
Abstract
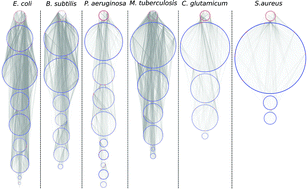
It is a current practice to organize biological data in a network structure where vertices represent biological components and arrows represent their interactions. A great diversity of graph theoretical notions, such as clustering coefficient, network motifs, centrality, degree distribution, etc., have been developed in order to characterize the structure of these networks. However, none of the existent characterizations allow us to determine global similarity among networks of different sizes. It is the aim of the present paper to introduce a mathematical tool to compare networks not only with regard to their topological structure, but also in their dynamical capabilities. For this reason we aim to propose a pseudo-distance between networks, built around the notions of determination and dominancy, concepts recently introduced in the context of regulatory dynamics on networks. We use our proposed pseudo-distance to compare networks from the following bacteria: E. coli, B. subtilis, P. aeruginosa, M. tuberculosis, S. aureus and C. glutamicum. We also use this pseudo-distance to compare these real bacterial networks with equivalent homogeneous, scale-free and geometric three dimensional random networks. We found that even when bacterial networks are characterized with different levels of detail, have different sizes and represent different aspects of the organisms, the proposed pseudo-distance captures all these characteristics, and indicates how similar they are or not from random networks.
- This article is part of the themed collection: Molecular BioSystems Network Biology

 Please wait while we load your content...
Please wait while we load your content...