Unravelling the antibacterial performances of a homochiral D-configured tetraphenylalanine appended 16-HPA derivative based mechanoresponsive and proteolytically stable hydrogel
Vaibhav
Shivhare
 a,
Anindya
Basu
a,
Anindya
Basu
 bc and
Anita
Dutt Konar
bc and
Anita
Dutt Konar
 *abc
*abc
aDept. of Chemistry, Rajiv Gandhi Technological University, Bhopal, India. E-mail: anitaduttkonar@rgpv.ac.in
bSchool of Pharmaceutical Sciences, Rajiv Gandhi Technological University, Bhopal, India
cUniversity Grants Commission, India
First published on 1st November 2025
Abstract
Microbial infection is one of the most pressing global challenges worldwide, and imposes significant economic burdens on healthcare systems. This work represents a rational combinatorial strategy that leverages hydrophobic harmony in multiple phenylalanine fragments, anchored to an amphiphile 16-hydroxy-palmitic acid at the N-terminus (16-HPA-D-Phe-D-Phe-OH, compound I; 16-HPA-D-Phe-D-Phe-D-Phe-OH, compound II; 16-HPA-D-Phe-D-Phe-D-Phe-D-Phe-OH, compound III), such that a viable therapeutic skeleton could be uncovered through this strategy. In pursuit of this objective, the minimum inhibitory concentrations of compounds I–III were investigated using four distinct microorganisms namely Staphylococcus aureus and B. subtilis (Gram positive), and E. coli and P. aeruginosa (Gram negative). Our systematic examination reflected that from a pool of three skeletons, compound III comprising of D-configured tetraphenylalanines displayed not only mechanoresponsive assisted hydrogelation propensities at physiological pH, but also excellent antibacterial activities in vitro, in the Gram positive micro-organisms backed by molecular modelling studies. Henceforth compound III was selected from the design and proceeded for its elaborate antibacterial activities using colony counting experiment, bacterial scanning electron microscopy and live–dead assay using flow cytometry. Furthermore, the β-sheet structured compound III, stabilized by weak non-covalent interactions, depicted optimum mechanical strength as well as proteolytic stability for 72 h when exposed to the proteolytic enzyme, proteinase K and chymotrypsin. Overall, our analysis highlights the potential of compound III as a promising candidate for future antimicrobial therapy. However, further experiments are necessary to validate these findings, and current claims are reflective of an early proof-of-concept until further preclinical data are available.
1. Introduction
Self-assembled mechanical stress responsive hydrogels derived from peptides have captivated immense impetus recently from diverse segments of therapeutic discovery.1–10 These hydrogels have more advantages over other stimuli responsive systems as they are devoid of drastic conditions and complex techniques that might affect the biocompatibility of the molecules. The propensity of the scaffolds to undergo a reversible phase change between gel/sol states upon application/withdrawal of mechanical stress demonstrates revolutionary advantages for clinical applications due to the exceptional tunability and reversible nature of supramolecular interactions, within a restricted time interval. However, these mechanoresponsive hydrogels often occupy the back seat due to weak mechanical integrity which impedes their suitability as preferential candidates for biomedical research.1–10 Although addition of a cross linking agent at times instigates the stability and longevity of hydrogels, they require repeated injection to maintain their efficacy.1–10 Therefore, there is an urgent prerequisite for the discovery of intelligent injectable materials that are user friendly, secured and enduring.Microbial infection is one of the most pressing global challenges worldwide, and imposes significant economic burdens on healthcare systems.11–20 This is because the rapid multiplication time of these micro-organisms through binary fission enhances continuous growth, thereby accentuating adaptability against antimicrobial agents. Thus, traditional antibiotic treatments are associated with over-prescription, culminating in the progressive rise of antibiotic resistance of various pathogens.
Along this perspective, heterocycles play a pivotal role as they are applied to boost the effectiveness and power, bioavailability and selectivity of target molecules.21 Ciprofloxacin (CPFX), a member of the second generation of fluoroquinolones, have shown strong antibacterial potential along with excellent pharmacokinetic properties. However, its adverse side effects make it a backbencher.22
In pursuit of this objective, in the past few years, there have been significant ongoing investigations that are focused on the use of diverse scaffolds that served as antibacterial agents, but each option has its own constraints.23–32 For example, the gels that incorporated noble metal nanoparticles exerted negative reactions encompassing changes in skin or eye color, oxidative DNA damage, and inflammation. Another example revealed that biomaterials loaded with antibiotics exhibited proteolytic degradation that required painful surgery for its removal. Few other charged scaffolds exerted cytotoxic effects in systems.23–25 Keeping this view in mind, we envisioned that if the benchmarks, namely (a) mechano-responsiveness, (b) injectability, and (c) significant mechanical strength, could be engineered under a single roof, and an appropriate skeleton with innate antibacterial potential could be extracted by hydrophobic harmony, our discovery would provide a solution to many unconquered stumbling blocks that remained unvalidated to date.
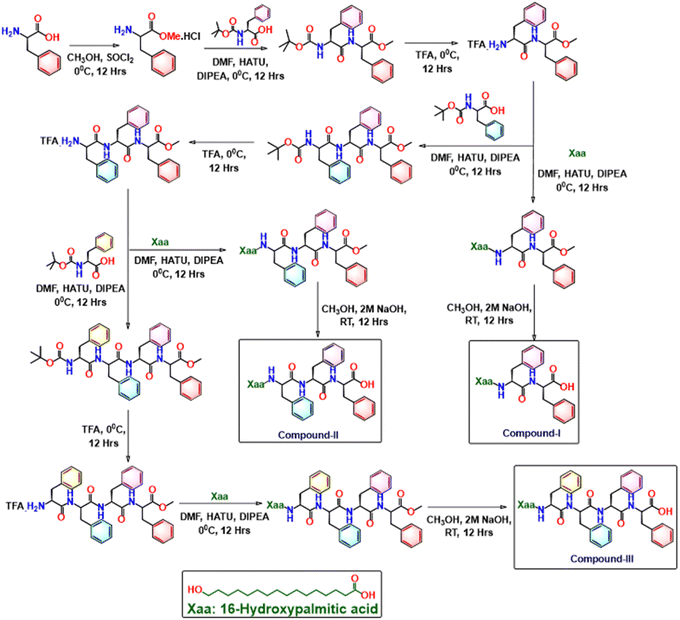
Henceforth, we initiated our voyage, inspired with the reported molecule: Boc-δ-Ava-D-Phe-Phe-OH comprising of homochiral D-configured diphenylalanine fragments, appended to an ω-amino acid, with four methylene units at the N-terminus, that showed moderate antibacterial activity.33 In search of better peptide based antibacterial agents, we replaced the δ-amino valeric acid with 16-HPA: 16-hydroxypalmitic acid (16-HPA: 16-hydroxypalmitic acid) and synthesized compound I: 16-HPA-D-Phe-D-Phe-OH, as the amphiphile is known to inculcate antibacterial propensities in the molecule (Fig. 1).34,35 Motivated by the information that an increase of residues enhances the overall amphiphilic character of the molecules as well as its activities, we enhanced the number of chiral units of compound I sequentially and synthesized compound II (with triphenylalanines 16-HPA-D-Phe-D-Phe-D-Phe-OH) and compound III (with tetraphenylalanines, 16-HPA-D-Phe-D-Phe-D-Phe-D-Phe-OH).36 Our systematic examination reflected that from a pool of three skeletons; compound III with the highest partition coefficient displayed not only mechanoresponsive assisted hydrogelation propensities at physiological pH, but also excellent antibacterial activities in vitro, in the Gram positive micro-organisms Staphylococcus aureus and B. subtilis backed by molecular modelling studies (Fig. 1). Henceforth compound III was selected from the series and proceeded for its elaborate antibacterial action using colony counting experiment, bacterial scanning electron microscopy and live–dead assay using flow cytometry. Indeed, the entire journey as to how the injectable compound III was deemed ideal for antibacterial use has been described in depth in later sections.
2. Experimental
2.1 Peptide synthesis
Compounds I–III were synthesized via a solution-phase method and purified by silica gel column chromatography using ethyl acetate/petroleum ether as the eluent. Structural characterization was performed using 1H NMR & IR spectroscopy, along with mass-spectrometry (Scheme 1 and Fig. S1–S3).36–392.2 Antimicrobial activity of compounds I–III
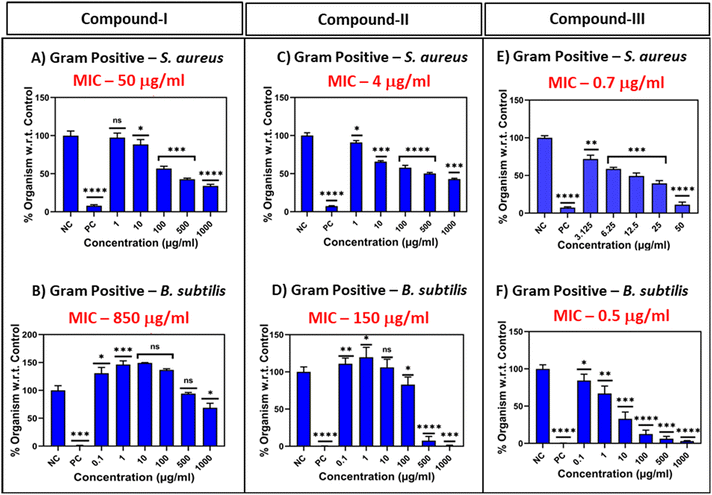
The in vitro antimicrobial activity of compounds I–III was studied against four reported micro-organisms, Staphylococcus aureus and B. subtilis (Gram positive), and E. coli and P. aeruginosa (Gram negative) by optical density methods.36–39 Fresh inocula of the organisms were made from their respective cultures, which were obtained from NCCS Pune as lyophilized powder, prior to the starting of the experiments and proceeded as described in ref. 36–39 (Fig. 2 and S4). Here, bacterial solution including ciprofloxacin was considered a positive control while only the nutrient broth was used as a negative control (Fig. 2 and S4).2.3 Hydrogel preparation
For the preparation of the hydrogels, 20 mg of each of compounds I–III were solubilized in 1 mL of phosphate buffer (4.2/7.5/9.2 pH) in a glass vial by gentle heating until a homogenous solution was obtained. The latter was left undisturbed for some time and gel forming propensity was noted. Compounds I & II formed gels only in pH 9.2 phosphate buffer, whilst compound III formed gels in all three pH buffers (Fig. S5). The xerogels were prepared by drying the gels slowly at room temperature using an anhydrous calcium chloride desiccator. Concentration dependent UV studies and stability measurement of a particular concentration of compound III were performed for 7 days (Fig. S6).392.4 Docking studies
To check out plausible internal targets apart from the membrane, we determined the binding ability of our molecule with some of the popular antibiotic targets within bacterial cells as described: A) a DNA gyrase receptor co-crystallized with ciprofloxacin (PDB ID: 2XCT, Fig. 7A),21,22 B) penicillin binding protein-class B (transpeptidase) co-crystallized with penicillin (β-lactam) (PDB ID: 6MKF, Fig. 7B),40 C) penicillin binding protein-class PBP1b co-crystallized with acyl ampicillin and moenomycin (acetylated β-lactam) (PDB ID: 5HL9, Fig. 7C),41 D) a 30S ribosomal subunit (bacterial 30S head) co-crystallized with tetracycline (PDB ID: 8CF1, Fig. 7D),42 and E) a TcaR-MarR-family transcriptional regulator co-crystallized with ampicillin and other aminoglycoside/β-lactam antibiotics (PDB ID: 6KP3, Fig. 7E).43,44Considering the docking scores and the corresponding binding energies (Table S1), it seems that our target molecule possesses higher affinity towards the DNA gyrase binding site compared to others considered.21,22,40–44
Henceforth, for gaining better explanation, compounds I–III were curtailed using the same constraints at the active site of DNA-gyrase taken from ref. 22, and docked using the protocol as described in ref. 37–39 (Fig. 3, S7 and S8, Tables S1 and S2).
 | ||
| Fig. 3 Illustrative diagrams showing the interaction between the receptor and: A) standard drug ciprofloxacin; B) compound I; C) compound II; D) compound III. | ||
2.5 Colony counting experiment of compound III
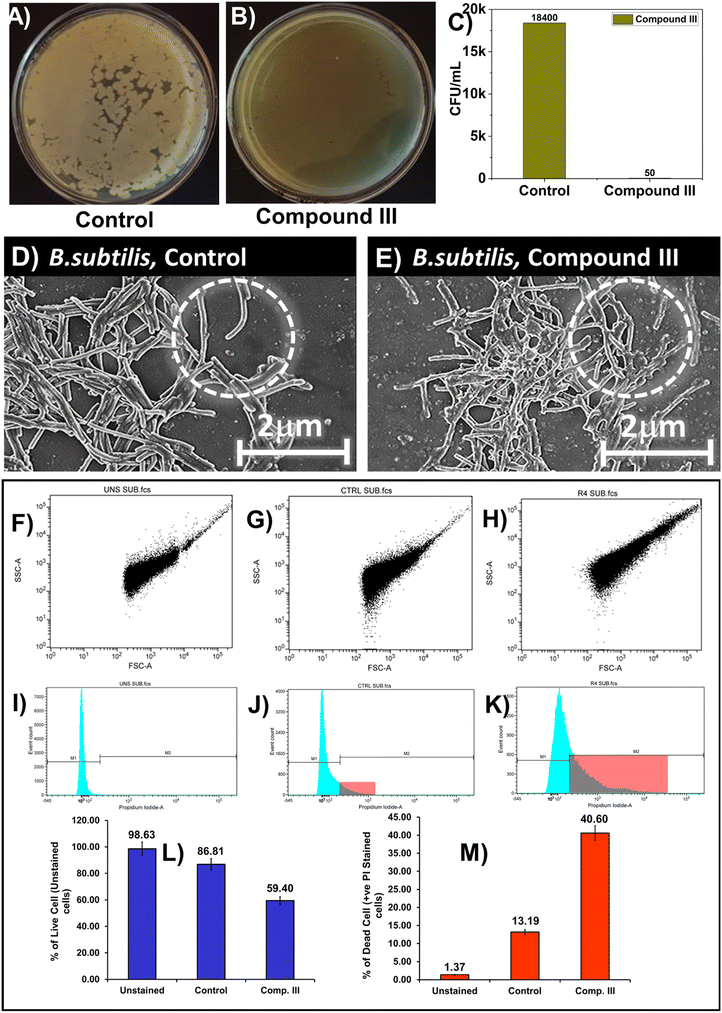
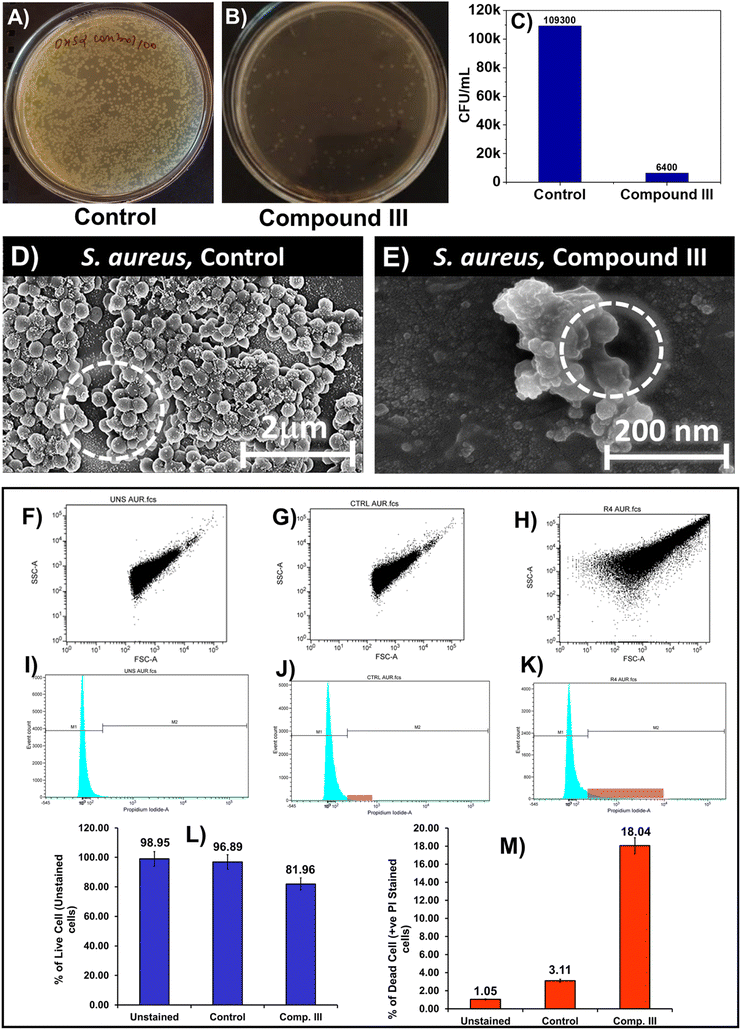
Bacterial culture samples, both untreated (control) and treated with compound III, collected at regular intervals, were subjected to 1000 dilutions and used for the quantification of live bacteria. For this, 20 mL of the diluted bacterial culture was spread on agar (1.5% w/v) plates containing media and incubated overnight at 37 °C temperature. The number of colonies obtained was determined using a colony counter. The concentration of live bacteria in the original culture was reported as cfu mL−1 upon consideration of the dilution factor (Fig. 4).392.6 Bacterial SEM of compound III
FESEM analysis of the control and test sample (treated with compound III) was performed using a field emission scanning electron microscope (JEOL scanning electron microscope model no. JSM-7600F) based on a reported protocol, to explore the changes in cell morphology when incubated with S. aureus and B. subtilis microorganisms (Fig. 4D and E and 5D and E).392.7 Live–dead assay of compound III using flow cytometry
The culture preparation for this experiment was done following the protocol described in ref. 33. In this experiment, 1 mL of cultures of respective micro-organisms were collected in fresh tubes, centrifuged and suspended in PBS buffer.A requisite amount of microbes nearly 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in number were taken for staining with propidium iodide (PI), dispensed in PBS and acquisition of the samples were done in a flow cytometer (BD FACS Caliber, USA) (Fig. 4F–K and 5F–K).45–49
000 in number were taken for staining with propidium iodide (PI), dispensed in PBS and acquisition of the samples were done in a flow cytometer (BD FACS Caliber, USA) (Fig. 4F–K and 5F–K).45–49
2.8 Rheological properties of compound III
Mechanical integrity of compound III was investigated using an Anton Paar Physica MCR 301 rheometer, where moduli were explored with respect to angular frequency as described in ref. 37–39. The injectability was studied by step-strain experiments at a constant frequency of 10 rad s−1, and strains introduced were altered from 0.1 to 40% (Fig. 6).37–392.9 Biocompatibility and proteolytic stability studies of the hydrogels
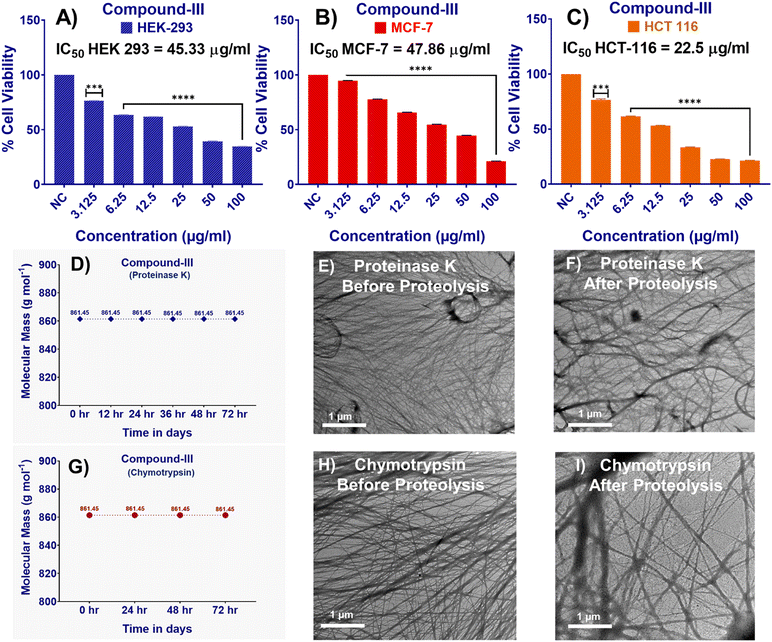
In order to ascertain the biocompatibility of compound III, the MTT assay protocol was followed in three different cell lines HEK293, MCF7 and HCT 116 according to ref. 37–39 (Fig. 7A–C).Proteolytic degradation studies of compound III was done as per ref. 37–39 using two different proteolytic enzymes, proteinase K and chymotrypsin (Fig. 7D, G and S9). Transmission electron micrographs before and after proteolysis were taken using a JEOL electron microscope (model: JEM-2100). Sample preparation was done following ref. 37–39 (Fig. 7E, F, H and I and S9).
A detailed protocol of haemolysis and plasma stability studies was given in the experimental section (Fig. 8, 9, S10 and S11).
2.10 Determination of the conformation of compound III
The conformation of compound III was determined using the following techniques described below with a concentration of 30 mg mL−1 for all the structural experiments (Fig. 10):37–393. Results and discussion
3.1 Rationale behind the design strategy
We commenced our journey, inspired with a previous report, from our lab, where we decided to replace the N-terminus ω-amino acid residue of Boc-δ-Ava-D-Phe-Phe-OH by 16-HPA: 16-hydroxypalmitic acid (16-HPA: 16-hydroxypalmitic acid) and designed compound I: 16-HPA-D-Phe-D-Phe-OH (Fig. 1).33 Motivated by the information that an increase of residue number enhances activities, we increased the number of chiral units sequentially and synthesized compound II (16-HPA-D-Phe-D-Phe-D-Phe-OH) and compound III (16-HPA-D-Phe-D-Phe-D-Phe-D-Phe-OH).36 We hypothesized that the amphiphilic character of the molecules are expected to facilitate their interaction with the microbial cell membranes, thereby contributing to enhanced antimicrobial activity. Furthermore, the strategic incorporation of D-amino acids into the peptide backbone is projected to improve both the mechanical robustness and resistance to enzymatic degradation of the resulting hydrogels—addressing a key limitation of peptide-based scaffolds relative to conventional polymers. Moreover, biocompatibility and in situ gelation are inherently addressed through a sol–gel transition mechanism that is triggered by mechanical stress, offering a more straightforward alternative to hydrogels that rely on external stimuli.The molecules were synthesized by solution phase methodology, purified by column chromatography and characterized using 1H NMR, IR and mass spectrometry (Scheme 1, Fig. S1–S3). Initially, all three compounds (I–III) demonstrated excellent gelation ability at room temperature at pH 9.2, with minimum gelation concentrations (MGCs) of 17.7, 21.0, and 23.2 mM (equivalent to 20 mg mL−1), respectively. Notably, only compound III exhibited gelation at all three pH values, acidic, basic and physiological pH conditions (Fig. S5).
3.2 Compound III exhibited the highest antibacterial efficacy in the Gram positive microorganisms
Aligned to the investigative workflow, the antibacterial efficacies of compounds I–III, with an increased number of phenylalanine units were studied using the optical density (OD595) method over a wide range of bacteria as described in Fig. 2 and S4.36–39The MIC values for the Gram-positive bacteria S. aureus and B. subtilis for the respective compounds were as follows: S. aureus; a) compound I: 50 μg mL−1; b) compound II: 4 μg mL−1; c) compound III: 0.7 μg mL−1; B. subtilis; a) compound I: 850 μg mL−1; b) compound II: 150 μg mL−1; c) compound III: 0.5 μg mL−1. Our results indicated that with the enhancement of D-phenylalanine units, the growth of B. subtilis/S. aureus was significantly inhibited by the compounds. We also performed experiments with Gram-negative organisms, namely E. coli and P. aeruginosa, but did not observe potency in them similar to that of the Gram-positive ones (Fig. 2 and S4).36–39 Our analysis demonstrated that for Gram-positive organisms, the tetraphenylalanine system exhibited significantly greater potency compared to its diphenylalanine and triphenylalanine analogues. This trend was supported by the partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) values measured in the n-octanol/water system for compounds I–III, which were found to be 1.058, 1.987, and 1.632, respectively, using DFT calculations.38 The enhanced potency observed in the tetrapeptide series might be attributed to reaching a threshold log
P) values measured in the n-octanol/water system for compounds I–III, which were found to be 1.058, 1.987, and 1.632, respectively, using DFT calculations.38 The enhanced potency observed in the tetrapeptide series might be attributed to reaching a threshold log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value—achieved through the incorporation of a fourth chiral residue—which aligns with our design principles.
P value—achieved through the incorporation of a fourth chiral residue—which aligns with our design principles.
So, compound III was selected and used for its detailed antibacterial activity.
Moreover, our experiments demonstrated that the antimicrobial activity against Gram-negative bacteria was weaker compared to Gram-positive bacteria. This observation is consistent with previous studies and can be attributed to structural differences in their cell envelopes. Gram-negative bacteria possess an additional outer membrane composed of lipopolysaccharides (LPSs), which acts as a permeability barrier. This outer layer can impede the penetration of many antimicrobial agents, thereby reducing their efficacy. In contrast, Gram-positive bacteria lack this outer membrane, making them more susceptible to such compounds.33
Indeed, concentration dependent UV studies exhibited an identical UV pattern with variation in concentration of compound III, confirming the significant stability of molecules. Also, compound III exhibited stability for 7 days, as an identical curve was obtained for each day, further emphasizing the stability of the molecule for a considerable number of days (Fig. S6).39
3.3 What could be the reason for better potency in compound III?
To check out plausible internal targets apart from the membrane, we determined the binding ability of our molecule with some of the popular antibiotic targets within bacterial cells as described:A) a DNA gyrase receptor co-crystallized with ciprofloxacin (PDB ID: 2XCT, Fig. S7A), B) penicillin binding protein-class B (transpeptidase) co-crystallized with penicillin (β-lactam) (PDB ID: 6MKF, Fig. S7B), C) penicillin binding protein-class PBP1b co-crystallized with acyl ampicillin and moenomycin (acetylated β-lactam) (PDB ID: 5HL9, Fig. S7C), D) 30S ribosomal subunit (bacterial 30S head) co-crystallized with tetracycline (PDB ID: 8CF1, Fig. S7D) and E) TcaR-MarR-family transcriptional regulator co-crystallized with ampicillin and other aminoglycoside/β-lactam antibiotics (PDB ID: 6KP3, Fig. S7E).37–44
Considering the docking scores and the corresponding binding energies (Table S1), it seems that our target molecule possesses higher affinity towards the DNA gyrase binding site compared to others considered. Henceforth for getting better explanation, compounds I–III were curtailed using the same constraints at the active site of DNA-gyrase as described in ref. 37–39 (Fig. 3, S7 and S8, Tables S1 and S2).21,22
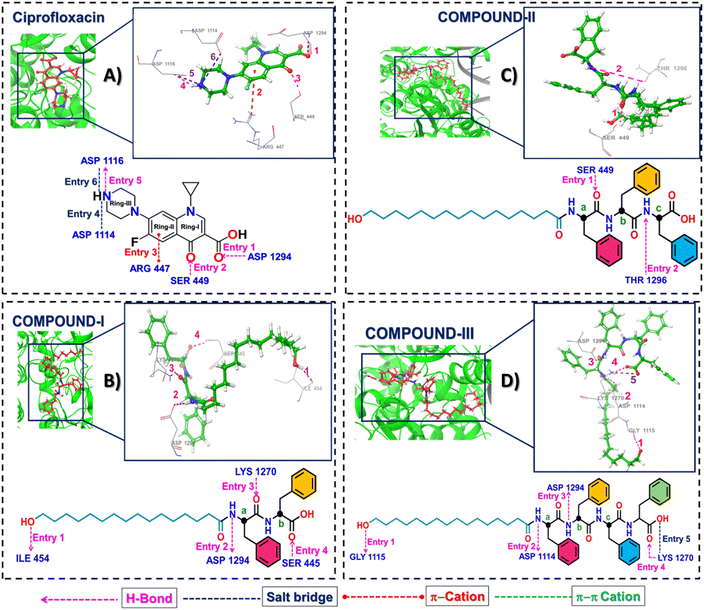
Closer inspection of the co-crystallized ligand ciprofloxacin with the receptor illustrated a docking score of −4.18, with a binding energy of −19.51 kcal mol−1 leading to 6 different interactions, 3 H-bonding, 2 salt bridges and a pi–cation bond respectively as described (Table S2, Fig. 3 and S8): entry 1) firstly, the ASP1294 sidechain –OH of carboxylate serves as a donor and CO of COOH of ring 1 as an acceptor; entry 2) secondly, SER 449 OH as a donor and CO of ring 1 as an acceptor; entry 3) thirdly, piperazine NH of ring 3 as a donor and ASP 1116 CO of the sidechain carboxylate as an acceptor. Moreover, the piperazine NH of ring 3 enters into two salt bridge interactions, one with ASP 1114 (entry 4) and the other with ASP 1116 (entry 5) (Table S2, Fig. 3 and S7). Finally, a pi–cationic correspondence was observed between ring 2 of ciprofloxacin and sidechain NH of ARG 447 (entry 6) (Table S2, Fig. 3 and S8).
Next, compound I, containing two D-phenylalanine residues, showed a docking score of −4.65 and a binding energy of −25.57 kcal mol−1, resulting in four hydrogen bonding interactions, as described below: entry 1) firstly, the sidechain –OH of 16-HPA functions as a donor and CO of ILE 454 as an acceptor; entry 2) secondly, PHE (1) NH as a donor and ASP1294 sidechain O– of –OH of carboxylate as an acceptor; entry 3) thirdly, LYS 1270 sidechain NH as a donor and PHE (1) CO as an acceptor; entry 4) fourthly, SER 445 sidechain OH as a donor and PHE (2) CO as an acceptor (Table S2, Fig. 3 and S8).
Next, the docking score of compound II, with D-configured triphenylalanines, was found to be −4.37, with a binding energy of −29.74 kcal mol−1 leading to 2 H-bonding interactions only: entry 1) firstly, SER 449 sidechain OH as a donor and PHE (1) CO as an acceptor; entry 2) secondly, PHE (2) NH as a donor and THR1296 sidechain O of –OH as an acceptor (Table S2, Fig. 3 and S8).
Finally, compound III, with D-configured tetraphenylalanines, depicted a docking score of −6.99 and a binding energy of −28.73 kcal mol−1 with 5 different interactions (4-H-bonding and 1 salt bridge). The H-bonding interactions were as follows: entry 1) firstly, the sidechain –OH of 16-HPA as a donor and GLY 1115 carbonyl as an acceptor; entry 2) secondly, NH of PHE(1) as a donor and –O– of OH of sidechain carboxylate of ASP 1114 as an acceptor; entry 3) thirdly, PHE(2) NH as a donor and –O– of OH of sidechain carboxylate of ASP 1294 as an acceptor; 4) fourthly, LYS 1270 sidechain NH as a donor and PHE(4) CO of carboxylate as an acceptor (Table S2, Fig. 3 and S8); fifthly, a salt bridge interaction between the same LYS 1270 sidechain NH and O– of carboxylate of PHE(4).
Detailed study of the docking results indicate that compound III, comprising of tetraphenylalanines, possessed an optimum partition coefficient of 1.68 that might have allowed the molecule to interact with the prime residue ASP1114 responsible for imparting higher antibacterial efficacy in the compound. However, compounds I and II, due to the presence of a lesser number of phenylalanine units, failed to display interactions with these two residues, the probable causative factor for lower potency in the microorganisms.21
Next, we moved towards the detailed antibacterial activity in the two micro-organisms B. subtilis and S. aureus as described below.
3.4 Detailed antibacterial activities of compound III
For both the microorganisms, in the control, the membranes were intact, but for the treated ones, membrane wrinkling, blister formation and rupture were noted.39
Since our molecule is a tetraphenylalanine–16-HPA amphiphilic derivative hydrogel comprising of a lipophobic/lipophilic part, it might exert antibacterial action via multiple mechanisms including membrane disruption and/or inhibition of some internal targets involving one or more cellular machineries. The detrimental impact of the peptide on the bacterial cells is clearly evident from the corresponding TEM images, where bacterial membrane disruption could be seen.
In this experiment, three groups were considered where microorganism cultures were present in all, but variation occurred in PI staining and categorized accordingly as a) unstained (without PI); b) control (with PI) and c) compound III (with PI). The pattern could be linked to different cellular states. As is typical for the LIVE/DEAD assay, the viable bacterial population in the dot plot demonstrates strong green fluorescence and weak red fluorescence, whilst a completely permeabilized population represents weak green fluorescence and strong red fluorescence.45–49 In the intermediate state, there occurs a significant shift of fluorescence signal from green to red.45–49Fig. 4 shows the flow cytometric qualitative and quantitative analysis of B. subtilis. The qualitative results obtained, in the form of dot plots (here forward scattering FSC is plotted (x-axis) against side scattering (y-axis) SSC), indicated that for unstained and control, the bacterial morphology remained in clusters affirming healthy intact cells (Fig. 4F–H).45–49 However for compound III, the treated sample, the dot plot suggests an increase in cell granularity which might be due to membrane disruption, characterised by enhanced red fluorescence.45–49
In the histograms, in the unstained case, all the bacterial populations remained in the M1 zone (live zone), indicating viable microorganisms, characterized by low fluorescence (Fig. 4I–K). In the control, similar observation to that of unstained was noticed, with a small tail towards M2 (dead zone), reflecting slightly enhanced fluorescence, suggesting the presence of a small population of dead bacteria (Fig. 4I–K).41–44 Surprisingly, in the case of compound III, a significant shift in population from M1 to M2 was observed with an increased PI fluorescence. Finally, the quantitative bar graph representation is in line with the qualitative analysis data, where 40.60% organisms were dead in the treated, and 13.19% were dead in the control, emphasizing the antibacterial efficacy of compound III (Fig. 4L and M).
We repeated the same experiment with S. aureus and found the dot plots and histograms to show similar inference to that of B. subtilis (Fig. 5F–K).
Although compound III was found to be active against S. aureus, its efficacy was found to be significantly lower than B. subtilis, as reflected by the bar graph plot; 18.04% organisms were dead in the treated one and 3.11% were dead in the control (Fig. 5L and M).
In a nutshell, the antibacterial experiments confirm the high efficacy of compound III as a potential antibacterial agent.
3.5 Compound III exhibited significant mechanical strength and mechanoresponsiveness
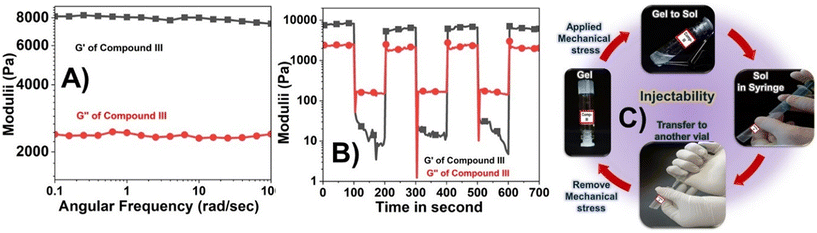
Next, we proceeded for the determination of mechanical integrity of the gels with a concentration of 30 mg mL−1, using rheological measurements, as it is one of the prime requirement for any gel to be used as a potential material. Frequency sweep measurements conducted at a fixed strain of 0.1% demonstrated that compound III possessed a storage modulus (G′) consistently greater than the loss modulus (G″) across the entire frequency range tested, indicating its viscoelastic character (Fig. 6A).To further evaluate its injectability under mechanical stress, a time-dependent step-strain experiment was performed (Fig. 6B and C).
The hydrogel was alternately exposed to low (0.1%) and high (40%) strain levels at a constant angular frequency of 10 rad s−1. As illustrated in Fig. 6B, under low strain (0.1%), G′ remained higher than G″ confirming the gel state. When the strain was increased to 40%, G′ decreased while G″ increased and exceeded G′, signifying a transition from gel to sol phase.
Upon reducing the strain back to 0.1%, G′ rapidly recovered and surpassed G″ once again, indicating the reformation of the gel network. The hydrogel showed nearly 100% recovery of its mechanical properties within approximately 100 seconds. This process was repeated over four cycles, demonstrating the material's reproducible and reversible mechanoresponsive behavior.
3.6 Compound III displayed excellent biocompatibility and proteolytic stability
For any biomedical application, proteolytic stability and bio-compatibility are two very important parameters that need to be taken care of. To check the biocompatibility, the cell viability experiment was performed by MTT assay, where compound III was incubated with the HEK 293 (human embryonic kidney) cell line. All the cell lines used for this study were purchased from NCCS Pune. The IC50 value was found to be 45.35 μg mL−1, which was optimum for biomedical applications (Fig. 7A).36–39 We have also added MTT assay experiments in two other different cell lines, B) MCF7 and C) HCT116, to diversify our claim. The IC50 values of compound III in different cell lines have been mentioned in the respective figure. As evident from Fig. 2, the MICs in the Gram positive micro-organisms were found to be 0.7 μg mL−1, which was found to be quite lower to the IC50 values (Fig. 7B and C). So, our compound III was found to be biocompatible in cell lines of diversified nature.To assess the proteolytic stability, compound III was incubated with the proteolytic enzyme proteinase K and chymotrypsin for 72 hours. Degradation was monitored using mass spectrometry at regular intervals—and subsequently up to 72 hours. The corresponding spectra are presented in Fig. S9, showing the data before proteolysis (spectrum A) and after treatment at different time points [(B) 12 h, (C) 24 h, (D) 36 h, (E) 48 h, (F) 60 h, (G) 72 h] for proteinase K and [(B) 24 h, (C) 48 h, (F) 72 h] for chymotrypsin. Across all spectra, the intact molecular ion peak of compound III ((M + H)+: 861.5) remained at 100%, confirming its resistance to enzymatic degradation. No peaks corresponding to expected fragments (fragments 1–5) were observed, further highlighting the compound's proteolytic stability.
To evaluate whether any structural changes occurred due to proteolysis, TEM was employed. The resulting micrographs consistently displayed a fine, delicate filamentous fibrillar morphology before and after treatment with both the proteolytic enzymes, proteinase K and chymotrypsin, suggesting no significant morphological alterations before and after proteolysis (Fig. 7D, E, H and I).
To access the haemocompatibility of the molecule, freshly isolated Swiss albino mouse red blood cells (RBCs) were used for the haemolysis experiment (vide order number: CPCSEA REG.NO.PH/VNS/2K23/37 in a meeting held on 03/11/2023).
The HC50 of compound III was 582.62 mg mL−1, whilst the MIC in the Gram positive micro-organisms was found to be 0.7 mg mL−1, nearly 800 times higher. So, our compound III was found to be haemocompatible too (Fig. 8 and S10).
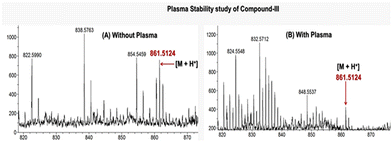
To check the plasma stability of compound III, we incubated the sample by following a standard protocol. Our mass spectra clearly indicated a peak of [M + H]+, before and after treatment, indicating significant plasma stability. The presence of plasma contaminants, despite protein precipitation with TCA–acetone following incubation, might be responsible for reducing the relative abundance of our target molecule (Fig. 9 and S11).50
Nevertheless, to draw more meaningful conclusions about the biomedical potential of compound III, a deeper investigation into its structural and morphological characteristics is warranted.
3.7 Compound III exists in supramolecular β-sheets
We next investigated the self-assembly behavior of compound III using a combination of spectroscopic techniques. Our initial focus was to assess the role of hydrogen bonding in its conformation through variable temperature-dependent NMR spectroscopy (VT-NMR) (Fig. 10A and S12).Temperature-dependent 1H NMR spectra revealed that the backbone amide protons of D-Phe(1), D-Phe(2), D-Phe(3), and D-Phe(4), which appeared at δ 7.89, 7.92, 8.10, and 8.32 ppm, respectively, at room temperature (25 °C), shifted to δ 7.69, 7.75, 7.89, and 8.04 ppm at 65 °C. The corresponding changes in the chemical shift (Δδ) were 0.20, 0.17, 0.21, and 0.28 ppm, respectively (Fig. 10A and S9).
These downfield shifts indicate that the NH protons are solvent-exposed, suggesting their involvement in intermolecular hydrogen bonding within the self-assembled structure.38,39
To further support this hypothesis, FT-IR spectroscopy was performed (Fig. 10B).38,39 Characteristic peaks observed at 3304, 2915, 2849, 1707, 1638, 1537, and 3388 cm−1 were attributed to hydrogen-bonded amide N–H groups and carbonyls. These spectral features are indicative of β-sheet-like arrangements stabilized by intermolecular hydrogen bonding and π–π stacking interactions.37–39
Finally, powder X-ray diffraction (PXRD) analysis was conducted to confirm the supramolecular architecture. Distinct diffraction peaks at 19.10° (d = 4.8 Å), 19.50° (d = 4.7 Å), 23.15° (d = 3.7 Å), and 25.04° (d = 3.4 Å) were observed, consistent with π–π stacking and hydrogen-bond-stabilized sheet-like assemblies. These findings corroborate the results obtained from NMR and FT-IR analyses (Fig. 10C).37–39
Conclusion
In summary, this work represents a rational combinatorial strategy that leverages hydrophobic harmony in multiple phenylalanine fragments, anchored to an amphiphile 16-hydroxy-palmitic acid at the N-terminus such that a viable therapeutic skeleton, addressing the microbial infection related challenges, could be uncovered through this approach. Our systematic examination reflected that from a pool of three skeletons, compound III, with a threshold partition coefficient, displayed not only mechanoresponsive assisted hydrogelation propensities at physiological pH, but also excellent antibacterial activities in vitro, in the Gram positive micro-organisms B. subtilis and S. aureus backed by molecular modelling studies.The potential interaction of compound III, to the residue ASP1114 in the receptor comprising of tetraphenylalanines, might be the driving factor for imparting higher antibacterial efficacy in the compound. Indeed, the colony counting experiment of compound III indicated that the colony forming units (cfu mL−1) were found to show nearly 100% reduction in B. subtilis and 94.14% in S. aureus. This observation was further affirmed by the bacterial SEM experiments where membrane wrinkling, blister formation and rupture were noted in the treated sample. Finally, the dot plot data suggested that the increase in cell granularity might be due to membrane disruption, characterised by enhanced red fluorescence. This observation was additionally supported by significant shift in population of micro-organisms from the M1 zone to the M2 zone with an increased PI fluorescence in the histograms.
Besides, the β-sheet-structured compound III, stabilized by intermolecular H-bonding and π–π interactions, depicted optimum mechanical strength as well as proteolytic stability for 72 h when incubated with the proteolytic enzymes, proteinase K and chymotrypsin. Moreover, compound III possessed all the benchmarks, namely (a) mechano-responsiveness, (b) injectability, and (c) significant mechanical strength, under one roof, inevitable for successful biomedical applications.
We anticipate that our work would spark further research in the realm of microbial infections for innovative therapeutic paradigms, further shedding light on the instrumental role of hydrophobic orchestration in fishing out intelligent biomimetic materials, adequate for need-based applications.
Author contributions
ADK conceived the project, designed the experiments and wrote the manuscript. VS carried out the synthesis, characterization, and in vitro analysis. AB contributed in writing the manuscript and analyzing the biological experiments.Conflicts of interest
The author declares no conflict of interest.Data availability
The data supporting this article have been included as part of the supplementary information (SI). 1H NMR, FT-IR, mass of compounds I–III, docking studies and detailed experimental protocols for all experiments carried out.Supplementary information: detailed experimental techniques and spectra data namely 1H NMR, IR, and mass. See DOI: https://doi.org/10.1039/d5md00728c.
Acknowledgements
VS gratefully acknowledges the RGPV Doctoral Fellowship for financial support. ADK thanks the University Grants Commission (UGC), New Delhi for funding under grant numbers F.4-(55)/2014(BSR)/FRP, SPG/2022/002214, and 01WS(006)/2023-24/EMR-II/ASPIRE. The authors also extend their sincere thanks to Mr. Rishabh Ahuja, Mr. Surendra Kumar Ahirwar, and Mr. Dipesh Barde for their valuable assistance during the preparation of the manuscript.Notes and references
- W. Yang, J. Chen, Z. Zhao, M. Wu, L. Gong, Y. Sun, C. Huang, B. Yan and H. Zeng, J. Mater. Chem. B, 2024, 12, 332–349 RSC.
- S. Halder, T. Das, R. Kushwaha, A. K. Misra, K. Jana and D. Das, Mater. Horiz., 2025, 12, 987–1001 RSC.
- R. Ahuja, V. Shivhare and A. D. Konar, Macromol. Rapid Commun., 2024, 45, 2400255 CrossRef CAS.
- Z. Rao, Y. Zhu, Z. Chen, Y. Luo, Z. Yang, W. Liu, C. Qiao, Y. Xia, P. Yang, D. Ye and Z. Wang, Adv. Sci., 2025, 12, 2408415 CrossRef CAS.
- A. Levin, T. A. Hakala, L. Schnaider, G. J. L. Bernardes, E. Gazit and T. P. J. Knowles, Nat. Rev. Chem., 2020, 4, 615–634 CrossRef CAS PubMed.
- B. Yin, R. Wang, Y. Guo, L. Li and X. Hu, Polymers, 2024, 16, 1221 CrossRef CAS PubMed.
- X. Ma, K. P. C. Sekhar, P. Zhang and J. Cui, Biomater. Sci., 2024, 12, 5468–5480 RSC.
- N. Kulkarni, P. Rao, G. S. Jadhav, B. Kulkarni, N. Kanakavalli, S. Kirad, S. Salunke, V. Tanpure and B. Sahu, ACS Omega, 2023, 8, 3551–3570 CrossRef CAS.
- B. Pramanik, Gels, 2022, 8, 569 CrossRef CAS.
- C. Liu, Q. Zhang, S. Zhu, H. Liu and J. Chen, RSC Adv., 2019, 9, 28299–28311 RSC.
- H. Zhao, J. Sun, Y. Cheng, S. Nie and W. Li, J. Mater. Chem. B, 2025, 13, 1518–1530 RSC.
- H. Wang, F. Gao, M. Rafiq, B. Yu, Q. Niu and H. Cong, J. Mater. Chem. B, 2025, 13, 2418–2430 RSC.
- H. Qu, Q. Yao, T. Chen, H. Wu, Y. Liu, C. Wang and A. Dong, Adv. Colloid Interface Sci., 2024, 325, 103099 CrossRef CAS.
- M. Guerrero, D. Filho, A. N. Ayala, D. Rafael, F. Andrade, A. Marican, S. Vijayakumar and E. F. Durán-Lara, Colloids Surf., B, 2025, 247, 114451 CrossRef PubMed.
- L. Shang, J. Liu, Y. Wu, M. Wang, C. Fei, Y. Liu, F. Xue, L. Zhang and F. Gu, ACS Appl. Mater. Interfaces, 2023, 15, 26273–26284 CrossRef CAS.
- A. L. Lakes, R. Peyyala, J. L. Ebersole, D. A. Puleo, J. Z. Hilt and T. D. Dziubla, Biomacromolecules, 2014, 15, 3009–3018 CrossRef CAS PubMed.
- Z. Yang, G. Liang, Z. Guo, Z. Guo and B. Xu, Angew. Chem., Int. Ed., 2007, 46, 8216–8219 CrossRef CAS PubMed.
- S. Mondal, S. Das and A. K. Nandi, Soft Matter, 2020, 16, 1404–1454 RSC.
- E. R. Cross, S. M. Coulter, S. Pentlavalli and G. Laverty, Soft Matter, 2021, 17, 8001–8021 RSC.
- S. H. Jeong, S. Cheong, T. Y. Kim, H. Choi and S. K. Hahn, ACS Appl. Mater. Interfaces, 2023, 15, 16471–16481 CrossRef CAS.
- I. Rehman, S. Gul, M. Akhtar, R. Amin and R. Fatima, J. Mol. Struct., 2024, 1295, 136810 CrossRef CAS.
- G.-F. Zhang, X. Liu, S. Zhang, B. Pan and M.-L. Liu, Eur. J. Med. Chem., 2018, 146, 599–612 CrossRef CAS PubMed.
- Y. Huang, N. Yang, D. Teng, R. Mao, Y. Hao, X. Ma, L. Wei and J. Wang, Appl. Microbiol. Biotechnol., 2022, 106, 3639–3656 CrossRef CAS PubMed.
- L. Li, Y. Zhou, P. Li, Q. Xu, K. Li, H. Hu, W. Bing and Z. Zhang, Front. Bioeng. Biotechnol., 2022, 10, 1066306 CrossRef PubMed.
- N. Nandi, K. Gayen, S. Ghosh, D. Bhunia, S. Kirkham, S. K. Sen, S. Ghosh, I. W. Hamley and A. Banerjee, Biomacromolecules, 2017, 18, 3621–3629 CrossRef CAS.
- A. K. Das and P. K. Gavel, Soft Matter, 2020, 16, 10065–10095 RSC.
- A. Dasgupta and D. Das, Langmuir, 2019, 35, 10704–10724 CrossRef CAS PubMed.
- S. Misra, S. Mukherjee, A. Ghosh, P. Singh, S. Mondal, D. Ray, G. Bhattacharya, D. Ganguly, A. Ghosh, V. K. Aswal, A. K. Mahapatra, B. Satpati and J. Nanda, Chem. – Eur. J., 2021, 27, 16744–16753 CrossRef CAS.
- S. Sharma, C. Kulkarni, M. M. Kulkarni, R. Ali, K. Porwal, N. Chattopadhyay, D. Tewari and S. Verma, Chem. Commun., 2020, 56, 3043–3046 RSC.
- W. Gao, D. Vecchio, J. Li, J. Zhu, Q. Zhang, V. Fu, J. Li, S. Thamphiwatana, D. Lu and L. Zhang, ACS Nano, 2014, 8, 2900–2907 CrossRef CAS PubMed.
- Y. Hu, W. Xu, G. Li, L. Xu, A. Song and J. Hao, Langmuir, 2015, 31, 8599–8605 CrossRef CAS PubMed.
- H. Wang, G. Zha, H. Du, L. Gao, X. Li, Z. Shen and W. Zhu, Polym. Chem., 2014, 5, 6489–6494 RSC.
- P. Tiwari, A. Gupta, D. N. Shukla, A. K. Mishra, A. Basu and A. Dutt Konar, ACS Appl. Bio Mater., 2021, 4, 4119–4130 CrossRef CAS PubMed.
- X. Wang, C. Zhang and N. Bao, Front. Oncol., 2023, 13, 1224125 CrossRef CAS PubMed.
- E. P. Ivanova, S. H. Nguyen, Y. Guo, V. A. Baulin, H. K. Webb, V. K. Truong, J. V. Wandiyanto, C. J. Garvey, P. J. Mahon, D. E. Mainwaring and R. J. Crawford, Acta Biomater., 2017, 59, 148–157 CrossRef CAS PubMed.
- P. Tiwari, A. Gupta, V. Shivhare, R. Ahuja, A. S. Mandloi, A. Mishra, A. Basu and A. D. Konar, ACS Appl. Bio Mater., 2024, 7, 332–343 CrossRef CAS PubMed.
- R. Ahuja, M. Singh and A. Dutt Konar, ACS Appl. Bio Mater., 2025, 8, 1108–1125 CrossRef CAS PubMed.
- V. Shivhare, A. Gupta, R. Ahuja, D. Barde, S. K. Ahirwar, A. Basu and A. D. Konar, Chem. Commun., 2025, 61, 9717–9720 RSC.
- A. Dutt Konar, V. Shivhare, R. Ahuja, A. Gupta, P. Tiwari, N. Khan, S. K. Ahirwar, A. S. Mandloi, A. K. Mishra, M. Singh and A. Basu, J. Mater. Chem. B, 2025, 13, 4594–4611 RSC.
- B. D. Bax, P. F. Chan, D. S. Eggleston, A. Fosberry, D. R. Gentry, F. Gorrec, I. Giordano, M. M. Hann, A. Hennessy, M. Hibbs, J. Huang, E. Jones, J. Jones, K. K. Brown, C. J. Lewis, E. W. May, M. R. Saunders, O. Singh, C. E. Spitzfaden, C. Shen, A. Shillings, A. J. Theobald, A. Wohlkonig, N. D. Pearson and M. N. Gwynn, Nature, 2010, 466, 935–940 CrossRef PubMed.
- T. M. Moon, É. D. D'Andréa, C. W. Lee, A. Soares, J. Jakoncic, C. Desbonnet, M. Garcia-Solache, L. B. Rice, R. Page and W. Peti, J. Biol. Chem., 2018, 293, 18574–18584 CrossRef CAS.
- D. T. King, G. A. Wasney, M. Nosella, A. Fong and N. C. J. Strynadka, J. Biol. Chem., 2017, 292, 979–993 CrossRef CAS PubMed.
- H. Paternoga, C. Crowe-McAuliffe, L. V. Bock, T. O. Koller, M. Morici, B. Beckert, A. G. Myasnikov, H. Grubmüller, J. Nováček and D. N. Wilson, Nat. Struct. Mol. Biol., 2023, 30, 1380–1392 CrossRef CAS PubMed.
- Y.-M. Chang, W.-Y. Jeng, T.-P. Ko, Y.-J. Yeh, C. K.-M. Chen and A. H.-J. Wang, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 8617–8622 CrossRef CAS PubMed.
- F. N. Yilmaz, M. Hacioglu and E. H. Aldogan, Curr. Microbiol., 2023, 80, 5 CrossRef CAS PubMed.
- B. Mishra, A. Basu, R. Saravanan, L. Xiang, L. K. Yang and S. S. J. Leong, RSC Adv., 2013, 3, 9534 RSC.
- R. S. Rasool, B. A. Padder, A. A. Wani, M. D. Shah, K. Z. Masoodi, N. A. Khan, A. Banoo and I. Khan, J. Microbiol. Methods, 2020, 171, 105885 CrossRef CAS PubMed.
- G. van de Geijn, V. van Rees, N. van Pul-Bom, E. Birnie, H. Janssen, H. Pegels, M. Beunis and T. Njo, Cytometry, Part A, 2011, 79A, 694–706 CrossRef PubMed.
- S. Kaya, O. Bedir, M. Baysallar, S. Ören, Ö. Koru and A. Albay, Diagn. Microbiol. Infect. Dis., 2024, 110, 116464 CrossRef CAS PubMed.
- A. Kohler, E.-M. Julke, J. Stichel and A. G. Beck-Sickinger, ACS Pharmacol. Transl. Sci., 2024, 7, 3618–3625 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |