LLDT-8 attenuates brain metastasis in non-small cell lung cancer via selective p53 activation
Junjie
Liu†
 a,
Lun
Liang†
bc,
Zhenning
Wang
d,
Kunsheng
Wei
a,
Zhixiong
Liang
a,
Junlei
Chang
a,
Lun
Liang†
bc,
Zhenning
Wang
d,
Kunsheng
Wei
a,
Zhixiong
Liang
a,
Junlei
Chang
 b,
Rongfeng
Lan
b,
Rongfeng
Lan
 *e,
Chunhua
Wang
*e,
Chunhua
Wang
 *a and
Min
Yu
*b
*a and
Min
Yu
*b
aSchool of Medicine, Foshan University, Foshan 528225, China. E-mail: pharmwch@126.com
bResearch Center for Protein and Cell-based Drugs, Institute of Biomedicine and Biotechnology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China. E-mail: min.yu@siat.ac.cn
cDepartment of Neurosurgery, The First Affiliated Hospital, Guangxi Medical University, Nanning 530021, China
dDepartment of Neurosurgery, The Tenth Affiliated Hospital, Southern Medical University (Dongguan People's Hospital), Dongguan 523018, China
eDepartment of Biochemistry & Cell Biology, College of Basic Medical Sciences, Shenzhen University Medical School, Shenzhen 518060, China. E-mail: lan@szu.edu.cn
First published on 16th December 2025
Abstract
Brain metastasis (BM) remains a leading cause of mortality in non-small cell lung cancer (NSCLC) owing to inadequate blood–brain barrier (BBB) penetration and therapy resistance. Here, we developed a brain-tropic A549-BrM2 subline that recapitulates aggressive BM progression and employed it to evaluate LLDT-8, a C14-hydroxylated derivative of triptolide for enhanced BBB permeability. In vitro, LLDT-8 exhibited attenuated cytotoxicity relative to triptolide, yet selectively induced p53-mediated apoptosis in BrM2 cells, an effect absent in parental lines. Transcriptomic profiling revealed that LLDT-8, unlike triptolide, upregulates Tp53 without robustly inducing its negative regulator Mdm2, thereby enabling preferential p53 protein accumulation and activation in the metastatic niche. In vivo, LLDT-8 significantly suppressed BM growth, achieving superior intracranial tumor control compared to triptolide, while eliciting no detectable systemic toxicity. These results identify LLDT-8 as a metastasis-selective agent that merges enhanced brain bioavailability with precise p53 pathway activation, providing a promising therapeutic strategy for NSCLC-derived BM.
1. Introduction
Lung cancer persists as the leading cause of cancer-related mortality worldwide, with non-small cell lung cancer (NSCLC) accounting for over 80% of cases.1–3 A critical clinical challenge lies in the propensity of NSCLC for brain metastases, occurring in 30–50% of advanced patients and correlating with a median survival of <6 months post-diagnosis.4,5 While targeted therapies (e.g., osimertinib) and immune checkpoint inhibitors have improved outcomes for some patients, their efficacy against BM is transient, limited by acquired resistance mutations (e.g., EGFR T790M/C797S) and suboptimal blood–brain barrier (BBB) penetration.6,7 Conventional chemotherapies further fail due to P-glycoprotein (P-gp)-mediated efflux and hydrophilicity, achieving intracranial response rates below 20%.8 This unmet need has revitalized interest in natural compounds capable of bypassing BBB exclusion while overcoming therapeutic resistance.Triptolide, a diterpenoid triepoxide from the plant thunder god vine, exhibits broad anti-tumor activity primarily by covalently binding to the XPB subunit of the general transcription factor TFIIH, thereby globally inhibiting RNA polymerase II-mediated transcription.9–11 This primary mechanism triggers downstream effects including induction of DNA replication stress, oxidative stress, and modulation of key pathways such as HSP70 inhibition and epithelial-mesenchymal transition (EMT) suppression via β-catenin reduction in NSCLC models.9,12 However, triptolide's clinical translation has been confronted by two major hurdles: dose-limiting multi-organ toxicity and unfavorable pharmacokinetic (PK) properties. Preclinical PK studies have revealed that while triptolide is rapidly absorbed with good oral bioavailability (∼72% in rats), it undergoes extensive metabolism, exhibits a very short elimination half-life (e.g., ∼17–22 min in rats), and is a confirmed substrate of P-glycoprotein (P-gp).13–15 These characteristics contribute to its rapid systemic clearance and low cumulative exposure, and critically, likely limit its distribution across the BBB, thereby compromising its efficacy against brain metastases. Furthermore, its narrow therapeutic index is evidenced by elevated liver toxicity in clinical trials and a low rodent LD50 (triptolide, ∼1.2 mg kg−1).16,17 Recent medicinal chemistry efforts have focused on structural modifications to mitigate toxicity while enhancing central nervous system (CNS) bioavailability. Among these derivatives, LLDT-8, a C14-hydroxylated analog with reduced epoxide reactivity, has shown improved safety profiles in rodent models (LD50 > 50 mg kg−1).18 While LLDT-8 likely shares this primary target due to its conserved core structure, the C14-hydroxylation may modulate its interaction with the transcriptional machinery or engage distinct signaling nodes, potentially underpinning its refined biological profile.9,19 Critically, the mechanistic basis for LLDT-8’s putative metastasis-selective activity and its detailed pharmacokinetic properties, especially its BBB penetration capability, remain uncharacterized.
While prior work established its NF-κB inhibitory properties,20 no studies have systematically compared its effects on parental versus brain-tropic NSCLC subpopulations or delineated its impact on cell cycle and apoptosis pathways in the context of brain metastasis. Addressing these gaps, our study employs a vertically integrated approach combining in vivo imaging methodologies and transcriptomics to quantify the differential anti-tumor efficacy of triptolide and LLDT-8 across NSCLC subtypes, to elucidate their distinct mechanisms of action through multi-omics profiling, and also to evaluate systemic toxicity in preclinical metastasis models. Our findings not only reposition LLDT-8 as a brain-metastasis-selective therapeutic but also provide a blueprint for optimizing natural product derivatives for CNS malignancies.
2. Materials and methods
2.1. Materials
Triptolide (S3604) was purchased from Selleck and its derivative LLDT-8 was obtained from Shanghai BioPro Bio-Tech Co., Ltd. Both compounds were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich) to prepare stock solutions at concentrations of 1 mM and 12.5 mg mL−1, respectively. The antibodies used were as follows: anti-Cyclin-A2 (#SC-751), anti-Cyclin-B1 (#SC-245), anti-Skp2 (#SC-7164), and anti-p27 (#SC-528) were purchased from Santa Cruz Biotechnology (Delaware, USA); anti-Beta-actin (#66009-1-Ig) were purchased from Proteintech (Wuhan, China); anti-p53 (#2524S) was purchased from Cell Signaling Technology (MA, USA); anti-Bcl-2 (#AF6139) and anti-BAX (AF0120) were purchased from Affinity (Jiangsu, China). A Cell Counting Kit-8 (CCK-8, #40203ES80) and an Annexin V-PE/7-AAD Apoptosis Detection Kit (#40310ES50) were obtained from YEASEN (Shanghai, China).2.2. Cell lines and culture conditions
The human lung cancer cell line A549 was obtained from American Type Culture Collection (ATCC). The A549-ZsGreen-Luci cell line was generated with lentiviruses expressing ZsGreen and luciferase. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin at 37 °C, 5% CO2 incubator. Cells were authenticated by Shanghai Biowing Biotechnology Co. Ltd., and mycoplasma was tested after cells were cultured for one week using a GMyc-PCR Mycoplasma Test Kit (YEASEN, Shanghai, China). Cells used for experiments were maintained for one month.2.3. In vivo and in vitro selection of brain metastatic subline
Procedures involving mice were approved by the Institutional Animal Care and Use Committee at the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences in compliance with the Guide for the Care and Use of Laboratory Animals. Female nude mice (5–6 weeks) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. All mice were housed with a maximum of 5 animals per cage on a 12 h light/dark cycle (light from 07:00 to 19:00) at 21 °C and 30–70% humidity.To establish the lung cancer brain metastasis model, female nude mice were anesthetized with isoflurane. For intracardiac injection, each mouse was properly anesthetized, positioned in the supine position, and the chest area was disinfected. A 1 mL syringe equipped with a 27–28 G needle was used to puncture the left ventricle at a site located 3 mm to the right of the second intercostal space, inserting the needle at a 45° angle to a depth of 6 mm. When clear blood return was observed, 0.8 × 106 tumor cells suspended in 100 μL PBS were injected slowly and intermittently into the cardiac chamber. The needle was then carefully withdrawn after completing the injection.
The IVIS system was used to measure the bioluminescence signal of metastatic tumor cells in the mice. After six weeks, brain tissues were harvested and brain-metastatic lung cancer cells were isolated and cultured, and the subpopulations of tumor (referred to as brain metastatic derivative 1, BrM1) were subjected to a second round of in vivo selection. BrM1 cells that metastasized to the brain were then isolated and grown in vitro for no more than two weeks (termed BrM2) (Fig. 1A).
2.4. Proliferation assays
To evaluate the cytotoxicity of triptolide and LLDT-8 in BrM2 cells, proliferation assays were performed using CCK-8 and crystal violet staining methods. During the exponential growth phase, parental and BrM2 cells were seeded in 96-well plates at a density of 750 cells per well. Following 24 hours of incubation, cells were treated with either triptolide or LLDT-8 (with equivalent DMSO concentrations as vehicle control) for 24, 48, or 72 hours. For the CCK-8 assay, 10 μL of CCK-8 solution was added to each well and incubated for 2 hours in the dark. Absorbance at 450 nm (OD450) was measured using a Multiskan GO microplate reader (Thermo Fisher Scientific). All experiments were performed in triplicate.Parental and BrM2 cells were plated onto 12-well plates at a density of 750 cells per well. After 24-hour incubation, cells were treated with or without triptolide or LLDT-8 at indicated concentrations for 24 hours. Following 7–14 days of culture, each well was washed three times with PBS at room temperature, fixed with 4% PFA for 10 minutes, and then stained with 0.3% crystal violet for 20 minutes at room temperature. All experiments were performed in triplicate.
2.5. RNA-seq
The RNA-seq data were generated from BrM2 cells which were treated with or without triptolide or LLDT-8 at indicated concentrations for 24 h. Each RNA-seq analysis was conducted on three biological replicates. Total RNA was extracted using an Arcturus PicoPure RNA Isolation Kit (Applied Biosystems). In brief, after sample QC, the mRNA was hybridized with an oligo (dT) probe and captured by magnetic beads. The mRNA was then reverse-transcripted to first strand DNA. First strand DNA was used as template to synthesis second strand DNA, obtaining dsDNA. The dTTP tailed adaptor was ligated to both ends of the dsDNA fragments. The ligation product was amplified by PCR, and circularized to get single-stranded circular (ssCir) library. The ssCir library was then amplified through rolling circle amplification (RCA) to obtain DNA nanoball (DNB). The DNB was then loaded to a flowcell, and sequenced by DNBSEQ Platform. Raw sequencing data were processed using SOAPnuke (v2.1.0) for quality control and adapter trimming. Cleaned reads were aligned to the GRCh38.p13 reference genome with HISAT2 (v2.2.1), followed by transcript quantification using StringTie (v2.2.1). Differential expression analysis was subsequently performed with DESeq2 (v1.38.3) under the R statistical framework. The RNA-seq raw data generated in this study have been deposited in the GSA-Human repository at the CNCB-NGDC (GSA-Human: HRA011961) and are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.21,222.6. Flow cytometry analysis of cell apoptosis
Cell apoptosis was assessed using Annexin V-PE/7-AAD double staining. Parental and BrM2 cells were treated with or without triptolide or LLDT-8 for 24 h. Cells were collected and washed with cold PBS twice and the precipitated cells were re-suspended in 100 μL binding buffer co-stained with 5 μL Annexin-V-PE and 10 μL 7-AAD, then incubated for 15–30 min in the dark. Stained cells were analyzed through flow cytometry (MA900, Sony).2.7. Western blot analysis
Cellular proteins were quantified, equally loaded, SDS polyacrylamide gel electrophoresis separated and transblotted. Membranes were blocked for 1 h at room temperature in 5% non-fat dry milk and then incubated overnight at 4 °C with primary antibodies diluted in TBS with 0.1% Tween20 (TBST). Membranes were washed in TBST and then incubated with secondary antibodies for 1 h at room temperature. Subsequently the membranes were exposed to enhanced chemiluminescence substrate detection solution (WBKLS0500, Millipore) and then detected by the instrument (Gel view, GV6000).2.8. BrM mouse model and treatment
Female nude mice (5–6 weeks) were obtained from Guangdong Medical Laboratory Animal Center. For stereotactic brain injections, the nude mouse skin was sterilized with 70% ethanol and a small midline incision was used to expose the skull. A 25 G hole was bored into the parietal bone 2 mm rostral to the central suture and right of the sagittal suture. 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were re-suspended in 5 μL PBS and injected at a depth of 2.8 mm at a rate of 1 μL min−1, allowing 5 min for pressure equilibration. The skull was closed with bone wax, and the skin with 3 M glue. After three weeks, metastatic tumor progression was monitored by measuring bioluminescent signals using the IVIS imaging system. Mice were then randomly allocated into experimental and control groups (n = 9 per group). Treatment groups received intraperitoneal (i.p.) injections of either 1 mg kg−1 triptolide or LLDT-8 every other day, while the control group was administered the vehicle alone (1
000 cells were re-suspended in 5 μL PBS and injected at a depth of 2.8 mm at a rate of 1 μL min−1, allowing 5 min for pressure equilibration. The skull was closed with bone wax, and the skin with 3 M glue. After three weeks, metastatic tumor progression was monitored by measuring bioluminescent signals using the IVIS imaging system. Mice were then randomly allocated into experimental and control groups (n = 9 per group). Treatment groups received intraperitoneal (i.p.) injections of either 1 mg kg−1 triptolide or LLDT-8 every other day, while the control group was administered the vehicle alone (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (v/v) DMSO in 0.9% NaCl). Following 24 days of treatment, all mice were euthanized for further analysis.
50 (v/v) DMSO in 0.9% NaCl). Following 24 days of treatment, all mice were euthanized for further analysis.
2.9. H&E staining and immunofluorescence analysis
Mice were deeply anaesthetized and perfused transcardially with 20 mL of ice-cold PBS. Mice were then rapidly decapitated, and the brain, heart, liver, spleen, lung, kidney tissues were dissected and immersed in ice-cold PBS and washed three times. Mice brain fresh tissues were placed in 4% PFA for 1 h at room temperature and then transferred to 15% and 30% sucrose in PBS overnight at 4 °C before embedding in OCT. The tissues of the heart, liver, spleen, lung, and kidneys were directly embedded in OCT after being weighed and then placed in a −80 °C freezer. Brain tissues were frozen in sections with 10 μm in thickness and were allowed to dry on Superfrost Plus slides (Fisher) at room temperature before being rehydrated in 1× PBS.For H&E staining using the hematoxylin–eosin HD Kit (Cat No. G1076, Servicebio), the frozen sections were restored to room temperature, fixed with tissue fixating solution (Cat No. G1101, Servicebio) for 15 min, and then rinsed with running water. The sections were subsequently treated with HD constant staining pretreatment solution for 1 minute. Next, the sections were immersed in hematoxylin solution for 3–5 minutes and rinsed with tap water. Then the sections were treated with hematoxylin differentiation solution and rinsed with tap water. Subsequently, the sections were treated with hematoxylin bluing solution and rinsed with tap water. Afterward, the sections were placed in 95% ethanol for 1 min, and eosin dye for 15 s. The sections were put into absolute ethanol I for 2 min, absolute ethanol II for 2 min, absolute ethanol III for 2 min, normal butanol I for 2 min, normal butanol II for 2 min, xylene I for 2 min and xylene II for 2 min, and sealed with neutral gum. Histopathological evaluation was conducted using bright field microscopy (Nikon Eclipse E2), with whole-slide digital imaging performed at 40× magnification.
For immunofluorescence analysis, brain tissues were blocked in 5% Normal Goat Serum (Jackson ImmunoResearch, West Grove, PA) in PBS/1% BSA/0.3% Triton X-100 for 30 min at room temperature. Samples were incubated at 4 °C with the following primary antibodies in 5% Normal Goat Serum in PBS/1% BSA/0.3% Triton X-100 + 0.1% NaN3: rabbit anti-Ki67 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, Cat No. 27309-1-AP, Proteintech). Excess primary antibody was removed by rinsing in PBS/0.1% Triton X-100 for 10 min 3 times. Samples were then incubated at room temperature for 2 h with the following secondary fluorescently labeled antibodies: Alexa Fluor 647 Goat Anti-Rabbit IgG (H + L) (1
50, Cat No. 27309-1-AP, Proteintech). Excess primary antibody was removed by rinsing in PBS/0.1% Triton X-100 for 10 min 3 times. Samples were then incubated at room temperature for 2 h with the following secondary fluorescently labeled antibodies: Alexa Fluor 647 Goat Anti-Rabbit IgG (H + L) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Cat No. 111-605-003, Jackson ImmunoResearch). Excess secondary antibodies were removed by rinsing in PBS/0.1% Triton X-100 for 10 min 3 times. Slides were mounted in anti-fade reagent with DAPI (cat. #S2110, Solarbio) and imaged with a laser scanning confocal microscope (Nikon A1R) or an inverted fluorescence microscope (OLYMPUS CKX53). Immunofluorescence signal area and integral optical density (IOD) were quantified by ImageJ.
1000, Cat No. 111-605-003, Jackson ImmunoResearch). Excess secondary antibodies were removed by rinsing in PBS/0.1% Triton X-100 for 10 min 3 times. Slides were mounted in anti-fade reagent with DAPI (cat. #S2110, Solarbio) and imaged with a laser scanning confocal microscope (Nikon A1R) or an inverted fluorescence microscope (OLYMPUS CKX53). Immunofluorescence signal area and integral optical density (IOD) were quantified by ImageJ.
2.10. Statistical analysis
Statistical analysis was conducted using GraphPad Prism software (v.8.0.2). Data are presented as mean ± standard error of the mean (S.E.M.). Statistical differences between two groups were determined using the t-test. Comparisons between more than two groups were performed by one-way ANOVA with Dunnett's multiple-comparison test. p values less than 0.05 were considered significant.3. Results
3.1. Brain metastatic A549-BrM obtained from in vivo and in vitro selection
To establish a longitudinal in vivo monitoring system for visualizing brain metastasis in non-small cell lung cancer (NSCLC), we employed lentiviral transfection to construct an A549-ZsGreen-Luci cell line stably co-expressing ZsGreen fluorescent protein and a luciferase reporter gene system (Fig. 1A). A brain metastasis mouse model was established using a modified stereotactic left ventricular injection approach, followed by dual in vivo/in vitro selection to obtain brain-tropic A549-BrM cell subpopulations (Fig. 1A and B). After two rounds of stringent selection, we successfully isolated the highly metastatic A549-BrM2 subline (designated BrM2), with its biological characteristics dynamically monitored using the IVIS Spectrum in vivo imaging system (Fig. 1C). Quantitative analysis revealed that BrM2 cells exhibited a significantly higher incidence of brain metastasis (66.7%, 6/9) compared to parental cells (25.0%, 7/28) (Fig. 1D). Standardized radiance measurements (photons per s cm−2 sr−1) demonstrated that BrM2-injected mice achieved 1.95 × 108 photons per s at the 42-day endpoint, representing a 49.56-fold increase over parental cell-derived metastases (3.94 × 106 photons per s) (p < 0.001, Student's t-test) (Fig. 1E). These analyses encompassing metastasis incidence and tumor burden collectively demonstrate that the systematically selected BrM2 subline possesses enhanced brain-tropic metastatic capacity, suggesting successful retention of critical molecular characteristics required for blood–brain barrier penetration and adaptation to the central nervous system microenvironment.3.2. Triptolide and LLDT-8 inhibit the proliferation of BrM2
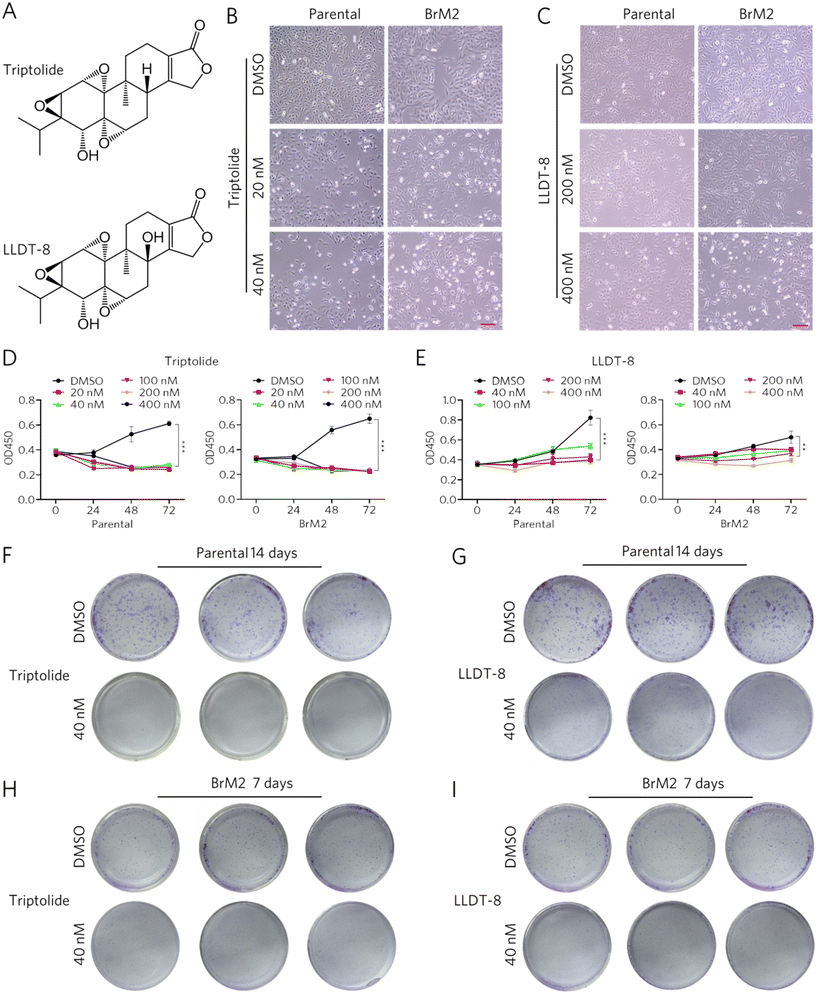
To investigate the anti-proliferative efficacy of triptolide and its structurally optimized derivative LLDT-8 (Fig. 2A) against brain-metastatic non-small cell lung cancer (NSCLC) cells, we performed analyses combining morphological, proliferation, and clonogenic assessments. Phase-contrast microscopy revealed that both triptolide and LLDT-8 induced concentration-dependent attenuation of cellular proliferation in parental A549 and brain-tropic BrM2 sublines, with visible reductions in cell density observed as early as 24 h post-treatment (Fig. 2B and C). CCK-8 assays demonstrated a dose-dependent suppression of proliferation by both compounds in all cell lines (Fig. 2D and E). Consistent with previous reports, LLDT-8 was effective; it exhibited a comparatively reduced cytotoxic potency relative to triptolide, a difference that was more pronounced in the BrM2 cells (Fig. 2D and E). This observed attenuation is consistent with the documented improved safety profile of LLDT-8 and underscores its context-dependent activity. Clonogenic formation assays further corroborated these findings, showing both triptolide and LLDT-8 reduced colony formation of parental and BrM2 cells (Fig. 2F–I). These analyses collectively establish that both triptolide and its derivative LLDT-8 effectively inhibit the proliferative capacity of NSCLC cells and their brain-metastatic subpopulations in vitro.3.3. Triptolide and LLDT-8 regulate the DNA replication and the process of the cell cycle in BrM2 cells
To elucidate the molecular mechanisms underlying the anti-proliferative effects of triptolide and LLDT-8 on BrM2 cells, we conducted RNA sequencing (RNA-seq) followed by systematic bioinformatics and biochemical validation. RNA-seq assays revealed that triptolide treatment induced significant expression changes in 7203 genes (adj p < 0.05, |log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC| > 2) compared to vehicle controls, with 6075 downregulated and 1128 upregulated genes (SI file 1, Vehicle_vs_Triptolide). Similarly, LLDT-8 treatment modulated 3185 genes (adj p < 0.05, |log2
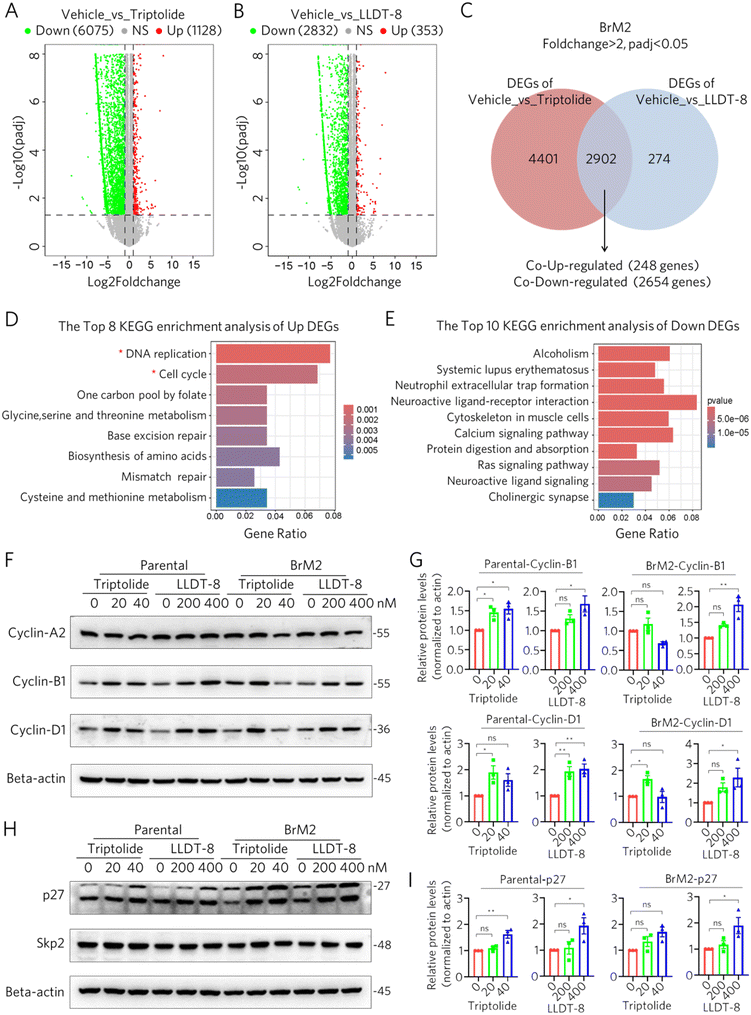
FC| > 2) compared to vehicle controls, with 6075 downregulated and 1128 upregulated genes (SI file 1, Vehicle_vs_Triptolide). Similarly, LLDT-8 treatment modulated 3185 genes (adj p < 0.05, |log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC| > 2), including 2832 downregulated and 353 upregulated genes (SI file 1, Vehicle_vs_LLDT-8) (Fig. 3A). Intersection analysis of the two treatment groups identified a common set of 2902 differentially expressed genes (SI file 2), of which 248 were upregulated and 2654 were downregulated in both treatments (Fig. 3B). KEGG pathway enrichment analysis of upregulated DEGs highlighted profound suppression of proliferation-associated pathways, with the most significantly enriched terms being “DNA replication” (p value = 8.55 × 10−10) and “Cell cycle” (p value = 0.0014), while down-regulated genes were enriched in inflammatory pathways including “Alcoholism” (p value = 9.81 × 10−15) and “Systemic lupus erythematosus” (p value = 5.15 × 10−13) (Fig. 3D and E). These findings demonstrate that both triptolide and LLDT-8 may suppress the proliferative capacity of parental and brain-metastatic cells in vitro through modulation of DNA replication machinery and disruption of cell cycle progression.
FC| > 2), including 2832 downregulated and 353 upregulated genes (SI file 1, Vehicle_vs_LLDT-8) (Fig. 3A). Intersection analysis of the two treatment groups identified a common set of 2902 differentially expressed genes (SI file 2), of which 248 were upregulated and 2654 were downregulated in both treatments (Fig. 3B). KEGG pathway enrichment analysis of upregulated DEGs highlighted profound suppression of proliferation-associated pathways, with the most significantly enriched terms being “DNA replication” (p value = 8.55 × 10−10) and “Cell cycle” (p value = 0.0014), while down-regulated genes were enriched in inflammatory pathways including “Alcoholism” (p value = 9.81 × 10−15) and “Systemic lupus erythematosus” (p value = 5.15 × 10−13) (Fig. 3D and E). These findings demonstrate that both triptolide and LLDT-8 may suppress the proliferative capacity of parental and brain-metastatic cells in vitro through modulation of DNA replication machinery and disruption of cell cycle progression.
Next, western blot analysis confirmed that triptolide and LLDT-8 treatment dose-dependently altered the expression of key cell cycle regulatory proteins in both parental and BrM2 cell lines. Specifically, both compounds modulated Cyclin-B1/D1 levels while showing no significant effect on Cyclin-A2 expression (Fig. 3F and G and S1A). Notably, although Skp2-mediated ubiquitin–proteasome degradation of cyclin-dependent kinase inhibitors (CDKIs) represents a major regulatory mechanism of cell cycle progression, we observed a dose-dependent accumulation of the CDKI p27 that occurred independently of Skp2 protein levels in both parental and BrM2 cells (Fig. 3H and I and S1B). These findings mechanistically link the proliferation–inhibitory effects of both compounds to their coordinated disruption of cell cycle progression through transcriptional repression of replication machinery and post-translational stabilization of cell cycle brakes, providing a molecular rationale for their efficacy against brain metastases of NSCLC.
3.4. Triptolide and LLDT-8 promote cell apoptosis by inducing the p53 signaling pathway
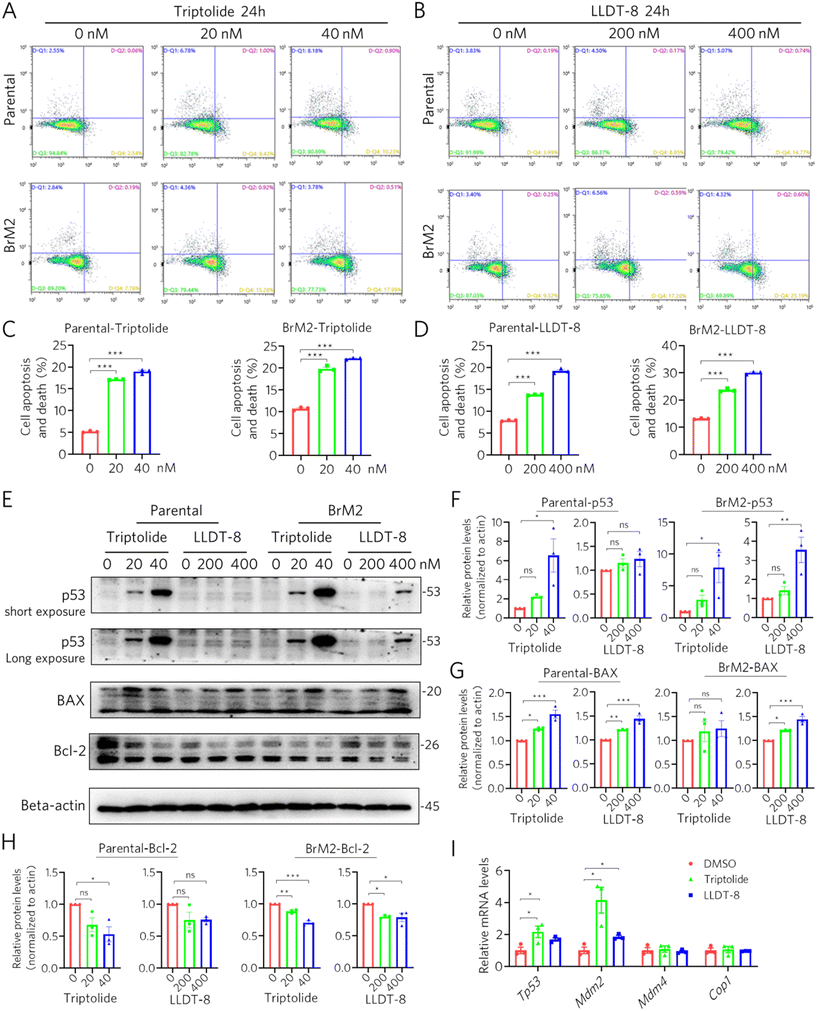
We next assessed the apoptotic effects of triptolide and LLDT-8 in both parental and brain-tropic BrM2 cells. Flow cytometry analysis with Annexin V-PE/7-AAD staining confirmed concentration-dependent apoptosis induction by both compounds following 24-hour treatment (Fig. 4A–D). Western blot analysis revealed that the triptolide dose dependently upregulated p53 protein expression and activated its downstream signaling in both cell types (Fig. 4E and F), accompanied by increased BAX and decreased Bcl-2 protein levels (Fig. 4G and H). Unexpectedly, LLDT-8 exhibited a distinct p53 regulation pattern. While it selectively induced p53 protein accumulation specifically in BrM2 cells, it showed no significant effect on p53 levels in parental cells (Fig. 4E and F). This metastasis-selective p53 activation occurred despite LLDT-8 consistently modulating BAX and Bcl-2 expression in both cell lines (Fig. 4G and H).To elucidate the mechanism underlying this differential p53 regulation, we analyzed transcriptomic data. Both compounds modestly upregulated Tp53 transcript levels in BrM2 cells. However, a critical divergence emerged in their regulation of Mdm2, its canonical ubiquitin ligase (Fig. 4I). While triptolide treatment triggered a substantial concurrent upregulation of Mdm2 transcription, LLDT-8 induced only a minimal increase in Mdm2 mRNA (Fig. 4I). This “Mdm2-sparing” transcriptional profile of LLDT-8 provides a mechanistic basis for its selective p53 protein accumulation in BrM2 cells. By elevating Tp53 expression without proportionally increasing its negative regulator, LLDT-8 creates a favorable environment for p53 protein stabilization and activation specifically in the brain-metastatic niche. These results demonstrate that while both compounds engage the p53 pathway, LLDT-8 exhibits a refined, metastasis-selective mechanism of p53 modulation, positioning it as a brain-metastasis-targeted apoptosis inducer with potentially reduced off-target effects on non-metastatic cells.
3.5. Triptolide and LLDT-8 inhibit the proliferation of brain metastatic lung cancer cells in vivo
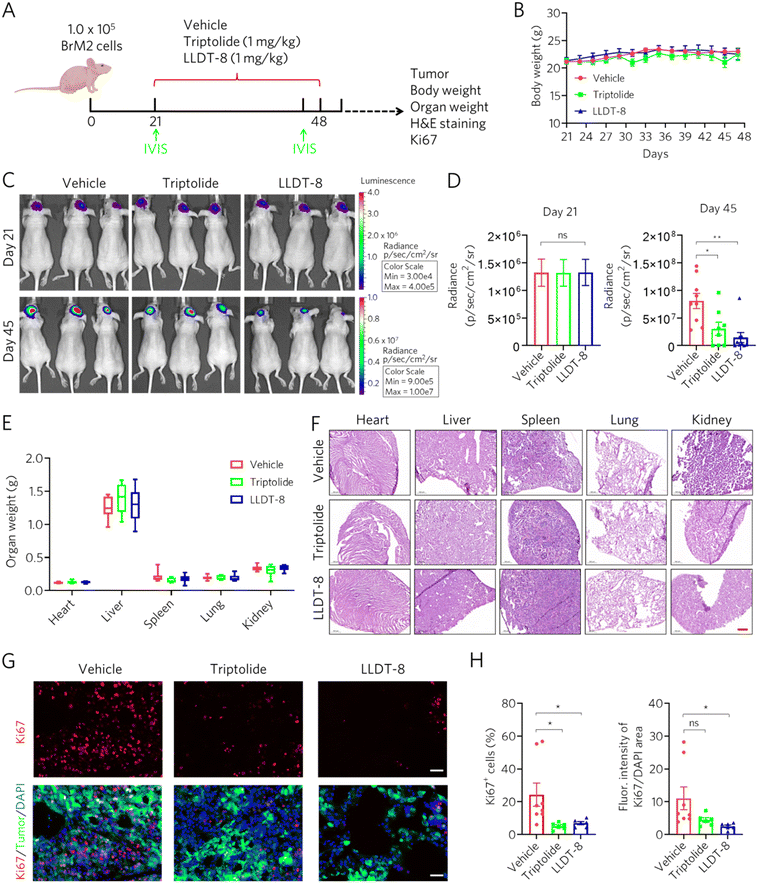
To evaluate the therapeutic efficacy and safety of triptolide and LLDT-8 against established brain metastases, we injected BrM2 cells into immunocompromised mice and initiated daily intraperitoneal treatments (1 mg kg−1 body weight) from day 21 post-inoculation (Fig. 5A). The bioluminescence imaging performed using the IVIS system revealed no significant intergroup differences in tumor burden at the initiation of treatment, but by day 45, both triptolide and LLDT-8-treated mice exhibited a dramatic reduction in tumor radiance compared to vehicle controls (Fig. 5C and D). After administration of triptolide and LLDT-8, no obvious weight loss or abnormal behavior was detected in vehicle and treatment groups (Fig. 5B). Moreover, comprehensive organ toxicity assessments revealed no significant alterations in organ/body weight indices or histopathological abnormalities in hematoxylin and eosin (H&E)-stained sections of the heart, liver, spleen, lung, and kidneys (Fig. 5E and F). These data demonstrated that the drug dosage we administered had no apparent toxic side effects on the mice. To determine whether LLTD-8 and triptolide inhibit the proliferation of brain metastasis cells in vivo, we assessed the proportion of Ki67-positive cells and Ki67 expression intensity via immunofluorescence staining (Fig. 5G). Compared to the vehicle control, triptolide significantly decreased the percentage of Ki67-positive cells but had no significant effect on Ki67 expression levels in tumor cells (Fig. 5H). In contrast, LLTD-8 not only markedly reduced the proportion of Ki67-positive cells but also significantly down-regulated Ki67 expression (Fig. 5H). These in vivo results align with our mechanistic investigations, confirming that both compounds effectively suppress BrM progression in the absence of systemic toxicity, with LLDT-8 exhibiting superior therapeutic efficacy.4. Discussion
Brain metastasis represents a critical therapeutic challenge in NSCLC, contributing substantially to patient mortality due to limited BBB penetrance of conventional therapies and inherent resistance mechanisms.23–25 To address this unmet need, our study established a novel brain-tropic A549-BrM2 subline through rigorous in vivo and in vitro selection protocols, which faithfully recapitulates key clinical features of human brain metastases including enhanced BBB transmigration capacity and neurotropic microenvironment adaptation.26 This validated model system provides a physiologically relevant platform for evaluating therapeutic agents against metastatic niches. Our investigations demonstrate that both the natural compound triptolide and its semi-synthetic derivative LLDT-8 exert potent anti-metastatic activity through coordinated multi-mechanistic actions.While prior studies have well-established that triptolide exerts its anti-tumor effects in NSCLC through concurrent modulation of multiple signaling pathways, including p53, NF-κB, and MAPKs,27–29 our work provides a novel and critical insight by demonstrating that its derivative, LLDT-8, achieves a refined, metastasis-selective activation of the p53 pathway. This context-dependent action represents a significant departure from the broad-spectrum mechanism of the parent compound.
Our findings indicate that both triptolide and LLDT-8 effectively inhibit the proliferation of NSCLC cells and their brain-metastatic subpopulations in vitro, with LLDT-8 exhibiting an inherently lower cytotoxicity (Fig. 2). RNA sequencing and RT-qPCR revealed a pivotal mechanistic divergence: while both compounds upregulated Tp53 transcription in BrM2 cells, triptolide concurrently induced a robust upregulation of its negative regulator, Mdm2. In contrast, LLDT-8 treatment resulted in a significantly weaker induction of Mdm2 mRNA (Fig. 3 and 4I). This unique Mdm2-sparing transcription profile of LLDT-8 creates a favorable kinetic window for p53 protein accumulation specifically in brain-metastatic cells. Pathway enrichment analysis highlighted the suppression of proliferation-associated pathways such as “DNA replication” and “Cell cycle”, suggesting that triptolide and LLDT-8 exert their anti-proliferative effects by disrupting these critical processes. The (5R)-hydroxylation at the C-5 position of the triptolide scaffold reduces P-gp binding by enhancing molecular polarity, disrupting hydrophobic interactions with the substrate-binding pocket of P-gp. This modification minimizes efflux-mediated resistance and improves intracellular retention of LLDT-8 in tumor cells.30 Importantly, LLDT-8 maintains this increased efficacy while showing no detectable systemic toxicity in vivo, a critical advancement given the narrow therapeutic window of many CNS-targeted agents.
Mechanistically, the compounds' ability to stabilize key cell cycle inhibitors while destabilizing pro-survival signals creates a multi-pronged attack on metastatic cells. The confluence of LLDT-8's distinct transcriptional signature (p53 activation without concomitant high Mdm2 induction) and its superior brain distribution creates a synthetic vulnerability in the BrM2 cells, leading to selective p53 protein accumulation and potent apoptosis. This metastasis-specific induction of tumor suppressor activity represents a significant therapeutic advantage, potentially creating a therapeutic window where LLDT-8 could preferentially target advanced metastatic diseases while minimizing off-target effects on normal tissues, a distinction not previously reported for triptolide derivatives.28 The differential regulation of apoptotic regulators between parental and metastatic cells further underscores the importance of molecular context in determining therapeutic outcomes.
In vivo studies in immunocompromised mice demonstrated that both triptolide and LLDT-8 significantly reduced tumor radiance in brain metastatic lesions without causing apparent systemic toxicity. Notably, LLDT-8 achieved superior efficacy in suppressing brain metastases compared to triptolide (Fig. 5), a finding that functionally validates its enhanced brain delivery and metastasis-selective mechanism of action. Immunofluorescence staining revealed that both compounds decreased the proportion of Ki67-positive cells, with LLDT-8 also down-regulating Ki67 expression levels. These results confirm the therapeutic efficacy of triptolide and LLDT-8 in inhibiting the progression of brain metastasis in vivo. Future research should focus on further elucidating the pharmacodynamics and pharmacokinetics of LLDT-8, as well as exploring its potential in combination with other therapeutic modalities to optimize treatment outcomes for NSCLC patients with brain metastasis.
5. Conclusion
Our study provides comprehensive evidence that triptolide and LLDT-8 exhibit potent anti-tumor activity against NSCLC brain metastasis. The mechanistic basis for their efficacy involves the disruption of DNA replication and cell cycle progression. A key finding is that LLDT-8 selectively induced p53 protein accumulation in BrM cells possibly by uncoupling p53 transcription from robust Mdm2 upregulation. The superior therapeutic efficacy and safety profile of LLDT-8 highlight its potential as a targeted therapy for brain metastasis.Author contributions
Junjie Liu: data curation, investigation, and methodology; Lun Liang: formal analysis, investigation and methodology; Zhenning Wang: funding acquisition and methodology; Kunsheng Wei: methodology and visualization; Zhixiong Liang: methodology and validation; Junlei Chang: supervision and validation; Rongfeng Lan: writing – original draft, writing – review & editing; Chunhua Wang: funding acquisition, supervision, and validation; Min Yu: funding acquisition, investigation, methodology, supervision, and writing – review & editing.Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.Data availability
The RNA-seq raw data generated in this study have been deposited in the GSA-Human repository at the CNCB-NGDC (GSA-Human: HRA011961) and are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human. The experimental procedures were described in 2.5 RNA-seq. The DEGs generated from RNA-seq and graphed in Fig. 3 are listed in supplementary information (SI) file 1 and 2.Supplementary information: the SI contains the following: 1) Files 1 and 2, containing the DEGs identified by RNA-seq in BrM2 cells; 2) Fig. S1, showing the statistical analysis of the relative protein expression levels of Cyclin A and Skp2 from Fig. 3F and H. See DOI: https://doi.org/10.1039/d5md00754b.
Acknowledgements
The work was funded by the National Natural Science Foundation of China (82203810, 82473099) to M. Yu; the Science Technology and Innovation Commission of Shenzhen Municipality (JCYJ20240813154847062) to M. Yu; the Basic and Applied Basic Research Foundation of Guangdong Province (2022B1515120015) to C. Wang; the Dongguan Social Development Science and Technology Project (20231800936072) to Z. Wang.References
- F. Bray, M. Laversanne, H. Sung, J. Ferlay, R. L. Siegel, I. Soerjomataram and A. Jemal, Ca-Cancer J. Clin., 2024, 74, 229–263 CrossRef PubMed.
- M. Wang, R. S. Herbst and C. Boshoff, Nat. Med., 2021, 27, 1345–1356 CrossRef CAS PubMed.
- C. Zappa and S. A. Mousa, Transl. Lung Cancer Res., 2016, 5, 288–300 CrossRef CAS PubMed.
- V. Ernani and T. E. Stinchcombe, J. Oncol. Pract., 2019, 15, 563–570 CrossRef PubMed.
- J. H. Suh, R. Kotecha, S. T. Chao, M. S. Ahluwalia, A. Sahgal and E. L. Chang, Nat. Rev. Clin. Oncol., 2020, 17, 279–299 CrossRef.
- S. Park, R. Baldry, H. A. Jung, J. M. Sun, S. H. Lee, J. S. Ahn, Y. J. Kim, Y. Lee, D. W. Kim, S. W. Kim, K. H. Lee, W. J. Lee, J. W. Choi, K. Chong, J. I. Lee, S. H. Gwon, N. H. Son and M. J. Ahn, J. Clin. Oncol., 2024, 42, 2747–2756 CrossRef CAS PubMed.
- A. Leonetti, S. Sharma, R. Minari, P. Perego, E. Giovannetti and M. Tiseo, Br. J. Cancer, 2019, 121, 725–737 CrossRef.
- P. R. Lockman, R. K. Mittapalli, K. S. Taskar, V. Rudraraju, B. Gril, K. A. Bohn, C. E. Adkins, A. Roberts, H. R. Thorsheim, J. A. Gaasch, S. Huang, D. Palmieri, P. S. Steeg and Q. R. Smith, Clin. Cancer Res., 2010, 16, 5664–5678 CrossRef CAS.
- D. V. Titov, B. Gilman, Q. L. He, S. Bhat, W. K. Low, Y. Dang, M. Smeaton, A. L. Demain, P. S. Miller, J. F. Kugel, J. A. Goodrich and J. O. Liu, Nat. Chem. Biol., 2011, 7, 182–188 CrossRef CAS PubMed.
- P. A. Phillips, V. Dudeja, J. A. McCarroll, D. Borja-Cacho, R. K. Dawra, W. E. Grizzle, S. M. Vickers and A. K. Saluja, Cancer Res., 2007, 67, 9407–9416 CrossRef CAS PubMed.
- B. Xu, X. Guo, S. Mathew, A. L. Armesilla, J. Cassidy, J. L. Darling and W. Wang, Cancer Lett., 2010, 291, 200–208 CrossRef CAS.
- Q. D. Deng, X. P. Lei, Y. H. Zhong, M. S. Chen, Y. Y. Ke, Z. Li, J. Chen, L. J. Huang, Y. Zhang, L. Liang, Z. X. Lin, Q. Liu, S. P. Li and X. Y. Yu, Acta Pharmacol. Sin., 2021, 42, 1486–1497 CrossRef CAS.
- F. Shao, G. Wang, H. Xie, X. Zhu, J. Sun and J. A, Biol. Pharm. Bull., 2007, 30, 702–707 CrossRef CAS PubMed.
- W. Song, M. Liu, J. Wu, H. Zhai, Y. Chen and Z. Peng, Curr. Drug Metab., 2019, 20, 147–154 CrossRef CAS PubMed.
- F. Shao, G. Wang, J. Sun, H. Xie, H. Li, Y. Liang, R. Zhang and X. Zhu, J. Pharm. Biomed. Anal., 2006, 41, 341–346 CrossRef CAS PubMed.
- C. Xi, S. Peng, Z. Wu, Q. Zhou and J. Zhou, Biomed. Pharmacother., 2017, 90, 531–541 CrossRef CAS.
- R. Chugh, V. Sangwan, S. P. Patil, V. Dudeja, R. K. Dawra, S. Banerjee, R. J. Schumacher, B. R. Blazar, G. I. Georg, S. M. Vickers and A. K. Saluja, Sci. Transl. Med., 2012, 4, 156ra139 Search PubMed.
- Z. L. Zhou, Y. X. Yang, J. Ding, Y. C. Li and Z. H. Miao, Nat. Prod. Rep., 2012, 29, 457–475 RSC.
- L. S. Zeng, P. Yang, Y. Y. Qin, W. H. He and L. Cao, Riv. Eur. Sci. Med. Farmacol., 2023, 27, 10181–10203 Search PubMed.
- Q. Tian, P. Zhang, Y. Wang, Y. Si, D. Yin, C. R. Weber, M. L. Fishel, K. E. Pollok, B. Qiu, F. Xiao and A. S. Chong, Elife, 2023, 12, e85862 CrossRef CAS PubMed.
- C.-N. Members and Partners, Nucleic Acids Res., 2022, 50, D27–D38 CrossRef.
- T. Chen, X. Chen, S. Zhang, J. Zhu, B. Tang, A. Wang, L. Dong, Z. Zhang, C. Yu, Y. Sun, L. Chi, H. Chen, S. Zhai, Y. Sun, L. Lan, X. Zhang, J. Xiao, Y. Bao, Y. Wang, Z. Zhang and W. Zhao, Genomics, Proteomics Bioinf., 2021, 19, 578–583 CrossRef.
- Q. Zhang, R. Abdo, C. Iosef, T. Kaneko, M. Cecchini, V. K. Han and S. S. Li, Nat. Commun., 2022, 13, 5983 CrossRef PubMed.
- P. Goldstraw, D. Ball, J. R. Jett, T. Le Chevalier, E. Lim, A. G. Nicholson and F. A. Shepherd, Lancet, 2011, 378, 1727–1740 CrossRef.
- F. R. Hirsch, G. V. Scagliotti, J. L. Mulshine, R. Kwon, W. J. Curran, Jr., Y. L. Wu and L. Paz-Ares, Lancet, 2017, 389, 299–311 CrossRef CAS.
- L. Miarka and M. Valiente, Neurooncol Adv., 2021, 3, v144–v156 Search PubMed.
- H. L. Wang, M. Guo, H. D. Wei and Y. H. Chen, Signal Transduct. Target. Ther., 2023, 8, 92 CrossRef CAS.
- Z. Wang, S. Zhang, L. M. Irakoze and Y. Zhao, Eur. J. Med. Chem., 2025, 297, 117909 CrossRef CAS.
- H. B. Xie, K. F. Ma, K. N. Zhang, J. C. Zhou, L. Li, W. P. Yang, Y. Q. Gong, L. Cai and K. Gong, Cell Death Differ., 2021, 12, 1 CAS.
- L. Wang, Y. Xu, L. Fu, Y. Li and L. Lou, Cancer Lett., 2012, 324, 75–82 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally: J. Liu and L Liang. |
| This journal is © The Royal Society of Chemistry 2026 |