Themed collection Proteomics

Inductive proteomics and large dataset collections
Inductive reasoning has been the corner stone of a large plethora of proteomics investigations, shifting the a priori paradigm of classical cellular and molecular biology to the experimental observation of protein modulation. These studies are now providing better definitions of their application framework.

Mol. BioSyst., 2015,11, 1485-1486
https://doi.org/10.1039/C5MB90021B
Why phosphoproteomics is still a challenge
Peptide-centric bottom-up proteomics can lead to ambiguous results.
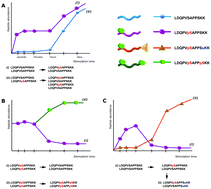
Mol. BioSyst., 2015,11, 1487-1493
https://doi.org/10.1039/C5MB00024F
Molecular effects of supraphysiological doses of doping agents on health
Supraphysiological doses of doping agents, such as T/DHT and GH/IGF-1, affect cellular pathways associated with apoptosis and inflammation.
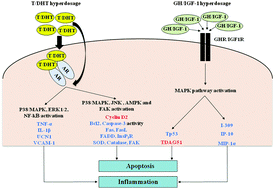
Mol. BioSyst., 2015,11, 1494-1506
https://doi.org/10.1039/C5MB00030K
Integrated proteomic platforms for the comparative characterization of medulloblastoma and pilocytic astrocytoma pediatric brain tumors: a preliminary study
The proteomic study of pediatric brain tumors tissues by top-down/bottom-up platforms revealed different expression profiles and potential malignancy biomarkers.
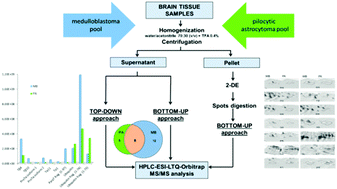
Mol. BioSyst., 2015,11, 1668-1683
https://doi.org/10.1039/C5MB00076A
Proteomic characterization of the qualitative and quantitative differences in cervical mucus composition during the menstrual cycle
The chemical composition of the cervical mucus (CM), its physical characteristics and the volume of secretion change cyclically throughout the menstrual cycle.
Mol. BioSyst., 2015,11, 1717-1725
https://doi.org/10.1039/C5MB00071H
Proteomic analysis of the cytotoxic effects induced by the organogold(III) complex Aubipyc in cisplatin-resistant A2780 ovarian cancer cells: further evidence for the glycolytic pathway implication
The cytotoxic mechanisms of the organogold(III) Aubipyc.

Mol. BioSyst., 2015,11, 1653-1667
https://doi.org/10.1039/C5MB00008D
Comparative membrane proteomics: a technical advancement in the search of renal cell carcinoma biomarkers
Set-up of a specific protocol for membrane protein analysis, applied to label free, comparative proteomics of renal cell carcinoma microdomains.
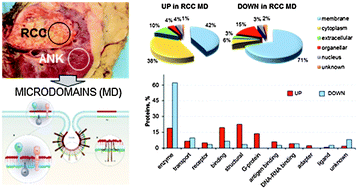
Mol. BioSyst., 2015,11, 1708-1716
https://doi.org/10.1039/C5MB00020C
Cigarette smoke alters the proteomic profile of lung fibroblasts
The protein identified here may offer a new insight into deciphering damage caused by cigarette smoke.
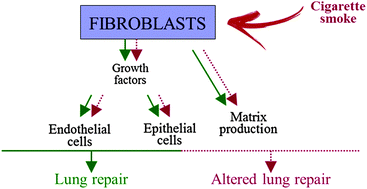
Mol. BioSyst., 2015,11, 1644-1652
https://doi.org/10.1039/C5MB00188A
Glucagon-like peptide 1 protects INS-1E mitochondria against palmitate-mediated beta-cell dysfunction: a proteomic study
Proteomic analysis of the protein expression profiles of enriched mitochondrial preparations of rat INS-1E β cells treated with palmitate in the presence and in the absence of GLP-1.
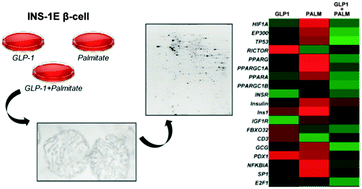
Mol. BioSyst., 2015,11, 1696-1707
https://doi.org/10.1039/C5MB00022J
Exploitation of complement regulatory proteins by Borrelia and Francisella
Borrelia and Francisella interact differently with complement regulatory proteins from various hosts, which influences the disease development and ecology of pathogens.
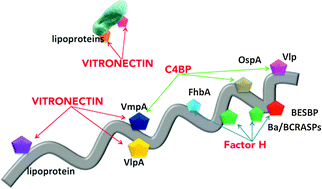
Mol. BioSyst., 2015,11, 1684-1695
https://doi.org/10.1039/C5MB00027K
Biocompatibility assessment of haemodialysis membrane materials by proteomic investigations
We examine and compare the protein adsorption capacity and coagulation profiles of different haemodialysis membrane biomaterials.

Mol. BioSyst., 2015,11, 1633-1643
https://doi.org/10.1039/C5MB00058K
Comparative proteomic analysis of two distinct stem-cell populations from human amniotic fluid
Characterization of two types of stem cells isolated from human amniotic fluid.

Mol. BioSyst., 2015,11, 1622-1632
https://doi.org/10.1039/C5MB00018A
Proteomic analysis of human glioblastoma cell lines differently resistant to a nitric oxide releasing agent
NO exposure of two human high grade glioma cell lines (CCF-STTG1 and T98G) characterized by a different proteomic profile shows differential ceramide distribution and proliferation.
Mol. BioSyst., 2015,11, 1612-1621
https://doi.org/10.1039/C4MB00725E
Variability assessment of 15N metabolic labeling-based proteomics workflow in mouse plasma and brain
Repeatability of the 15N metabolic labeling workflow for quantitative proteomics in mouse plasma and brain specimens.
Mol. BioSyst., 2015,11, 1536-1542
https://doi.org/10.1039/C4MB00702F
Reversible redox modifications in the microglial proteome challenged by beta amyloid
Reversible redox modifications of the microglial proteome contribute to switching of these neuronal sentinel cells toward a neuroinflammatory phenotype.
Mol. BioSyst., 2015,11, 1584-1593
https://doi.org/10.1039/C4MB00703D
Proteomic analysis of human sonic hedgehog (SHH) medulloblastoma stem-like cells
First proteomic characterization of sonic hedgehog human medulloblastoma stem-like cells.
Mol. BioSyst., 2015,11, 1603-1611
https://doi.org/10.1039/C5MB00034C
Comparative proteomic analyses of urine from rat urothelial carcinoma chemically induced by exposure to N-butyl-N-(4-hydroxybutyl)-nitrosamine
Bladder cancer is estimated to be the ninth most common malignancy with a high rate of recurrence and progression despite therapy, early diagnosis being crucial for timely intervention.
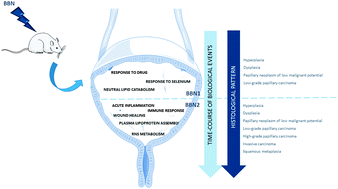
Mol. BioSyst., 2015,11, 1594-1602
https://doi.org/10.1039/C4MB00606B
Differential proteome–metabolome profiling of YCA1-knock-out and wild type cells reveals novel metabolic pathways and cellular processes dependent on the yeast metacaspase
A combined proteomic and metabolomic approach revealed new non-apoptotic roles of the metacaspase YCA1 gene in Saccharomyces cerevisiae, highlighting its involvement in the cell metabolism and stress response.
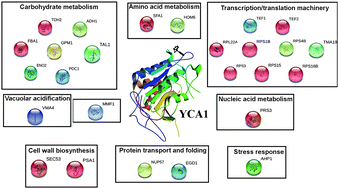
Mol. BioSyst., 2015,11, 1573-1583
https://doi.org/10.1039/C4MB00660G
Targeted metabolomics in the expanded newborn screening for inborn errors of metabolism
This paper highlights the importance of metabolic profiling by LC-MS/MS and GC-MS of biological fluids for diagnosis of inborn errors of metabolism and confirms a high incidence of these disorders.
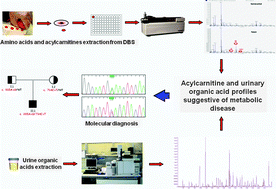
Mol. BioSyst., 2015,11, 1525-1535
https://doi.org/10.1039/C4MB00729H
An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis
(1) Lipid profiling in MuS and OND patients. (2) Search of alterations associated with MuS. (3) Characterization of differences.
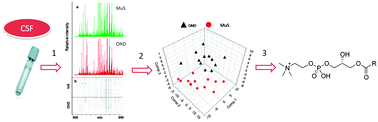
Mol. BioSyst., 2015,11, 1563-1572
https://doi.org/10.1039/C4MB00700J
The salivary proteome profile in patients affected by SAPHO syndrome characterized by a top-down RP-HPLC-ESI-MS platform
Salivary proteomic investigation in SAPHO syndrome reveals significant variation of cystatins, histatins, aPRP, S100A12, suggesting their potential role as biomarkers.
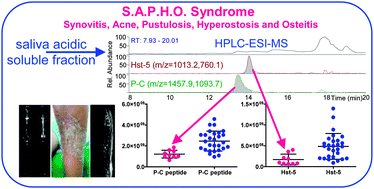
Mol. BioSyst., 2015,11, 1552-1562
https://doi.org/10.1039/C4MB00719K
Early markers of Fabry disease revealed by proteomics
Discovery of early urinary biomarkers for Fabry disease in male and female adult patients revealed by proteomics.
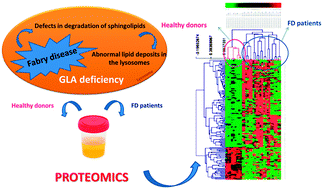
Mol. BioSyst., 2015,11, 1543-1551
https://doi.org/10.1039/C4MB00707G
A rapid and simple pipeline for synthesis of mRNA–ribosome–VHH complexes used in single-domain antibody ribosome display
A simple and rapid technique for construction of single domain antibody (VHH) library for ribosome display is described in this study.
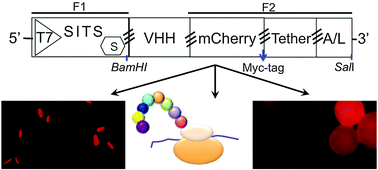
Mol. BioSyst., 2015,11, 1515-1524
https://doi.org/10.1039/C5MB00026B
A MALDI-Mass Spectrometry Imaging method applicable to different formalin-fixed paraffin-embedded human tissues
The paper shows a new method for the application of Matrix Assisted Laser Desorption/Ionisation (MALDI) Mass Spectrometry Imaging (MSI) technology on formalin-fixed paraffin-embedded (FFPE) tissue samples.
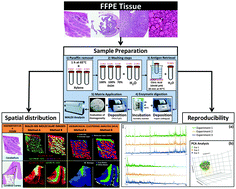
Mol. BioSyst., 2015,11, 1507-1514
https://doi.org/10.1039/C4MB00716F
About this collection
Molecular BioSystems is delighted to present this collection of articles from the themed issue Proteomics 2015, guest edited by Professors Massimo Castagnola and Andrea Urbani, President of the Italian Proteomics Association.
We hope you enjoy this selection!