Themed collection The free energy landscape: from folding to cellular function

A second molecular biology revolution? The energy landscapes of biomolecular function
This themed issue showcases a collection of work from leading scientists, demonstrating the significance of the free energy landscape in the field of physiochemical biology.
Phys. Chem. Chem. Phys., 2014,16, 6321-6322
https://doi.org/10.1039/C4CP90027H
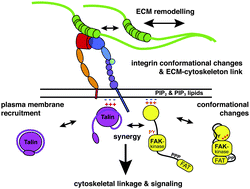
Protein conformation as a regulator of cell–matrix adhesion
Conformational changes within proteins play key roles in the regulation of cell–matrix adhesion. We discuss the mechanisms involved in conformational regulation, including mechanical signals, posttranslational modifications and intrinsically disordered proteins.
Phys. Chem. Chem. Phys., 2014,16, 6342-6357
https://doi.org/10.1039/C3CP54884H
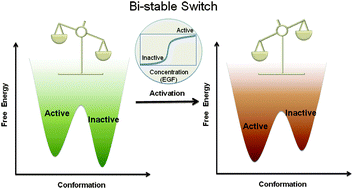
The free energy landscape in translational science: how can somatic mutations result in constitutive oncogenic activation?
The free energy landscape theory has transformed the field of protein and related it to function.
Phys. Chem. Chem. Phys., 2014,16, 6332-6341
https://doi.org/10.1039/C3CP54253J
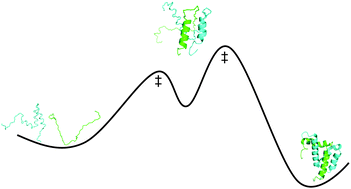
The binding mechanisms of intrinsically disordered proteins
Intrinsically disordered protein domains display a range of binding mechanisms, from apparent two-state to multistate.
Phys. Chem. Chem. Phys., 2014,16, 6323-6331
https://doi.org/10.1039/C3CP54226B
Fast recovery of free energy landscapes via diffusion-map-directed molecular dynamics
An adaptive sampling algorithm is proposed to rapidly reconstruct free-energy landscapes of macromolecular systems.
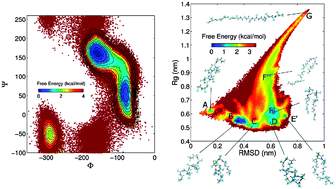
Phys. Chem. Chem. Phys., 2014,16, 19181-19191
https://doi.org/10.1039/C3CP54520B
From structure to function: the convergence of structure based models and co-evolutionary information
A combination of physical models and co-evolutionary information helps to improve our understanding of biomolecular structure and function.
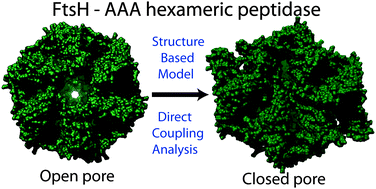
Phys. Chem. Chem. Phys., 2014,16, 6496-6507
https://doi.org/10.1039/C3CP55275F
Predicted disorder-to-order transition mutations in IκBα disrupt function
Single predicted disorder-to-order mutations impair the ability of IκBα to dissociate NFκB from the DNA.
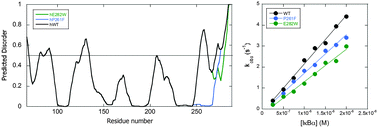
Phys. Chem. Chem. Phys., 2014,16, 6480-6485
https://doi.org/10.1039/C3CP54427C
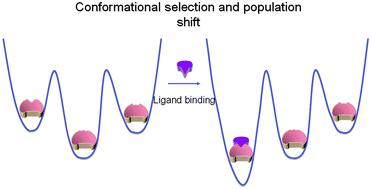
NMR mapping of protein conformational landscapes using coordinated behavior of chemical shifts upon ligand binding
The CONCISE statistical analysis of chemical shifts measures the population shifts and collectiveness of protein response associated with ligand titrations.
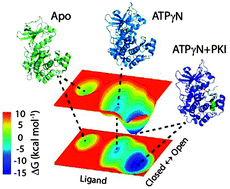
Phys. Chem. Chem. Phys., 2014,16, 6508-6518
https://doi.org/10.1039/C4CP00110A
Diffuse transition state structure for the unfolding of a leucine-rich repeat protein
Φ-value analysis of the folding mechanism of the leucine-rich repeat domain from TAP reveals a diffusely structured transition state.
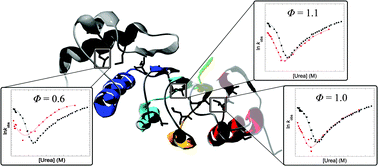
Phys. Chem. Chem. Phys., 2014,16, 6448-6459
https://doi.org/10.1039/C3CP54818J
Exploring multi-dimensional coordinate-dependent diffusion dynamics on the energy landscape of protein conformation change
Two-dimensional diffusion coefficiencies on the free energy landscape.
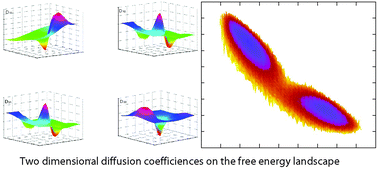
Phys. Chem. Chem. Phys., 2014,16, 6486-6495
https://doi.org/10.1039/C3CP54476A
A hybrid MD-kMC algorithm for folding proteins in explicit solvent
We present a novel hybrid MD-kMC algorithm and apply it to the folding of the Trp-Cage protein. The algorithm presented not only captures experimentally observed folding intermediates and kinetics, but yields insights into the relative roles of local and global interactions in determining folding mechanisms and rates.
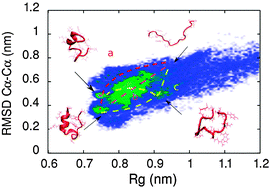
Phys. Chem. Chem. Phys., 2014,16, 6430-6440
https://doi.org/10.1039/C3CP55251A
Effects of desolvation barriers and sidechains on local–nonlocal coupling and chevron behaviors in coarse-grained models of protein folding
Coarse-grained protein chain models with desolvation barriers or sidechains lead to stronger local–nonlocal coupling and more linear chevron plots.
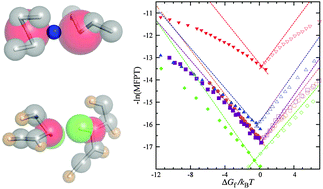
Phys. Chem. Chem. Phys., 2014,16, 6460-6479
https://doi.org/10.1039/C3CP54866J
Equilibrium thermodynamics and folding kinetics of a short, fast-folding, beta-hairpin
Equilibrium thermodynamics of a short beta-hairpin are studied using unbiased all-atom replica exchange molecular dynamics simulations in explicit solvent.
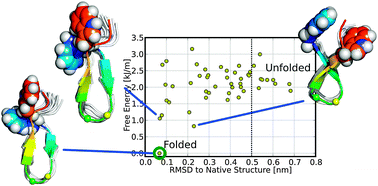
Phys. Chem. Chem. Phys., 2014,16, 6422-6429
https://doi.org/10.1039/C3CP54336F
Free energy landscape of G-protein coupled receptors, explored by accelerated molecular dynamics
G-protein coupled receptors (GPCRs) mediate cellular responses to various hormones and neurotransmitters and are important targets for treating a wide spectrum of diseases.
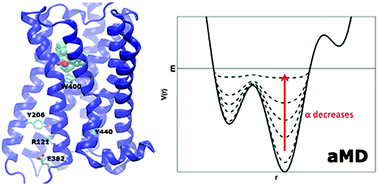
Phys. Chem. Chem. Phys., 2014,16, 6398-6406
https://doi.org/10.1039/C3CP53962H
The energy landscape of a protein switch
Protein switches are made of highly similar sequences that fold to dramatically different structures.
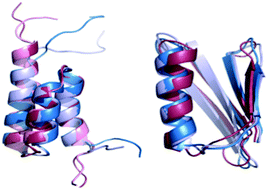
Phys. Chem. Chem. Phys., 2014,16, 6407-6421
https://doi.org/10.1039/C3CP55209H
The kinetics of folding of frataxin
The role of the denatured state in protein folding represents a key issue for the proper evaluation of folding kinetics and mechanisms.
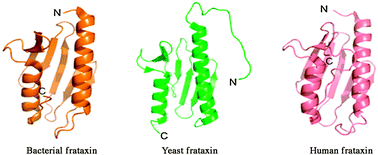
Phys. Chem. Chem. Phys., 2014,16, 6391-6397
https://doi.org/10.1039/C3CP54055C
Conformational flexibility of loops of myosin enhances the global bias in the actin–myosin interaction landscape
Increase in the compaction–expansion fluctuation of loop 2, the stronger binding of myosin to actin, and the biased Brownian motion of myosin S1 are coupled with each other and should take place in a concurrent way.
Phys. Chem. Chem. Phys., 2014,16, 6441-6447
https://doi.org/10.1039/C3CP54464H
A kinetic study of domain swapping of Protein L
NMR real-time kinetic experiments show that domain swapping of Protein L proceeds through a compact transition state.
Phys. Chem. Chem. Phys., 2014,16, 6383-6390
https://doi.org/10.1039/C3CP54126F
Sequence-dependent folding landscapes of adenine riboswitch aptamers
Multi-scale simulations show that a point mutation eliminates the differences in the folding landscapes of pbuE and add A-riboswitches.
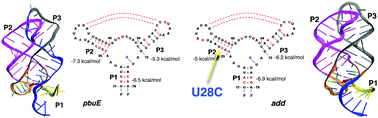
Phys. Chem. Chem. Phys., 2014,16, 6376-6382
https://doi.org/10.1039/C3CP53932F
Exploring the electrostatic energy landscape for tetraloop–receptor docking
It has long been appreciated that Mg2+ is essential for the stabilization of RNA tertiary structure.
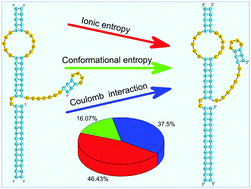
Phys. Chem. Chem. Phys., 2014,16, 6367-6375
https://doi.org/10.1039/C3CP53655F
Effect of interactions with the chaperonin cavity on protein folding and misfolding
Attractive interactions between a substrate protein can modulate the folding rate and stability, and help to prevent domain-swapped misfolding.
Phys. Chem. Chem. Phys., 2014,16, 6358-6366
https://doi.org/10.1039/C3CP52872C
About this collection
This themed issue showcases a collection of work from leading scientists, demonstrating the significance of the free energy landscape in the field of physiochemical biology.