DOI:
10.1039/D5MD00481K
(Research Article)
RSC Med. Chem., 2026,
17, 531-548
Synthesis and evaluation of HFIP bearing triazolo-amides as amyloid-β aggregation inhibitors and suppressors of aggregation induced neuroinflammation
Received
27th May 2025
, Accepted 29th October 2025
First published on 30th October 2025
Abstract
Alzheimer's disease (AD) is a complex neurodegenerative disease with biological signatures of amyloid beta (Aβ) aggregated plaques and increased levels of bio-metals like copper (Cu), zinc (Zn), and iron (Fe). Aβ-induced lysosomal membrane permeabilization is a key event in neuronal injury in AD. Aβ aggregation also modulates mitochondria membrane potential (MMP), activates interleukin 1β and NLRP3 inflammasome eventually leading to increased reactive oxygen species (ROS) production, neuronal apoptosis and mitochondrial dysfunction. Here, we report a multi-functional compound (2f) identified through structure–activity relationship study from a series of polyfluorinated triazole compounds. Compound 2f suppressed metal induced aggregation, downregulated NLRP3 inflammasome and IL-1β expression. It has maintained the lysosomal acidic pH and restored mitochondrial membrane potential. HFIP bearing triazolo amide (2f) was found to chelate with Cu(II) and Zn(II) selectively in the presence of a range of other physiologically relevant metals. Further, a molecular dynamics (MD) simulation study revealed 2f disrupted the aggregation via interacting with chain A of pentameric Aβ. Therefore the HFIP bearing triazole amides may serve as potential scaffolds for drug development towards the treatment of AD.
1. Introduction
Alzheimer's disease (AD) is a complicated neurodegenerative disease which is the leading cause of death among the elderly.1 The disease symptoms include gradual memory loss, dementia, cognitive impairment, and behavioural changes.2 As per current reports, AD associated dementia has affected approximately 50 million people and is expected to rise thrice in the next three decades.3 Although the exact molecular mechanism behind the onset of AD is elusive, the expected pathophysiology suggests that amyloid beta (Aβ) aggregation, hyperphosphorylated tau protein, dysregulated metal ion homeostasis, mitochondrial dysfunction and oxidative stress initiated programmed neuronal cell death that contributes to the inception of AD.4–7 Existing literature reports based on mechanistic investigations have demonstrated that deposition of Aβ plaques due to insoluble oligomer aggregation (by the deposition of Aβ peptide; 39–43 amino acid residues) plays a significant role in the pathophysiology of AD. Aβ oligomers interact with neurons and glial cells, activating NLRP3-IL1β-like proinflammatory pathways, leading to neuronal apoptosis and cell death.8
Molecules that restrict Aβ aggregation can be an effective therapy for AD treatment.9 Furthermore, Aβ deposits and neuroinflammation are strongly linked to dyshomeostasis of brain metal ions such as iron, copper, and zinc.10–14 Thus, it has been suggested that metal chelators are crucial for enhancing metal homeostasis, inhibiting Aβ aggregation, and reducing neuroinflammation.15–17 FDA-approved medications such as rivastigmine, donepezil, galantamine, and memantine provide transient and inadequate symptom mitigation by targeting neurotransmitters, however, they have no effect on the underlying etiology.18–20 Furthermore, the recent failure of several molecules such as PPI-1019, NAP (NAPVSIPQ), tarenflurbil, semagacestat, avagacestat, LY288672, bapineuzumab, solanezumab, PBT1 and PBT2 in clinical trials has projected the need for identification of novel drug entities for treatment of AD.21–25 Owing to the complex pathophysiology, AD targeting pharmacophores must possess broad-spectrum activities as multi-target directed ligands (MTDLs).26–28 Triazoles are biologically active scaffolds and often explored in developing molecules targeting AD.29 For example, a library of ditriazole-based compounds has been evaluated as prospective therapeutic molecules against AD. Qu et al. utilized radiolabelled diphenyl triazole derivatives (b) to target Aβ plaques.30 Vajragupta et al. identified three series of substituted 1,2,3-triazole derivatives (c) as agonists of α7 nicotinic acetylcholine receptor which is found to be affected during the onset of AD.31 Jones et al. developed 1,2,3-triazole derivatives (d–e) that reduced metal-induced Aβ aggregation by exhibiting metal chelation and antioxidant properties.32 Jiaranaikulwanitch et al. reported triazole derivatives containing a tryptamine moiety (f–g) as multifunctional ligands for the treatment of AD (Fig. 1-A).33
 |
| | Fig. 1 A; Reported triazole derivatives for AD and rational design of the triazolo amide/amine scaffold B; synthetic scheme for compounds 1(a–b). Reagents and reaction conditions: (i) NH2OH·HCl, NaOAc·3H2O, MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (9 H2O (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), rt, 3 h. (ii) NCS, ethyl propiolate, DABCO, CAN, rt, 18 h. (iii) LiOH·H2O, THF 1), rt, 3 h. (ii) NCS, ethyl propiolate, DABCO, CAN, rt, 18 h. (iii) LiOH·H2O, THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1 H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), rt, 5 h. (iv) HOBt, DCC, DMF, rt, 18 h. C; synthetic scheme for compounds 2(r–t). Reagents and reaction conditions: (i) MsCl, DCM, 0 °C, 4 h. (ii) morpholine, Et3N, DCM, 0 °C → rt, 3 h. D; structure of all synthesized molecules. 4), rt, 5 h. (iv) HOBt, DCC, DMF, rt, 18 h. C; synthetic scheme for compounds 2(r–t). Reagents and reaction conditions: (i) MsCl, DCM, 0 °C, 4 h. (ii) morpholine, Et3N, DCM, 0 °C → rt, 3 h. D; structure of all synthesized molecules. | |
In a similar approach, our group has previously designed, synthesized and evaluated the activity of fluorinated triazoles as MTDLs against AD. The triazole amide bearing hexafluoroisopropanol (HFIP) group has emerged as the potential hit that could reduce Aβ aggregation and chelate with Fe, Zn and Cu.34
In this work, we have synthesized a series of different substituted HFIP bearing triazole amides and investigated their structure–activity-relationship (SAR). SAR analysis coupled with in vitro mechanistic studies reveals compound 2f as the potential amyloid aggregation inhibitor with neuroprotective, antioxidant, anti-inflammatory and metal chelating properties. It has also modulated aggregation induced activated NRPL3 inflammasome, mitochondria membrane potential, and lysosomal pH as well as downregulated inflammatory interleukin 1β expression in N2a cells.
2. Results and discussion
2.1 Design and synthesis of compounds
Aβ peptide aggregation is primarily driven by the core residue penta-peptide fragment (K16LVFF20). The phenylalanine dipeptide sequence (F19F20) promotes self-aggregation through π–π-stacking between phenyl rings. Numerous aromatic compounds, including curcumin, have been found to effectively prevent Aβ aggregation, as they possess a common feature of having two aromatic rings connected through an optimal spacer.35–40 In anticipation that aromatic rings may engage through π–π interaction with the phenyl alanine residue of the KLVFF motif of the Aβ peptide and prevent aggregation, aromatic residues are included in our scaffold design. The hydroxyl group of hexafluoroisopropanol (HFIP), which is flanked by two CF3 groups, is an intriguing pharmacophore that has been studied previously for its ability to prevent Aβ aggregation.41,42 Fluorine improves the lipophilicity and metabolic stability of molecules, which can significantly increase their ability to cross the BBB—a major hurdle in AD drug design.43,44 Fluorinated scaffolds often show increased binding affinity to amyloid-β due to electronic effects, hydrogen bonding, and other intermolecular interactions.45 HFIP is also a well-established solvent utilized to break down the Aβ peptide and inclusion of HFIP into the molecule may boost its disaggregation capabilities, as well as the drug's cellular permeability and plasma half-life. As an isoxazole moiety plays an important role in inhibiting Aβ aggregation during AD pathophysiology, few representative molecules bearing isoxazole by isosteric replacement of triazole are included46,47 (Fig. 1-A). Iso-oxazoles were synthesized by converting corresponding aldehydes into oximes and in situ-converting them to chloro-oximes. Treatment of the intermediate with ethylpropiolate and further cyclization leads to the corresponding isoxazole ester. Isoxazole ester was hydrolysed into carboxylic acid which was coupled with HFIP containing aniline to yield the final molecule 1(a–b). Triazolo-amides were prepared via synthetic routes previously reported.34 For synthesizing compounds 2(r–t), mesylation of the hydroxy group followed by addition of morpholine yielded 2(s) and the chloride generated in situ gives 2(t). Compounds 3(a–d) were synthesized following the reported procedure.34 A library of twenty-six compounds was characterized by 1H, 13C NMR and HRMS. The presence of HFIP was corroborated through 19F NMR. The purity of the compounds was established through reverse phase HPLC.
2.2 Biological evaluation
2.2.1 Structure–activity relationship study on cell viability.
Understanding the cytocompatibility of bioactive compounds is crucial for their potential advancement as lead agents. Thus, we used the Alamar blue assay to evaluate the library of synthesized molecules for cell viability on neuronal cells (Neuro-2a).48,49 A single concentration test of 50 μM was used to screen for quick identification of compounds that have acceptable cell viability. Assessment on replacing the triazole ring with an isoxazole ring yielded 1a–1b molecules. Although replacement of triazole with isoxazole (1a) did not affect the cell viability, substitution of a p-methyl group on aryl (1b) exhibited cytotoxicity. Since our prior investigations yielded potent amyloid β aggregation inhibitors with 3,4-diOMe and 3,4-diMe substitutions on the phenyl ring connected with triazole,33 3,4-diOMe and 3,4-diMe substitutions on the phenyl ring linked with triazole/isoxazole with varying substitution on HFIP bearing aryl amides were investigated. From the 3,4-diOMe substituted series (2a–2g and 3a–3d), p-Me group substitution (2a) was found to be more cytocompatible than the o-Me (2b) substitution. Surprisingly di-Me substituted compounds (2c–2d) were found to be non-cytotoxic. Replacement of the aryl with heteroaryl group (2e) leads to substantial cytotoxicity. When the hydroxyl group of the HFIP moiety was O-alkylated, compound bearing OMe (2f) was found non-toxic along with higher alkyl chain bearing 2h. Surprisingly, replacement of OMe with OEt (2g) leads to toxicity. Replacement of amide with amine (3a–3d) also demonstrated cytotoxicity. When 3,4-di OMe was substituted to 3,4-diMe, in compounds 2a and 2c resulting analogues 2i and 2k were found to be toxic. Meanwhile analogues of 2b were found to be cytocompatible. Heteroaryl substitution (2n) did not improve the cytocompatibility. Replacement of the hydroxyl group with mesityl (2r) and morpholine (2s) retained the cytocompatibility, while halo substitution (2t) leads to cytotoxicity. Compounds (thirteen) which have demonstrated good cell viability (Fig. 2-A1) were selected for further studies.
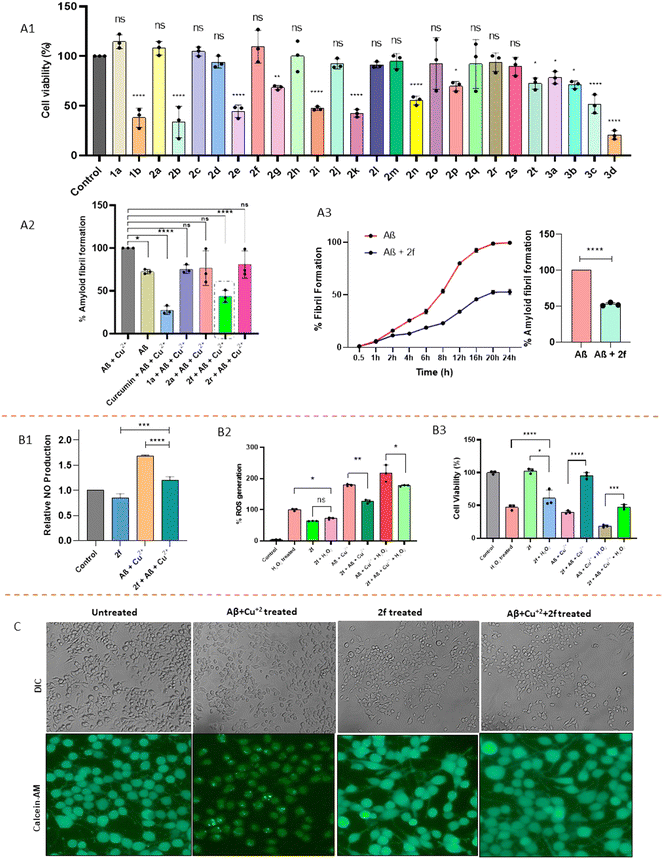 |
| | Fig. 2 (A1) Cell viability of compounds at 50 μM; (A2) bar graph showing the % amyloid fibril formation in the presence of 1a, 2a, 2f and 2r; (A3) kinetics of ThT fluorescence assay. [Aβ] = 5 μM, 2f = 50 μM. (B1) Bar graph showing the relative nitric oxide production in the presence of 2f; (B2) bar graph showing % ROS generation of 2f in the presence or absence of H2O2 induced cellular ROS generation with Aβ at 50 μM respectively; (B3) bar graph showing the % cell viability of 2f in the presence or absence of H2O2 induced cell toxicity with Aβ at 50 μM. (C) Neuronal outgrowth in N2a cells in the presence of 2f and with Aβ and Cu2+; fluorescence image of calcein-AM stained N2a cells showing neurite outgrowth (statistical significance was calculated using a paired t-test for the data of n = 3 in triplicate). | |
2.2.2 Aβ disaggregation property determination.
After confirming the cellular viability, all 13 compounds (50 μM, 24 h) were examined for their ability to prevent Aβ peptide aggregation by the thioflavin T (ThT) assay. ThT is a cationic benzothiazole dye utilized to quantify Aβ peptide aggregation, as it exhibits a significant increase in emission intensity upon binding to β-sheet-rich structures. Conversely, a low ThT emission intensity indicates a reduced level of amyloid aggregation.50,51 It has been reported that free Cu in the brain accelerates the aggregation of Aβ peptides, hence, we evaluated the prevention of Aβ peptide aggregation by the synthesized compounds against Cu2+-induced Aβ peptide aggregation.12,52–54
After monomerization Aβ compounds were incubated with Cu2+ and Aβ peptide (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, respectively). The samples were treated with ThT and fluorescence at 450 nm excitation and 485 nm emission wavelengths was recorded. The percent inhibition of aggregation was calculated in comparison to the control. According to preliminary screening, out of 13 molecules (Fig. S8), compounds 1a, 2a, 2f, and 2r showed anti-aggregation properties. These selected compounds were tested further in triplicate and compared with the standard amyloid β aggregation inhibitor curcumin, Fig. 2-A2. In the presence of compound 2f, the Cu2+ induced Aβ peptide aggregation was significantly reduced (IC50 value 16.8 ± 1.8 μM) (SI S139). Furthermore, a time-dependent ThT fluorescence assay was conducted to assess the effect of compound 2f on the growth kinetics of Aβ aggregation from monomeric to aggregated species.55 The fibrillation of Aβ exhibited a characteristic sigmoidal growth curve, encompassing an initial nucleation phase, a subsequent rapid growth phase, and a final saturation phase, reached after approximately 20 hours of incubation. In the absence of compound 2f, a substantial increase in ThT fluorescence intensity was observed for the untreated Aβ peptide, indicating significant aggregation. In contrast, the presence of 2f significantly suppressed the aggregation of Aβ, as evidenced by the reduced ThT fluorescence. Fig. 2-A3 shows that 2f inhibits Aβ42 aggregation with an efficiency of 52.73%, drastically reducing the growth phase after 4 h of incubation. Hence, compound 2f was further used for subsequent cellular studies.
2 ratio, respectively). The samples were treated with ThT and fluorescence at 450 nm excitation and 485 nm emission wavelengths was recorded. The percent inhibition of aggregation was calculated in comparison to the control. According to preliminary screening, out of 13 molecules (Fig. S8), compounds 1a, 2a, 2f, and 2r showed anti-aggregation properties. These selected compounds were tested further in triplicate and compared with the standard amyloid β aggregation inhibitor curcumin, Fig. 2-A2. In the presence of compound 2f, the Cu2+ induced Aβ peptide aggregation was significantly reduced (IC50 value 16.8 ± 1.8 μM) (SI S139). Furthermore, a time-dependent ThT fluorescence assay was conducted to assess the effect of compound 2f on the growth kinetics of Aβ aggregation from monomeric to aggregated species.55 The fibrillation of Aβ exhibited a characteristic sigmoidal growth curve, encompassing an initial nucleation phase, a subsequent rapid growth phase, and a final saturation phase, reached after approximately 20 hours of incubation. In the absence of compound 2f, a substantial increase in ThT fluorescence intensity was observed for the untreated Aβ peptide, indicating significant aggregation. In contrast, the presence of 2f significantly suppressed the aggregation of Aβ, as evidenced by the reduced ThT fluorescence. Fig. 2-A3 shows that 2f inhibits Aβ42 aggregation with an efficiency of 52.73%, drastically reducing the growth phase after 4 h of incubation. Hence, compound 2f was further used for subsequent cellular studies.
2.2.3 Compound 2f effects on Aβ-Cu2+ induced nitric oxide generation.
It has been reported that β-amyloid stimulation triggers the expression of iNOS, leading to a surge in NO production.56–58 This excess production of NO causes oxidative stress in cells, and consequently, cell death. To investigate the effect of compound 2f on Aβ + Cu2+ induced NO production, we employed Griess reagents to detect NO levels in different treated groups. Interestingly, we observed that the absorbance of Aβ + Cu2-treated N2a cells is higher which indicates higher NO production (Fig. 2-B1). The control group and 2f-treated cells do not show any significant increase in NO generation. Interestingly, we observed that compound 2f reduced NO production when cotreated with Aβ + Cu2. Therefore, compound 2f reduces NO production and inhibits oxidative stress.
2.2.4 Neuroprotective properties of 2f.
From the Alzheimer's disease perspective, Aβ peptide aggregation leads to the inhibition of neuritogenesis, subsequently leading to neuronal cell death. Hence, we were keen to evaluate whether compound 2f was able to reverse the inhibition of neuritogenesis. Our observations confined that in the presence of compound 2f, the inhibition of neuronal outgrowth imposed by Aβ peptide aggregation was reversed in the co-treated N2A cells (Fig. 2-B2). In addition, Aβ peptide aggregation generally generates ROS in the neuronal cells which eventually fuels cell death induction. Hence, we evaluated the ROS scavenging property of compound 2f in N2a cells. The level of ROS generation in Aβ peptide treated-N2a cells was evaluated using the 2′-7′-dichlorodihydrofluorescein diacetate (DCFDA) assay.59 We observed that compound 2f was able to scavenge the ROS generated in the Aβ-Cu2+ treated N2a cells. This data was further validated by treatment of H2O2 (200 μM, 5 h), where 2f was able to scavenge the ROS generated by H2O2. The ROS scavenging efficacy of compound 2f was further validated in the Aβ-Cu2+–H2O2 treated N2a cells. Interestingly, compound 2f was able to scavenge the ROS generated in the co-treated group (Fig. 2-B2). Furthermore, it has also been reported that ROS generation is critical for neuronal cell death induction. Under unregulated redox homeostasis, the neuronal cells undergo cell death. Hence, we checked whether compound 2f can reverse cell death (Fig. 2-B3) induction caused by Aβ peptide aggregation-related ROS generation. We observed that in the presence of compound 2f, the cell viability was found to be increased even in the Aβ-Cu2+ and Aβ-Cu2+–H2O2 treated N2a cells as compared to their non-treated counterparts. Our findings highlighted the neuroprotective effect of compound 2f in neuronal cells (Fig. 2-C), which can be further implemented as a therapeutic agent against AD.
2.2.5
2f inhibits Aβ42-induced cell death.
Aβ42, a key protein implicated in AD, causes cell death, particularly in neurons.60 As 2f has shown neuroprotective effects, to evaluate the effect of 2f on cell death caused by Aβ42, we performed a live dead assay. The experimental outcomes of this assay are shown in (Fig. 3-A). Microscopy images exhibit significantly higher dead cells in the case of Aβ + Cu2+ in comparison with the cells treated with compound 2f and Aβ + Cu2+ cotreatment. This finding suggests that 2f is able to inhibit cell death caused by Aβ + Cu2+.
 |
| | Fig. 3 (A) Fluorescence image of calcein-AM and ethidium homodimer-1 stained N2a cells showing live and dead cells in different treated groups, respectively and the bar graph (bottom) shows the % live and dead cell viability vs. different treatments; (B) Fluorescence microscopy images of N2a cells showing cells with acidic vacuoles in different treated groups stained with acridine orange (AO) dye and relative fluorescence intensity. (C) Fluorescence microscopy images of N2a cells with LysoTracker Red with different treated groups and the relative fluorescence intensity. | |
2.2.6
2f restores Aβ42-induced dysregulated lysosomal acidification.
Aβ-induced lysosomal membrane permeabilization is a key event in neuronal injury in AD.61,62 Lysosomal dysfunction generally leads to oxidative stress by virtue of impaired clearance of amyloidogenic peptides and ROS producing mitochondria which is one of the earliest pathogenic features of AD.63–67
Hence, to check the lysosomal acidity, we employed LysoTracker™ Deep Red and acridine orange dyes in Aβ + Cu2+ treated N2A cells, in the presence of 2f. The experimental outcomes of our hypothesis suggested that compound 2f was able to restore the lysosomal acidity, which was found to be reduced in treatment with Aβ + Cu2+. The red fluorescence intensity of acridine orange dye (Fig. 3-B) as well as LysoTracker™ Deep Red was found to be increased in 2f treated conditions as compared with Aβ + Cu2+ treatment alone (Fig. 3-C). In brief, we can speculate that compound 2f can enhance lysosomal acidification which is required for the clearance of amyloidogenic peptides and ROS producing mitochondria via selective autophagic degradation supporting neuronal cell survival.
2.2.7
2f inhibits neuroinflammation by restoring mitochondrial membrane potential and downregulating the NLRP3-IL-1β signalling cascade.
Disruption of mitochondrial membrane homeostasis and fission–fusion dynamics impairs redox homeostasis and energy metabolism which fuels AD progression.68
During the progressive stages of AD, the mitochondrial membrane potential (MMP) is found to be decreased indicating loss of mitochondrial homeostasis. During AD pathogenesis, it has been reported that Aβ aggregation potentiates MMP, resulting in mitochondrial dysfunction, eventually leading to increased reactive oxygen species (ROS) production and neuronal apoptosis.69 Hence, restoration of MMP is a critical factor to be considered for AD treatment. Based on this, we checked the MMP in the Aβ + Cu2+-treated N2A cells using Rhodamine 123 staining. Interestingly, we observed that compound 2f was able to restore the MMP caused by Aβ + Cu2+ in the N2a cells, red in Fig. 4-A. The representative microscopy images revealed that co-treatment of Aβ + Cu2+ with 2f had higher fluorescence intensity as compared to the Aβ + Cu2+ treatment alone. Our results suggested that compound 2f mediated MMP restoration subsequently decreased the mitochondrial ROS generation preventing neuronal apoptosis.
 |
| | Fig. 4 (A) Representative images of N2A cells with NLRP3 expression varying with different treatment groups as observed by fluorescence microscopy and the bar graph of relative fluorescence intensity of NLRP3 expression in different treated groups with 2f; (B) representative images of N2A cells with IL-1β expression varying with different treatment groups as observed by fluorescence microscopy and the bar graph of relative fluorescence intensity of IL-1β expressions in different treated groups with 2f. (C) Representative images of N2A cells with Rhodamine 123 staining with different treatment groups as observed by fluorescence microscopy and the bar graph of relative fluorescence intensity. | |
Mitochondrial ROS generation is a key precursor for neuroinflammation which is closely associated with AD pathophysiology. The NLRP3 inflammasome in microglia is considered a key component in the pro-inflammatory response of immune cells. Several studies have reported that Aβ activates the NLRP3 inflammasome in microglia,70 with evident mitochondrial dysfunction and mitochondrial ROS generation.71–74 Further, the NLRP3 inflammasome complex activates interleukin 1β (IL 1β), a key neuroinflammatory interleukin in AD pathogenesis via the NLRP3-CASP1 signalling cascade.75–77 To investigate the anti-neuroinflammatory activity of compound 2f, we checked the expression of NLRP3 (Fig. 4-A) and IL-1β (Fig. 4-B). Interestingly, we observed that compound 2f was able to reduce the expression of both inflammatory markers when cotreated with Aβ + Cu2+. The outcomes of our experimentation suggested that compound 2f mitigates neuroinflammation via reduced mitochondrial ROS generation, inducing MMP and subsequent inhibition of neuroinflammation via downregulating the NLRP3-IL-1β signalling cascade.
2.3
ln silico studies
2.3.1 Molecular docking and simulation studies.
Based on the effective outcome of compound 2fin vitro experiments, in silico analysis was conducted to investigate the molecular interaction of compound 2f with the Aβ peptide. Tomaselli et al. showed that in an aqueous environment, Aβ(1–42) retains its N terminal α helix while the shorter C terminal helix converts to β structure, a helix to sheet transition that drives early Aβ42 aggregation; key driver regions include the central hydrophobic KLVFF core, C terminal residues 29–36, and a turn segment, making the central helix (13–26) and KLVFF (16–20) prime targets for helix stabilizing inhibitors that reduce oligomer and fibril formation.78,79 Ligands binding to the central helix region of Aβ can stabilize its α helical structure, preventing self-association.80,81 This stabilization blocks the α helix to β sheet conformational change, thereby inhibiting the formation of fibrils and toxic oligomeric Aβ species.82,83 Molecular docking studies showed that curcumin binds to Phe19 of chain A in the Aβ pentamer (2BEG) via aromatic π–π stacking engaging the dimethoxy-substituted phenyl ring (binding energy −6.0 kcal mol−1) (Fig. S1). Compound 2f demonstrated similar binding affinity towards chain A of 2BEG. The triazole and HFIP bearing aryl group has shown T-shaped π–π interaction and conventional π–π stacking with the phenyl ring of Phe19 of the Aβ pentamer (Fig. 5-A).
 |
| | Fig. 5 (A–D) represents the pre and post-MD complex of 2f-2BEG after 100, 200 and 300 ns; (E) graph represents apo and holo RMSD of the 2f-2BEG complex after 300 ns simulation; (F) graph represents apo and holo RMSF of the 2f-2BEG complex after 300 ns simulation; (G) graph represents apo and holo SASA of the 2f-2BEG complex after 300 ns simulation; (H) graph represents the apo and holo radius of gyration of the 2f-2BEG complex after 300 ns simulation; (I) graph represents T-shape interaction of 2f with 2BEG; (J) graph represents inter-hydrogen of holo of the 2f-2BEG complex after 300 ns simulation. | |
The optimal binding conformation of the compound 2f–Aβ fibril complex was evaluated through molecular dynamics (MD) simulations, as conventional docking provides only a static snapshot of ligand–target interactions. MD simulations enable the assessment of the stability and persistence of binding interactions observed in docking. For comparison, simulations of the apo (ligand-free) system were also conducted. The simulation study was conducted for 300 ns, using a cubic simulation box defined such that the minimum distance between the solute and the box boundary was at least 2.8 nm. Post-MD complex analyses revealed that the T-shaped π–π interaction and conventional π–π interaction between the triazole ring of compound 2f and Phe19 were retained throughout the simulation, indicating stable ligand engagement (Fig. 5-I).
Root mean square deviation (RMSD) is used to measure the average change in displacement of a selection of atoms for a particular frame with respect to a reference frame. The RMSD profile of the backbone atoms at 100 ns in the apo state (red) shows a more stable and lower RMSD profile (around 0.8–1.0 nm), suggesting a rigid nature of the protein and limited conformational changes. Conversely, the holo complex (black) exhibits larger fluctuation and raised RMSD values (about 1.1–1.3 nm), suggesting that ligand binding causes initial destabilization and enhanced backbone mobility. The apo form maintains a continuously low RMSD up to 200 ns while the RMSD difference between the forms is more noticeable between 100–200 ns where the apo maintains its stability, whereas holo exhibits much higher and more fluctuating RMSD values (near or over 1.5 nm). These findings support the fact of long-term ligand–protein connections destabilize the backbone, increasing flexibility and decreasing overall structural rigidity.
The radius of gyration (Rg) of the 2BEG-compound 2f complex was monitored at 100, 200, and 300 ns during the simulation. At 100 ns, the Rg was stable at approximately 1.42–1.45 nm, denoting a compact structure. This stability persisted at 200 ns, with minor fluctuations. However, by 300 ns, a gradual increase in Rg was observed, with values exceeding 1.45 nm and isolated peaks above 1.50 nm. These results indicate a time-dependent relaxation of structural compactness, which may be associated with ligand-induced conformational changes or incipient unfolding in the protein. This trend corresponds to a modest loss of compactness, potentially reflecting partial unfolding or rearrangement within the protein–ligand complex at this late simulation stage.
Inter-molecular hydrogen bond analysis of the 2BEG-compound 2f complex throughout the 300 ns simulation indicates dynamic but steadily stabilizing interactions. In the early simulation phase (up to 100 ns), the number of hydrogen bonds ranged from 0 to 2, occasionally reaching 3. At 200 ns, the complex displayed an increase in interaction frequency, with the formation of 2–3 hydrogen bonds. By 300 ns, in (Fig. 5-J) hydrogen bonds were sustained mainly in the 1–3 range, suggesting moderate yet stable binding. The data demonstrate that hydrogen bonding between 2BEG and 2f is dynamic but progressively stabilizes over time, supporting the formation of a robust, adaptable interaction interface.
2.3.2 ADMET properties of compound 2f.
Drug likeness is a crucial criterion for any novel chemical entity since it shows the molecule's absorption, distribution, metabolism, elimination, and toxicity.84,85 An in silico analysis was conducted using the tool QikProp in Schrödinger Suite software to predict parameters such as pKa, total polar surface area, hydrogen bond donor, hydrogen bond acceptor, molecular weight, etc. Lipinski's rule of five, which indicates drug likeness, has not been violated by compound 2f (M.W. 504.3880), except for the molecular weight rule. Furthermore, good oral absorption was discovered using in silico prediction (% human oral absorption). Furthermore, toxicity, cell, and brain permeability were studied by predicting the IC50 value for the blockage of HERG K+ channels (QPlogHERG), apparent Caco-2 cell permeability in nm s−1 (QPPCaco), and brain/blood partition coefficient (QPlogBB). Importantly, 2f was predicted to have good Caco-2 permeability and acceptable hERG inhibitory activity. The QPlogBB value of 2f continues to be within the approved range, indicating the compound's potential to penetrate the blood–brain barrier. Overall, compound 2f was projected to exhibit good BBB permeability, acceptable toxicity, and drug-like characteristics (Table S1).
2.4 Metal scavenging properties
2.4.1 Metal chelation studies.
Metals are crucial in the pathophysiology of AD, as the combination of Aβ with metals like copper, zinc, and iron leads to the creation of toxic aggregated complexes.11,12 Hence, various compounds possessing metal chelating properties like desferrioxamine (DFO), DP-109, clioquinol, etc., have been examined and shown effectiveness in animal studies. Hence, compound 2f was tested for its potential to bind to metals like copper, zinc, and iron. Compound 2f demonstrated absorption maximum around 291 nm (Fig. 6). A notable increase in absorbance was observed for Zn and Cu metals when exposed to compound 2f in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and 1
3 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio with CuSO4 and ZnSO4 at a concentration of 50 μM for 30 minutes at room temperature in MeOH. Hyperchromic shift was observed for interaction of 2f with Cu2+, whereas hypochromic shift was observed for the interaction with Zn2+. A small change in absorbance observed for Fe3+ and 2f interaction suggesting limited interaction between 2f and Fe3+.
5 ratio with CuSO4 and ZnSO4 at a concentration of 50 μM for 30 minutes at room temperature in MeOH. Hyperchromic shift was observed for interaction of 2f with Cu2+, whereas hypochromic shift was observed for the interaction with Zn2+. A small change in absorbance observed for Fe3+ and 2f interaction suggesting limited interaction between 2f and Fe3+.
 |
| | Fig. 6 UV-vis spectra of 2f with (A) Cu2+, (B) Zn2+ and (C) Fe3+. (D) Tyr fluorescence dequenching upon the addition of 10 μM 2f into Aβ-Cu2+. (E) Competitive metal binding studies of 2f with Cu2+, Zn2+ and Fe3+ with other bio-metals like Ca2+, K+, Mg2+, Mn2+ and Na+. | |
2.4.2 Tyrosine fluorescence assay.
Since Tyr10 is located in the proximity of three histidine residues (6, 13, 14), it is expected to be involved in metal coordination, thereby resulting in metal-induced chemical changes. Tyr10 is the main fluorophore for the intrinsic fluorescence of Aβ and this intrinsic fluorescence of Aβ introduced by Tyr10 has been utilized to investigate the formation of a complex between Aβ and Cu2+ ions. The tyrosine intrinsic fluorescence of Aβ is quenched when Cu2+ ion binds to the Aβ peptide, whereas the fluorescence is regained on adding the chelators. This transformation in the tyrosine intrinsic fluorescence assay was utilized to exploit the capability of 2f for chelating Cu2+ aggregates.86 The changes in the fluorescence of Tyr10 in Aβ were observed by introducing 5 and 10 μM Cu2+ chelator ions from Aβ-Cu2+ to 5 μM Aβ in 10 mM HEPES buffer at pH 7.4. Aβ showed the highest relative fluorescence intensity at 305 nm (Fig. 6-D, black line). On adding 5 and 10 μM concentration of Cu2+ ions into the Aβ solution, the fluorescence of Tyr10 at 305 nm (Fig. 6-D, red line and blue line) decreased rapidly due to the formation of the Aβ-Cu2+ complex and fluorescence quenching. The fluorescence quenching of Tyr10 occurred due to the significant increase in the electron density at the Cu2+ centre which further results in charge transfer to other groups nearby such as the Tyr10 residue. The quenched intrinsic fluorescence of Tyr was rapidly restored upon the addition of 5 and 10 μM of compound 2f (Fig. 6-(D), pink line and green line). These results exhibit that 2f would clear Cu2+ in the Aβ-Cu2+ aggregates and thus inhibit neurotoxicity.
3. Experimental
3.1 Synthesis of compounds
N-(4-(1,1,1,3,3,3-Hexafluoro-2-hydroxypropan-2-yl)phenyl)-3-phenylisoxazole-5-carboxamide (1a).
According to the general procedure, the corresponding acid (50 mg) was dissolved in dry DMF (100 μl) and cooled in an ice-bath. To the DMF solution, HOBt (1 equiv.) and DCC (1.2 equiv.) were added. The reaction mixture was stirred in the ice-bath for about an hour. After 1 hour, the corresponding amine (1 equiv.) was added, and the reaction was allowed to warm up to room temperature and then stirring was continued for 24 hours. The progress of the reaction was monitored by TLC. After completion of the reaction, the mixture was diluted with ice-water (10% v/v) and extracted with ethyl acetate. Combined organic layers were dried over anhydrous Na2SO4 and the solvent was evaporated under vacuum to obtain the crude product. The crude mixture was purified on silica gel (100–200 mesh) column chromatography by using ethyl acetate and petroleum ether as eluent to obtain pure product 1a (13.1 mg, 23%) as a pinkish white solid. Melting point: 169–171 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 11.07 (s, 1H), 8.74 (s, 1H), 8.01–7.96 (m, 2H), 7.96–7.92 (m, 2H), 7.87 (s, 1H), 7.71 (d, J = 8.6 Hz, 2H), 7.60–7.54 (m, 3H); 13C NMR (125 MHz, DMSO-d6) δ 164.3, 163.2, 154.8, 139.9, 131.3, 129.8, 128.1, 128.0, 127.3, 127.0, 124.6, 122.3, 121.0, 106.3, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5; 19F NMR (470 MHz, DMSO-d6) δ −73.98; HRMS (Q-TOF, ESI) m/z calculated for C19H12F6N2O3 [M + H]+ 431.0825, found: 431.0863.
N-(4-(1,1,1,3,3,3-Hexafluoro-2-hydroxypropan-2-yl)phenyl)-3-(p-tolyl)isoxazole-5-carboxamide (1b).
A similar procedure was followed as in the case of 1a. Isolated (26.2 mg, 65%) as a white solid. Melting point: 196–198 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 11.05 (s, 1H), 8.73 (s, 1H), 7.96–7.92 (m, 2H), 7.89–7.85 (m, 2H), 7.82 (s, 1H), 7.71 (d, J = 8.6 Hz, 2H), 7.40–7.35 (m, 2H), 2.39 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 164.1, 163.1, 154.8, 141.1, 139.9, 130.3, 128.0, 127.2, 126.9, 125.3, 124.6, 122.3, 121.0, 106.2, 76.9, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.8, 39.6, 39.5, 21.5; 19F NMR (470 MHz, DMSO-d6) δ −73.94; HRMS (Q-TOF, ESI) m/z calculated for C20H14F6N2O3 [M + H]+ 445.0981, found: 445.0983.
1-(3,4-Dimethoxyphenyl)-N-(3-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-4-methylphenyl)-1H-1,2,3-triazole-4-carboxamide (2a).
A similar procedure was followed as in the case of 1a. Isolated (19.4 mg, 40%) as a white solid. Melting point: 207–209 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.19 (s, 1H), 9.43 (s, 1H), 8.71 (s, 1H), 7.69 (d, J = 8.5 Hz, 1H), 7.59 (d, J = 12.1 Hz, 2H), 7.54 (d, J = 7.9 Hz, 2H), 7.17 (d, J = 8.8 Hz, 1H), 3.89 (s, 3H), 3.85 (s, 3H), 2.36 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.7, 149.8, 149.8, 143.6, 137.9, 133.3, 130.1, 129.2, 128.2, 126.0, 126.0, 125.1, 124.6, 122.3, 113.1, 112.5, 105.5, 77.2, 56.4, 56.3, 40.5, 40.3, 40.2, 40.0, 39.8, 39.7, 39.5, 18.5; 19F NMR (470 MHz, DMSO-d6) δ −73.85; HRMS (Q-TOF, ESI) m/z calculated for C21H18F6N4O4 [M + H]+ 505.1305, found: 505.1311.
1-(3,4-Dimethoxyphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-2-methylphenyl)-1H-1,2,3-triazole-4-carboxamide (2b).
A similar procedure was followed as in the case of 1a. Isolated (18 mg, 37%) as a white solid. Melting Point: 208–210 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.17 (s, 1H), 9.42 (s, 1H), 8.70 (s, 1H), 7.68 (d, J = 8.5 Hz, 1H), 7.59 (d, J = 2.3 Hz, 1H), 7.57 (d, J = 2.5 Hz, 1H), 7.54 (d, J = 2.7 Hz, 1H), 7.53 (d, J = 2.5 Hz, 1H), 7.17 (d, J = 8.7 Hz, 1H), 3.88 (s, 3H), 3.84 (s, 3H), 2.35 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.7, 149.8, 149.8, 143.6, 137.9, 133.3, 130.1, 129.2, 128.2, 126.1, 125.1, 113.1, 112.5, 105.5, 56.5, 56.3, 40.6, 40.5, 40.4, 40.3, 40.3, 40.2, 40.1, 40.0, 39.9, 39.8, 39.7, 39.5, 38.7, 18.5; 19F NMR (470 MHz, DMSO-d6) δ −73.83; HRMS (Q-TOF, ESI) m/z calculated for C21H18F6N4O4 [M + H]+ 505.1305, found: 505.1276.
1-(3,4-Dimethoxyphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-2,5-dimethylphenyl)-1H-1,2,3-triazole-4-carboxamide (2c).
A similar procedure was followed as in the case of 1a. Isolated (25 mg, 28%) as a white solid. Melting point: 203–205 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.12 (s, 1H), 9.40 (s, 1H), 8.49 (s, 1H), 7.57 (d, J = 2.6 Hz, 1H), 7.54 (dd, J = 8.6, 2.5 Hz, 1H), 7.47 (s, 1H), 7.36 (s, 1H), 7.17 (d, J = 8.7 Hz, 1H), 3.89 (s, 3H), 3.85 (s, 3H), 2.56 (s, 3H), 2.28 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.7, 149.8, 149.8, 143.6, 137.6, 137.4, 130.9, 130.3, 130.1, 129.8, 126.2, 126.0, 125.0, 122.7, 113.1, 112.5, 105.5, 80.0, 56.5, 56.3, 40.6, 40.5, 40.4, 40.3, 40.3, 40.2, 40.1, 40.0, 39.9, 39.8, 39.7, 39.5, 22.5, 18.1; 19F NMR (470 MHz, DMSO-d6) δ −72.43; HRMS m/z: calcd. for C22H20F6N4O4 [M + H]+ 519.1462, found: 519.1471.
1-(3,4-Dimethoxyphenyl)-N-(2-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-4,5-dimethylphenyl)-1H-1,2,3-triazole-4-carboxamide (2d).
A similar procedure was followed as in the case of 1a. Isolated (18 mg, 35%) as a white solid. Melting point: 181–183 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 11.37 (s, 1H), 10.21 (s, 1H), 9.43 (s, 1H), 8.36 (s, 1H), 7.58 (d, J = 2.6 Hz, 1H), 7.54 (dd, J = 8.6, 2.5 Hz, 1H), 7.25 (s, 1H), 7.17 (d, J = 8.7 Hz, 1H), 3.88 (s, 3H), 3.84 (s, 3H), 2.28 (s, 3H), 2.25 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 157.6, 149.8, 144.0, 139.6, 130.0, 125.9, 113.3, 112.5, 105.6, 56.5, 56.3, 40.5, 40.4, 40.3, 40.2, 40.2, 40.1, 40.0, 39.9, 39.8, 39.7, 39.5, 20.0, 19.6; 19F NMR (470 MHz, DMSO-d6) δ −72.68; HRMS (Q-TOF, ESI) m/z calculated for C22H20F6N4O4 [M + H]+ 519.1462, found: 519.1472.
1-(3,4-Dimethoxyphenyl)-N-(3-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-1H-indol-5-yl)-1H-1,2,3-triazole-4-carboxamide (2e).
A similar procedure was followed as in the case of 1a. Isolated (18.2 mg, 28%) as a white solid. Melting point: 293–295 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 11.58 (s, 1H), 10.44 (s, 1H), 9.32 (s, 1H), 8.45–8.22 (m, 2H), 7.81 (d, J = 2.4 Hz, 1H), 7.70 (dd, J = 8.1, 2.4 Hz, 1H), 7.62 (dd, J = 8.8, 2.0 Hz, 1H), 7.54 (s, 1H), 7.41 (dd, J = 17.8, 8.5 Hz, 2H), 2.35 (s, 3H), 2.32 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.3, 144.5, 138.7, 138.1, 134.7, 134.0, 131.5, 131.1, 126.8, 125.6, 125.3, 121.7, 118.2, 117.4, 113.9, 112.0, 105.1, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −74.76; HRMS (Q-TOF, ESI) m/z calculated for C22H17F6N4O4 [M + H]+ 530.1257, found: 530.1245.
1-(3,4-Dimethoxyphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-methoxy propan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (2f).
A similar procedure was followed as in the case of 1a. Isolated (35 mg, 30%) as a white solid. Melting point: 183–185 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.92 (s, 1H), 9.46 (s, 1H), 8.13–8.04 (m, 2H), 7.60–7.51 (m, 4H), 7.17 (dd, J = 8.7, 1.3 Hz, 1H), 3.89 (d, J = 1.4 Hz, 3H), 3.85 (d, J = 1.4 Hz, 3H), 3.47 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 159.1, 149.8, 143.7, 141.1, 130.0, 128.9, 126.4, 121.6, 121.2, 113.2, 112.4, 105.4, 56.4, 56.3, 54.7, 40.4, 40.3, 40.2, 40.2, 40.1, 39.9, 39.7, 39.6, 39.4; 19F NMR (470 MHz, DMSO-d6) δ −70.41; HRMS (Q-TOF, ESI) m/z calculated for C21H18F6N4O4 [M + H]+ 505.1305, found: 505.1325.
1-(3,4-Dimethoxyphenyl)-N-(4-(2-ethoxy-1,1,1,3,3,3-hexafluoro propan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (2g).
A similar procedure was followed as in the case of 1a. Isolated (25 mg, 32%) as a white solid. Melting point: 161–163 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.92 (s, 1H), 9.47 (s, 1H), 8.09 (d, J = 8.9 Hz, 2H), 7.60–7.53 (m, 4H), 7.18 (d, J = 8.7 Hz, 1H), 3.90 (s, 3H), 3.85 (s, 3H), 3.61 (q, J = 6.9 Hz, 2H), 1.32 (t, J = 6.9 Hz, 3H); 13C NMR (125 MHz, DMSO-d6) δ 159.1, 149.8, 149.8, 143.7, 141.0, 130.0, 128.7, 126.4, 122.3, 121.2, 113.2, 112.5, 105.4, 82.7, 62.8, 56.5, 56.3, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.8, 39.6, 39.5, 15.5; 19F NMR (470 MHz, DMSO-d6) δ −70.49; HRMS (Q-TOF, ESI) m/z calculated for C22H20F6N4O4 [M + H]+ 519.1462, found: 519.1476.
1-(3,4-Dimethoxyphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-(isopentyl oxy)propan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (2h).
A similar procedure was followed as in the case of 1a. Isolated (21 mg, 26%) as a yellowish white solid. Melting point: 149–151 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.91 (s, 1H), 9.46 (d, J = 1.3 Hz, 1H), 8.11–8.07 (m, 2H), 7.60–7.52 (m, 4H), 7.18 (dd, J = 8.7, 1.3 Hz, 1H), 3.89 (d, J = 1.3 Hz, 3H), 3.85 (d, J = 1.2 Hz, 3H), 3.58 (t, J = 6.6 Hz, 2H), 1.76 (dt, J = 13.3, 6.6 Hz, 1H), 1.61 (q, J = 6.6 Hz, 2H), 0.91 (d, J = 1.3 Hz, 3H), 0.90 (d, J = 1.3 Hz, 3H); 13C NMR (125 MHz, DMSO-d6) δ 159.0, 149.8, 149.8, 143.7, 141.0, 130.0, 128.8, 126.4, 122.2, 121.2, 113.2, 112.4, 105.4, 65.2, 56.4, 56.3, 49.1, 40.5, 40.5, 40.4, 40.3, 40.2, 40.1, 40.0, 40.0, 39.9, 39.8, 39.6, 39.5, 38.3, 24.8, 22.8; 19F NMR (470 MHz, DMSO-d6) δ −70.40; HRMS (Q-TOF, ESI) m/z calculated for C25H26F6N4O4 [M + H]+ 561.1931, found: 561.1928.
1-(3,4-Dimethylphenyl)-N-(3-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-4-methylphenyl)-1H-1,2,3-triazole-4-carboxamide (2i).
A similar procedure was followed as in the case of 1a. Isolated (70 mg, 40%) as a white solid. Melting point: 215–217 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 11.46 (s, 1H), 10.33 (s, 1H), 9.41 (s, 1H), 8.49 (d, J = 8.5 Hz, 1H), 7.81 (d, J = 2.4 Hz, 1H), 7.71 (dd, J = 8.1, 2.4 Hz, 1H), 7.39 (dd, J = 13.7, 8.5 Hz, 2H), 7.32 (s, 1H), 2.35 (s, 3H), 2.34 (s, 3H), 2.31 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 157.7, 144.0, 138.7, 138.3, 136.5, 134.6, 133.4, 131.7, 131.0, 128.3, 126.9, 125.8, 124.6, 123.6, 122.3, 121.9, 120.0, 118.3, 118.2, 80.6, 80.3, 80.1, 79.9, 40.5, 40.4, 40.3, 40.2, 40.1, 40.0, 39.9, 39.9, 39.8, 39.6, 39.4, 21.1, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −72.57; HRMS (Q-TOF, ESI) m/z calculated for C21H18F6N4O2 [M + H]+ 473.1407, found: 473.1407.
1-(3,4-Dimethylphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-2-methylphenyl)-1H-1,2,3-triazole-4-carboxamide (2j).
A similar procedure was followed as in the case of 1a. Isolated (35 mg, 32%) as a white solid. Melting point: 212–215 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.19 (s, 1H), 9.39 (s, 1H), 8.71 (s, 1H), 7.82 (s, 1H), 7.74–7.66 (m, 2H), 7.59 (s, 1H), 7.54 (d, J = 7.6 Hz, 1H), 7.38 (d, J = 8.2 Hz, 1H), 2.36 (s, 3H), 2.34 (s, 3H), 2.31 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.6, 143.7, 138.7, 138.2, 137.9, 134.6, 133.3, 131.1, 129.2, 128.2, 126.0, 125.8, 125.1, 124.6, 122.3, 121.8, 118.2, 79.4, 77.2, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5, 19.9, 19.5, 18.5; 19F NMR (470 MHz, DMSO-d6) δ −73.85; HRMS (Q-TOF, ESI) m/z calculated for C21H18F6N4O2 [M + H]+ 473.1407, found: 473.1400.
1-(3,4-Dimethylphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-2,5-dimethylphenyl)-1H-1,2,3-triazole-4-carboxamide (2k).
A similar procedure was followed as in the case of 1a. Isolated (20 mg, 22%) as a white solid. Melting point: 291–293 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.15 (s, 1H), 9.38 (s, 1H), 7.82 (d, J = 2.4 Hz, 1H), 7.71 (dd, J = 8.1, 2.4 Hz, 1H), 7.47 (s, 1H), 7.41–7.33 (m, 2H), 2.56 (s, 3H), 2.34 (s, 3H), 2.31 (s, 3H), 2.28 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.6, 143.7, 138.7, 138.2, 137.6, 137.3, 134.6, 131.1, 130.9, 130.3, 129.8, 125.7, 125.1, 121.8, 118.2, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.8, 39.6, 39.5, 22.5, 19.9, 19.5, 18.1; 19F NMR (470 MHz, DMSO-d6) δ −70.39; HRMS (Q-TOF, ESI) m/z calculated for C22H20F6N4O2 [M + H]+ 487.1563, found: 487.1560.
1-(3,4-Dimethylphenyl)-N-(2-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-4,5-dimethylphenyl)-1H-1,2,3-triazole-4-carboxamide (2l).
A similar procedure was followed as in the case of 1a. Isolated (22 mg, 25%) as a white solid. Melting point: 248–250 °C, HPLC purity: 96.72%; 1H NMR (500 MHz, DMSO-d6) δ 11.38 (s, 1H), 10.22 (s, 1H), 9.38 (s, 1H), 8.37 (s, 1H), 7.82 (d, J = 2.4 Hz, 1H), 7.72 (dd, J = 8.1, 2.4 Hz, 1H), 7.38 (d, J = 8.2 Hz, 1H), 7.25 (s, 1H), 2.34 (s, 3H), 2.31 (s, 3H), 2.28 (s, 3H), 2.25 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 157.6, 138.7, 138.3, 134.6, 131.0, 128.6, 125.7, 124.6, 121.9, 118.3, 115.7, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5, 20.0, 19.9, 19.6, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −72.68; HRMS (Q-TOF, ESI) m/z calculated for C22H20F6N4O2 [M + H]+ 487.1563, found: 487.1555.
1-(3,4-Dimethylphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-2-methoxyphenyl)-1H-1,2,3-triazole-4-carboxamide (2m).
A similar procedure was followed as in the case of 1a. Isolated (20 mg, 18%) as a white solid. Melting point: 203–205 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 9.68 (s, 1H), 9.47 (s, 1H), 8.80 (s, 1H), 8.37 (d, J = 8.5 Hz, 1H), 7.82 (d, J = 2.4 Hz, 1H), 7.72 (dd, J = 8.2, 2.4 Hz, 1H), 7.40–7.36 (m, 2H), 7.33 (d, J = 8.6 Hz, 1H), 3.98 (s, 3H), 2.34 (s, 3H), 2.31 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 157.9, 149.0, 143.4, 138.7, 138.4, 134.6, 131.1, 127.1, 125.8, 121.8, 120.3, 118.3, 109.9, 56.7, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.8, 39.6, 39.5, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −73.90; HRMS (Q-TOF, ESI) m/z calculated for C21H18F6N4O3 [M + H]+ 489.1356, found: 489.1365.
1-(3,4-Dimethylphenyl)-N-(3-(1,1,1,3,3,3-hexafluoro-2-hydroxy propan-2-yl)-1H-indol-5-yl)-1H-1,2,3-triazole-4-carboxamide (2n).
A similar procedure was followed as in the case of 1a. Isolated (20 mg, 20%) as a white solid. Melting point: 278–280 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 11.58 (s, 1H), 10.44 (s, 1H), 9.32 (s, 1H), 8.45–8.22 (m, 2H), 7.81 (d, J = 2.4 Hz, 1H), 7.70 (dd, J = 8.1, 2.4 Hz, 1H), 7.62 (dd, J = 8.8, 2.0 Hz, 1H), 7.54 (s, 1H), 7.41 (dd, J = 17.8, 8.5 Hz, 2H), 2.35 (s, 3H), 2.32 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.3, 144.5, 138.7, 138.1, 134.7, 134.0, 131.5, 131.1, 126.8, 125.6, 125.3, 121.7, 118.2, 117.4, 113.9, 112.0, 105.1, 94.1, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −74.72; HRMS (Q-TOF, ESI) m/z calculated for C22H17F6N4O2 [M + H]+ 498.1359, found: 498.1363.
1-(3,4-Dimethylphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-methoxypropan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (2o).
A similar procedure was followed as in the case of 1a. Isolated (40 mg, 33%) as a white solid. Melting point: 200–202 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.91 (s, 1H), 9.41 (s, 1H), 8.12–8.05 (m, 2H), 7.82 (d, J = 2.3 Hz, 1H), 7.71 (dd, J = 8.1, 2.4 Hz, 1H), 7.57 (d, J = 8.5 Hz, 2H), 7.38 (d, J = 8.2 Hz, 1H), 3.46 (s, 3H), 2.34 (s, 3H), 2.31 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 159.0, 143.8, 141.1, 138.7, 138.3, 134.6, 131.1, 128.9, 126.1, 123.9, 121.8, 121.6, 121.6, 121.2, 118.2, 82.8, 79.7, 54.7, 40.6, 40.5, 40.4, 40.3, 40.2, 40.2, 40.1, 40.0, 39.9, 39.8, 39.7, 39.5, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −70.41; HRMS (Q-TOF, ESI) m/z calculated for C21H18F6N4O2 [M + H]+ 473.1407, found: 473.1415.
1-(3,4-Dimethylphenyl)-N-(4-(2-ethoxy-1,1,1,3,3,3-hexafluoro propan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (2p).
A similar procedure was followed as in the case of 1a. Isolated (20 mg, 25%) as a white solid. Melting point: 194–196 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.93 (s, 1H), 9.42 (s, 1H), 8.09 (d, J = 8.5 Hz, 2H), 7.82 (d, J = 2.4 Hz, 1H), 7.72 (dd, J = 8.1, 2.4 Hz, 1H), 7.57 (d, J = 8.5 Hz, 2H), 7.38 (d, J = 8.2 Hz, 1H), 3.61 (q, J = 6.9 Hz, 2H), 2.34 (s, 3H), 2.31 (s, 3H), 1.31 (t, J = 6.9 Hz, 3H); 13C NMR (125 MHz, DMSO-d6) δ 159.0, 143.8, 141.0, 138.7, 138.3, 134.6, 131.1, 128.7, 126.1, 124.0, 122.2, 121.8, 121.7, 121.2, 118.2, 62.8, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5, 19.9, 19.5, 15.5; 19F NMR (470 MHz, DMSO-d6) δ −70.51; HRMS (Q-TOF, ESI) m/z calculated for C22H20F6N4O2 [M + H]+ 487.1563, found: 487.1563.
1-(3,4-Dimethylphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-(isopentyl oxy)propan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (2q).
A similar procedure was followed as in the case of 1a. Isolated (60 mg, 36%) as a white solid. Melting point: 166–168 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.92 (s, 1H), 9.42 (s, 1H), 8.11–8.06 (m, 2H), 7.83 (d, J = 2.3 Hz, 1H), 7.72 (dd, J = 8.1, 2.4 Hz, 1H), 7.56 (d, J = 8.5 Hz, 2H), 7.39 (d, J = 8.2 Hz, 1H), 3.57 (t, J = 6.6 Hz, 2H), 2.35 (s, 3H), 2.31 (s, 3H), 1.76 (dp, J = 13.3, 6.7 Hz, 1H), 1.61 (q, J = 6.7 Hz, 2H), 0.90 (d, J = 6.6 Hz, 6H); 13C NMR (125 MHz, DMSO-d6) δ 159.0, 143.8, 141.1, 138.8, 138.3, 134.6, 131.1, 128.8, 126.1, 124.0, 122.1, 121.8, 121.1, 118.2, 82.5, 65.1, 40.5, 40.5, 40.4, 40.3, 40.2, 40.1, 40.0, 40.0, 39.9, 39.8, 39.6, 39.5, 38.3, 24.8, 22.8, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −70.39; HRMS (Q-TOF, ESI) m/z calculated for C25H26F6N4O2 [M + H]+ 529.2033, found: 529.2032.
1-(3,4-Dimethylphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-morpholino propan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (2s).
According to the general procedure, 1-(3,4-dimethylphenyl)-N-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl)-1H-1,2,3-triazole-4-carboxamide (50 mg) was dissolved in dichloromethane (200 μl) at 0 °C, to which triethylamine (1.5 equiv.) was added. Following which, methane sulfonyl chloride (5 equiv.) was added dropwise to the above solution and kept stirring for 30 min at 0 °C and room temperature for 2 h. Completion of the reaction was monitored by TLC. Morpholine (1.5 equiv.) was added to the reaction mixture at room temperature and it was allowed to stir further for another 5 h. The reaction mixture was quenched with a saturated ammonium chloride solution and extracted with DCM. Organic layers were dried over Na2SO4 (anhyd.) and concentrated under vacuum. The crude mixture was purified by silica gel (100–200 mesh) column chromatography by using ethyl acetate and petroleum ether as eluent to isolate 2s, 2r and 2t.
Compound 2s (20 mg, 20%) as a white solid crystal. Melting point: 194–196 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.98 (s, 1H), 9.43 (s, 1H), 8.14–8.07 (m, 2H), 7.85–7.81 (m, 3H), 7.72 (dd, J = 8.1, 2.4 Hz, 1H), 7.39 (d, J = 8.2 Hz, 1H), 2.35 (s, 3H), 2.31 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 159.1, 143.7, 141.6, 138.7, 138.3, 134.6, 131.1, 129.2, 126.2, 121.8, 121.6, 121.0, 118.2, 40.6, 40.5, 40.4, 40.3, 40.2, 40.2, 40.1, 40.0, 39.9, 39.8, 39.7, 39.5, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −69.56; HRMS (Q-TOF, ESI) m/z calculated for C20H15ClF6N4O [M + H]+ 477.0911, found: 477.0902.
2-(4-(1-(3,4-Dimethylphenyl)-1H-1,2,3-triazole-4-carboxamido) phenyl)-1,1,1,3,3,3-hexafluoropropan-2-yl methanesulfonate (2r).
(5 mg, 10%) as a yellowish white solid. Melting point: 201–203 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 11.03 (s, 1H), 9.44 (s, 1H), 8.14 (d, J = 9.3 Hz, 2H), 7.86 (d, J = 8.7 Hz, 2H), 7.83 (d, J = 2.4 Hz, 1H), 7.72 (dd, J = 8.1, 2.5 Hz, 1H), 7.39 (d, J = 8.2 Hz, 1H), 3.18 (s, 3H), 2.35 (s, 3H), 2.31 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 159.1, 143.7, 141.7, 138.8, 138.3, 134.6, 131.1, 130.7, 126.3, 121.8, 121.2, 118.2, 42.7, 40.6, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −59.19; HRMS m/z: calcd. for C21H18F6N4O4S [M + Na]+ 559.4386, found: 559.4158.
N-(4-(2-Chloro-1,1,1,3,3,3-hexafluoropropan-2-yl)phenyl)-1-(3,4-dimethylphenyl)-1H-1,2,3-triazole-4-carboxamide (2t).
(12% ethyl acetate/petroleum ether) to give compound 2t (6 mg, 8%) as a white solid. Melting point: 193–195 °C, HPLC purity:; 1H NMR (500 MHz, DMSO-d6) δ 10.86 (s, 1H), 9.41 (s, 1H), 8.03 (d, J = 8.9 Hz, 2H), 7.82 (d, J = 2.4 Hz, 1H), 7.71 (dd, J = 8.2, 2.3 Hz, 3H), 7.39 (d, J = 8.2 Hz, 1H), 3.68 (s, 4H), 2.80 (s, 4H), 2.34 (s, 3H), 2.31 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 158.9, 143.8, 140.4, 138.7, 138.3, 134.6, 131.1, 129.4, 126.1, 125.8, 121.8, 121.0, 118.2, 67.3, 49.2, 40.5, 40.4, 40.3, 40.2, 40.1, 40.1, 40.0, 39.9, 39.8, 39.6, 39.5, 19.9, 19.5; 19F NMR (470 MHz, DMSO-d6) δ −62.83; HRMS (Q-TOF, ESI) m/z calculated for C24H23F6N5O2 [M + H]+ 528.1829, found: 528.1829.
2-(5-(((1-(3,4-Dimethoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)amino)-2-methylphenyl)-1,1,1,3,3,3-hexafluoropropan-2-ol (3a).
According to the general procedure, a mixture of Et3N (2 equiv.), 1.2 mmol of corresponding amine (1.2 equiv.) and propargyl halide (1.2 equiv.) in sodium dioctyl sulfosuccinate (2% w/v) in water (100 μl) was stirred vigorously for 60 min at room temperature. Then, azide (1 equiv., 50 mg) and 10 mol% of CuI were added into the mixture. The crude mixture was purified by column chromatography (22% ethyl acetate/petroleum ether) to give compound 3a (6 mg, 13%) as a brownish white solid. Melting point: 166–168 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 10.59 (s, 1H), 8.64 (s, 1H), 7.45 (d, J = 2.5 Hz, 1H), 7.38 (dd, J = 8.6, 2.5 Hz, 1H), 7.20 (dd, J = 8.4, 2.0 Hz, 1H), 7.14 (d, J = 8.8 Hz, 1H), 7.10 (s, 1H), 6.99 (d, J = 8.4 Hz, 1H), 6.50 (s, 1H), 4.36 (d, J = 4.4 Hz, 2H), 3.86 (s, 3H), 3.83 (s, 3H), 2.23 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 149.8, 149.4, 146.3, 145.8, 132.0, 130.5, 128.3, 127.6, 121.8, 117.7, 114.9, 112.6, 112.5, 105.1, 79.7, 56.3, 56.3, 41.1, 40.6, 40.5, 40.4, 40.3, 40.2, 40.2, 40.1, 40.0, 39.8, 39.7, 39.5, 20.7; 19F NMR (470 MHz, DMSO-d6) δ −73.17; HRMS (Q-TOF, ESI) m/z calculated for C21H20F6N4O3 [M + H]+ 491.1512, found: 491.1521.
2-(4-(((1-(3,4-Dimethoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)amino)-2,5-dimethylphenyl)-1,1,1,3,3,3-hexafluoropropan-2-ol (3b).
A similar procedure was followed as in the case of 3a. The crude mixture was purified by column chromatography (22% ethyl acetate/petroleum ether) to give compound 3b (7 mg, 14%) as a brown solid. Melting point: 83–85 °C, HPLC purity: 95.72%; 1H NMR (500 MHz, DMSO-d6) δ 8.63 (s, 1H), 8.00 (s, 1H), 7.45 (d, J = 2.5 Hz, 1H), 7.40 (dd, J = 8.6, 2.5 Hz, 1H), 7.12 (d, J = 8.8 Hz, 1H), 7.03 (s, 1H), 6.52 (s, 1H), 5.78 (t, J = 5.9 Hz, 1H), 4.47 (d, J = 5.8 Hz, 2H), 3.85 (s, 3H), 3.82 (s, 3H), 2.43 (s, 3H), 2.10 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 149.8, 149.2, 147.2, 146.9, 138.0, 134.5, 130.6, 129.1, 121.6, 119.5, 115.6, 114.5, 112.5, 105.0, 56.3, 56.3, 40.6, 40.5, 40.4, 40.3, 40.2, 40.2, 40.1, 40.0, 39.8, 39.7, 39.5, 22.9, 18.0; 19F NMR (470 MHz, DMSO-d6) δ −74.74; HRMS (Q-TOF, ESI) m/z calculated for C22H22F6N4O3 [M + H]+ 505.1669, found: 505.1676.
2-(4-(((1-(3,4-Dimethoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)amino)-3-methylphenyl)-1,1,1,3,3,3-hexafluoropropan-2-ol (3c).
A similar procedure was followed as in the case of 3a. The crude mixture was purified by column chromatography (30% ethyl acetate/petroleum ether) to give compound 3c (7.4 mg, 22%) as a brownish white solid. Melting point: 174–176 °C, HPLC purity: 90%; 1H NMR (500 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.23 (s, 1H), 7.44 (d, J = 2.6 Hz, 1H), 7.39 (dd, J = 8.6, 2.5 Hz, 1H), 7.29–7.24 (m, 2H), 7.12 (d, J = 8.8 Hz, 1H), 6.72 (d, J = 8.6 Hz, 1H), 5.90 (t, J = 5.8 Hz, 1H), 4.49 (d, J = 5.7 Hz, 2H), 3.85 (s, 3H), 3.82 (s, 3H), 2.18 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 149.8, 149.2, 147.7, 147.1, 130.6, 128.4, 125.9, 124.8, 122.2, 121.6, 117.6, 112.5, 112.4, 109.3, 105.0, 79.7, 56.3, 56.3, 40.6, 40.5, 40.4, 40.3, 40.2, 40.2, 40.1, 40.0, 39.8, 39.7, 39.5, 39.0, 18.5; 19F NMR (470 MHz, DMSO-d6) δ −74.16; HRMS (Q-TOF, ESI) m/z calculated for C21H20F6N4O3 [M + H]+ 491.1512, found: 491.1514.
2-(((1-(3,4-Dimethoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)amino)-5-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenol (3d).
A similar procedure was followed as in the case of 3a. The crude mixture was purified by column chromatography (22% ethyl acetate/petroleum ether) to give compound 3d (5 mg, 12%) as a dark greyish solid. Melting point: 87–89 °C, HPLC purity: 100%; 1H NMR (500 MHz, DMSO-d6) δ 8.90 (s, 1H), 8.30 (s, 1H), 7.46 (d, J = 2.5 Hz, 1H), 7.41 (dd, J = 8.7, 2.5 Hz, 1H), 7.20 (d, J = 2.1 Hz, 1H), 7.16 (d, J = 8.7 Hz, 1H), 7.03–7.00 (m, 1H), 6.72 (d, J = 8.4 Hz, 1H), 6.55 (s, 1H), 5.19 (s, 2H), 3.87 (s, 3H), 3.83 (s, 3H); 13C NMR (125 MHz, DMSO-d6) δ 149.8, 149.4, 144.7, 144.4, 140.4, 130.5, 123.1, 121.0, 117.5, 113.8, 112.7, 112.5, 112.1, 105.1, 62.7, 56.3, 56.3, 40.6, 40.5, 40.4, 40.3, 40.2, 40.2, 40.1, 40.0, 39.8, 39.7, 39.5; 19F NMR (470 MHz, DMSO-d6) δ −74.1; HRMS (Q-TOF, ESI) m/z calculated for C20H18F6N4O4 [M + H]+ 493.1305, found: 493.1293.
3.2 Biological evaluation
3.2.1 Alamar blue assay for cell viability.
The toxicity of the compounds was assessed against N2A mouse neuroblasts cells (purchased from ATCC) by Alamar blue assay by using the protocol. The cells were allowed to grow at 37 °C in the presence of CO2 (5%) by providing DMEM (high glucose) medium, which is also supplemented with 5% FBS, 100 U ml−1 penicillin G, and 100 μg mL−1 streptomycin. The cells were cultured in T-25 flasks and further incubated for 48 h and then allowed to adhere at 37 °C. The cells were sub-cultured once in a week's time and two-thirds of the growth medium was replaced after every 3–4 days. The stock solution of synthesized compounds (3 mM) was prepared in DMSO. Synthesized compounds were incubated with the cells in DMEM media deprived of FBS serum, at 50 μM concentration respectively in 96 well plates in triplicate.
Initially, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded into each 96 well plate and cells were allowed to adhere. After 24 h, the cells were then treated with 50 μM of the synthesized compound (dissolved in DMSO and diluted in DMEM media) for the next 24 h. Thereafter, the entire medium was removed and 10% volume of Alamar blue reagent was then added (from stock solution of 1 mg ml−1) and incubated for an additional 4 h at 37 °C. The fluorescence measurements were performed at an excitation wavelength of 560 nm and emission wavelength of 590 nm using a Varioskan Lux multiplate reader (Thermo Fischer). For the positive control, cells were treated with 1.66% v/v DMSO, cultured in DMEM media only, and considered as 100% viable, and the cell viability was calculated based on this.
000 cells were seeded into each 96 well plate and cells were allowed to adhere. After 24 h, the cells were then treated with 50 μM of the synthesized compound (dissolved in DMSO and diluted in DMEM media) for the next 24 h. Thereafter, the entire medium was removed and 10% volume of Alamar blue reagent was then added (from stock solution of 1 mg ml−1) and incubated for an additional 4 h at 37 °C. The fluorescence measurements were performed at an excitation wavelength of 560 nm and emission wavelength of 590 nm using a Varioskan Lux multiplate reader (Thermo Fischer). For the positive control, cells were treated with 1.66% v/v DMSO, cultured in DMEM media only, and considered as 100% viable, and the cell viability was calculated based on this.
3.2.2 Thioflavin T assay.
Thioflavin T (ThT) assay was performed to check the amyloid β anti-aggregation effect. The Aβ42 peptide (0.1 mg) was purchased from Sigma Aldrich and was dissolved in 100 μl 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP). Further, the HFIP was evaporated overnight in a fume hood and the peptide was used freshly. Aβ42 (221.5 μM) stock solution was prepared by dissolving 0.1 mg Aβ42 in 100 μl DMSO. 1 mM stock solution of ThT was then prepared by dissolving it in 50 mM glycine–NaOH buffer (pH 8.0), filtered and then diluted to 200 μM stock.
To perform the Cu2+-induced aggregation assay, a mixture of Aβ42 (5 μM) with Cu2+ (10 μM) and ThT (20 μM) in the presence or absence of test compounds (50 μM) was diluted to reach a final volume of 100 μL with 50 mM PBS buffer (pH 7.4) in a black 96-well plate and incubated for 24 h at 37 °C with constant agitation (180 rpm). Thereafter, the fluorescence intensity measurements were performed using a Varioskan Lux spectrophotometer with 450 nm as the excitation wavelength and 485 nm emission wavelength. The percentage Aβ42 fibril formation capacity was calculated using the fluorescence intensity from the Cu2+ induced Aβ peptide without any treatment as 100%. The following procedure has been employed at different timepoints with a duration of 24 h.
3.2.3 NO level detection by Griess reagent.
The procedure of the nitrite assay kit (Griess reagent) (catalogue no. MAK367, Merck) was performed as per the protocol provided by the manufacturer. The absorbance of the treated cells in a 96 well plate was taken at 540 nm in a microplate reader.
3.2.4 Measurement of reactive oxygen species (ROS).
The intracellular ROS level was determined using the 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) staining technique, as earlier reported. After 24 h acclimatization, N2a (1 × 104 cells per wells) cells were pre-treated with H2O2 (200 μM) for 5 h before incubation, with 50 μM compounds in the presence or absence of Aβ (5 μM) with Cu2+ (10 μM) for 24 h. Cells were then treated with 25 μM H2DCFDA and incubated for 45 min in the dark at 37 °C. Fluorescence intensity (Ex 495 nm, Em 520 nm) was measured in a microplate reader. The ROS level was calculated as a percentage of the H2O2 treated control cells (100%) in triplicate measurements.
3.2.5 Aβ and Cu2+ induced cell toxicity.
To determine the effect of 2f on Aβ and Cu2+-induced cell toxicity, Alamar blue assay was performed. N2a (1 × 104 cells per wells) cells were seeded in a black 96-well plate. After 24 h, pretreatment of H2O2 has been done for 5 h and then a mixture of Aβ (5 μM) with Cu2+ (10 μM) was incubated in the presence or absence of 2f (50 μM) (diluted in DMEM media) to reach a final volume of 100 μL and incubated for 24 h at 37 °C with constant agitation (180 rpm). Thereafter, the fluorescence intensity measurements were performed using a Varioskan Lux spectrophotometer with 450 nm as the excitation wavelength and 485 nm emission wavelength. The percentage Aβ fibril formation capacity was calculated using the fluorescence intensity from the Cu2+ induced Aβ peptide without any treatment as 100%.
3.2.6 Immunocytochemistry analysis.
N2A (∼50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) cells were cultured in chamber slides followed by treatment with Aβ42 + Cu2+ and 2f (24 h). Post treatment, the cells were fixed with 4% PFA (15 min at RT). Then, the cells were blocked with immunofluorescence blocking buffer (5% BSA in PBS and 0.3% Triton-X). Further, the cells were incubated with primary antibodies: NLRP3 (MA5-52041, Sigma-Aldrich; 1
000) cells were cultured in chamber slides followed by treatment with Aβ42 + Cu2+ and 2f (24 h). Post treatment, the cells were fixed with 4% PFA (15 min at RT). Then, the cells were blocked with immunofluorescence blocking buffer (5% BSA in PBS and 0.3% Triton-X). Further, the cells were incubated with primary antibodies: NLRP3 (MA5-52041, Sigma-Aldrich; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) and IL-1β (P420B, Sigma-Aldrich; 1
250) and IL-1β (P420B, Sigma-Aldrich; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) at 4 °C overnight followed by secondary antibody anti-rabbit Alexa Fluor 488 (1
250) at 4 °C overnight followed by secondary antibody anti-rabbit Alexa Fluor 488 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, A11008, Abcam) and goat anti-rat Alexa Fluor 647 (A-21434, Thermo Fisher Scientific; 1
500, A11008, Abcam) and goat anti-rat Alexa Fluor 647 (A-21434, Thermo Fisher Scientific; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) incubation (4–6 h at RT). Next, the cells were washed with PBS 3 times. DAPI was used as a counterstain before visualizing in a confocal microscope (SP8 Leica Light Microscope).
500) incubation (4–6 h at RT). Next, the cells were washed with PBS 3 times. DAPI was used as a counterstain before visualizing in a confocal microscope (SP8 Leica Light Microscope).
For determination of lysosomal pH and lysosome enzyme activity, Aβ42 + Cu2+-treated N2A cells with the absence or presence of 2f (24 h) then were incubated in LysoTracker® Deep Red (50 nM, 15 min) and acridine orange (5 μM, 15 min, 37 °C) in separate sets of treatments, followed by washing with PBS twice. The images were captured immediately post-washing using a fluorescence microscope. For the quantification for cell fluorescence, ImageJ software used.
3.2.7 LysoTracker® Deep Red staining.
For determination of lysosomal pH and lysosome enzyme activity, Aβ42 + Cu2+-treated N2A cells with the absence or presence of 2f (24 h) then were incubated in LysoTracker® Deep Red (50 nM, 15 min), followed by washing with PBS twice. The images were captured immediately post-washing using a fluorescence microscope. For the quantification for cell fluorescence, ImageJ software used.
3.2.8 Acridine orange staining.
For this staining, Aβ42 + Cu2+-treated N2A cells with the absence or presence of 2f (24 h) then were incubated in acridine orange (5 μM, 15 min, 37 °C), followed by PBS washing. The images were taken immediately after PBS washing using a fluorescence microscope. For the quantification for cell fluorescence, ImageJ software used.
3.2.9 Live dead assay.
The procedure of live dead assay (catalogue no. L3224, Invitrogen) was performed as per the manufacturer's protocol. A fluorescence microscope was used for taking the images. For the quantification for cell number, ImageJ software used.
3.3
In silico studies
3.3.1 Molecular docking studies.
In order to study the effective outcome of compound 2f in in vitro experiments, an in silico analysis was carried out to investigate the molecular interaction of compound 2f with the Aβ42 peptide as a fibril structure. The Aβ42 peptide as a β-sheet pentamer was taken directly from the Protein Data Bank (both structures determined by NMR spectroscopy; PDB code 2BEG). The structure selected was a U-shaped model consisting of two beta strands. Strand 1 (residue range 17 to 26) and strand 2 (residue range 31 to 42) are connected by a bend having residues ranging from 27 to 30. The model is a pentamer consisting of five identical chains denoted as A to E. The U-shaped model (2BEG) has been part of most of the previously reported molecular simulation studies.87 The three-dimensional structure of the synthesised compound 2f was generated by using the ChemDraw 20.0 tool.
The Auto Dock 4.2 tool was employed for molecular docking. The PDBQT file for the Aβ42 pentamer was generated by adding polar hydrogen, and Kollman charges were initially applied to the protein using ADT v.1.5. Then, Gasteiger partial charges were applied to the ligand. The torsions were set to be rotatable for the pentamer during the docking procedure. The molecular docking was performed using the Lamarckian genetic algorithm. The grid was generated around the whole 2BEG pentamer using blind dimensions. The grid dimensions for the X, Y, and Z coordinates were defined as follows: X-axis = 72 Å, Y-axis = 60 Å, and Z-axis = 98 Å. The central point of this grid was established at specific coordinates: X-axis: −8.869 Å, Y-axis: −3.71 Å, and Z-axis: −5.94 Å. A uniform grid spacing of 0.375 Å was employed. All other parameters related to the molecular docking procedure were maintained at their default settings. The grid dimensions, gaps, and characteristics were selected to allow for the full expansion of both ligands and protein. The Auto Dock Tools interaction determination module was utilised to examine the binding interaction and various bonds implicated in the binding of the docked complexes. The resulting docked complexes were further analysed using the BIOVIA Discovery Studio 4.5 Visualizer88 (BIOVIA, San Diego, CA, USA).
3.3.2 MD simulation setup.
The CHARMM36 force field89 used for topologies and parameters for the molecules generated through CGENFF90 in the GROMACS version 5.1.4 molecular dynamics (MD) simulation package was used to analyse the apo (protein only) and holo (protein–ligand) states, which represented the highest binding affinity from our docking analysis and were further processed for MD simulations to explore their inhibitor specificity, dynamic behaviour and mode of binding activity. Subsequently, the complexes were immersed in a pre-equilibrated periodic water box, with counterions added to maintain system neutrality. However, the Aβ42 protofibril and Aβ42 protofibril 2f complex were kept in a cubic box with dimensions of 10.4 nm × 10.4 nm × 10.4 nm and 2.8 nm was the minimal distance between the protein and the periodic box boundary.
The free 2BEG and 2BEG-2f systems were enveloped in cubic boxes, utilising the SPC (spc216) water model for aqueous environments. In each system, NaCl was added in the cubic box along with the required number of Na+ and Cl− counter ions to keep the system overall neutral at physiological pH. The particle mesh Ewald (PME) approach was used to calculate long-range electrostatic interactions, and the Fourier grid spacing was 0.16 nm (ref. 91) and a cut-off of 1.0 nm was used to evaluate short-range van der Waals interactions. All bonds involving hydrogen atoms of the Aβ42 monomer were constrained using the LINCS algorithm.92 The steepest descent method with a 1000 kJ mol−1 tolerance was used for energy minimisation. During equilibration, the system temperature was gradually raised from 0 to 300 K, maintaining constant volume under periodic boundary conditions and a stable pressure of 1 bar.
Energy was optimized by the steepest descent method, followed by energy minimisation with the conjugate gradient method, equilibrated for 100, 200 and 300 ns, followed by at 300 K under NVT, then maintained the desired pressure (1 atm), the V-scaling method.93,94 The temperature was maintained at 300 K using a modified Berendsen thermostat95 and a Parrinello–Rahman barostat96 have been employed respectively. The periodic boundary conditions were applied in all simulations. Trajectory analysis encompassed the evaluation of the 2BEG-2f complex stability across the simulation duration. Systematic parameter calculations were conducted to monitor and quantify the structural changes and fluctuations occurring within the complex. The ‘pdb2gmx’ programme of the GROMACS package was used for the generation of the topology file. T. Visual MD (VMD 1.9.1)97,98 was used to analyse the resultant trajectories, which are inbuilt in GROMACS. The root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), total energy and solvent accessible surface area (SASA) were analysed99 using gmx_rmsd, gmx_rmsf, gmx_gyrate, gmx_tenergy and gmx_sasa. All 2D plots were graphed using Graphing, Advanced, Computation and Exploration 5.1.23 version (https://www.its.hku.hk/services/research/hpc/software/grace) for data analysis of MD simulations.
3.4 Metal scavenging
3.4.1 Metal chelation assay.
To investigate whether the compounds can chelate bio-metals such as Cu2+, Zn2+, and Fe3+, we employed UV-vis spectroscopy utilizing a Thermo-Fisher UV-visible spectrophotometer EVO 300 PC. The test chemical stock solution was created in methanol and then diluted in HEPES buffer (pH 7.4) to attain the appropriate concentration (50 μM). The metal salts (CuSO4·5H2O, ZnSO4·7H2O, FeCl3) were dissolved in double distilled water. Metal salt solutions of Cu2+, Zn2+, and Fe3+ were added in the desired ratio to compound solutions and samples were incubated at 37 °C for 30 minutes. All UV-vis spectra were acquired with a UV spectrometer in a 1 cm quartz cell. In scan mode, spectral changes were recorded throughout a 200 to 700 nm wavelength range. The above technique was followed for competitive metal binding assay in various metal salts such as CaCl2/FeCl2/KCl/MgSO4·7H2O/MnCl2/NaCl/NiCl2. The absorbance of the samples was monitored at the λmax of the complex of the compounds with metal ions (Cu(II)/Zn(II)/Fe(III)).
3.4.2 Tyrosine intrinsic fluorescence assay.
For the studies, 5 μM Aβ42 with 5 μM and 10 μM Cu2+ and 2f solution were utilized as needed. The sample was prepared in a black 96-well plate with a final volume of 100 μL in HEPES (10 mM, pH 7.4), and the dequenching sample was incubated for 24 hours at 37 °C in a water bath. In 10 mM HEPES buffer (pH 7.4), fluorescence measurements were carried out using excitation at 274 nm and emission detection at 305 nm. The binding of Cu2+ to Aβ was observed using tyrosine fluorescence spectroscopy. The fact that Cu2+ binds to Aβ and suppresses tyrosine fluorescence is well known.
4. HPLC analysis of compounds
The compounds were analysed using an RP-HPLC (LC model 1260 Infinity II, Agilent Technologies, Singapore) equipped with an autosampler, a diode array detector (DAD), and a quaternary pump. Samples were prepared in HPLC grade methanol and run on an (Xbridge BEH C18 peptide column (300 Å, 10 μM, 1.6 mm × 150 mm)) at 40 °C. The chromatograms were examined at a wavelength of 280 nm. The mobile phase was 0.1% formic acid in Milli-Q H2O–acetonitrile at a flow rate of 1 mL min−1, the same as that used in HRMS. For a duration of 13 minutes, the injection volume for every sample was maintained at 10 μL. The solvent system was gradient, equilibrating back to aqueous 90% at the end of the run after moving from A) aqueous 90% to B) organic 90%. Using Open Lab CDS EZ Chrome software, the results were interpreted using the same LC model.
5. Conclusions
By combining with a metal chelating and anti-amyloid aggregation pharmacophore, a SAR study was conducted with HFIP bearing triazole amides. Amongst the synthesized compound library, compound 2f displayed the most potent inhibitory activity against metal induced Aβ aggregation with IC50 value = 16.8 ± 1.8 μM. Cu-induced Aβ aggregation that leads to activation of the inflammatory NLRP3-IL-1β signalling cascade found to be suppressed in the presence of compound 2f. Moreover, 2f successfully controlled the generation of cellular ROS in N2a cell lines and shown neuritogenesis in N2a cells. Modulation of mitochondria membrane potential (MMP), downregulation the NLRP3-IL-1β signalling cascade with neuroprotective effects and predicted ADMET revealed the desirable drug like properties of compound 2f corroborating its potential development as a hit candidate in drug discovery for the treatment of AD.
Author contributions
Bhaskar Dewangan: methodology, investigation, validation, formal analysis, main draft writing – review and editing, Parijat Swain: methodology, investigation, writing – review & editing, Srimant Patra: methodology, investigation, writing – review & editing, Praveen Reddy Bodhe: methodology, investigation, Neeraj Kulkarni: writing – review & editing, investigation, validation, Bichismita Sahu: supervision, funding acquisition, project administration, conceptualization, visualization, validation, formal analysis, writing – review & editing.
Conflicts of interest
There are no conflicts to declare.
Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5md00481k.
Acknowledgements
Dr. Bichismita Sahu and all authors are thankful to the Director of National Institute of Pharmaceutical Education and Research (NIPER)-Ahmedabad and Department of Pharmaceuticals (DoP), Ministry of Chemicals and Fertilizers, India for giving support such as student fellowships and required facilities to complete this work.
References
- N. Kumar, K. Jangid, V. Kumar, R. P. Yadav, J. Mishra, S. Upadhayay, V. Kumar, B. Devi, V. Kumar, A. R. Dwivedi, P. Kumar, S. Baranwal, J. S. Bhatti and V. Kumar, ACS Chem. Neurosci., 2024, 15, 2565–2585 CrossRef CAS PubMed.
- K. Wasnik, P. S. Gupta, G. Singh, S. Maity, S. Patra, D. Pareek, S. Kumar, V. Rai, R. Prakash, A. Acharya, P. Maiti, S. Mukherjee, Y. Mastai and P. Paik, J. Mater. Chem. B, 2024, 12, 6221–6241 RSC.
- D. F. Zhang, M. Xu, R. Bi and Y. G. Yao, ACS Chem. Neurosci., 2019, 10, 890–901 CrossRef CAS.
- H. D. Nguyen, W. K. Kim and H. Huong Vu, Mech. Ageing Dev., 2024, 219, 111930 CrossRef CAS.
- B. Bai, D. Vanderwall, Y. Li, X. Wang, S. Poudel, H. Wang, K. K. Dey, P. C. Chen, K. Yang and J. Peng, Mol. Neurodegener., 2021, 16, 55 CrossRef.
- J. Q. Li, J. H. Song, J. Suckling, Y. J. Wang, C. T. Zuo, C. Zhang, J. Gao, Y. Q. Song, A. M. Xie, L. Tan and J. T. Yu, Neurobiol. Aging, 2024, 134, 106–114 CrossRef CAS.
- M. B. Abubakar, K. O. Sanusi, A. Ugusman, W. Mohamed, H. Kamal, N. H. Ibrahim, C. S. Khoo and J. Kumar, Front. Aging Neurosci., 2022, 14, 742408 CrossRef CAS PubMed.
- D. Gouilly, M. Rafiq, L. Nogueira, A. S. Salabert, P. Payoux, P. Péran and J. Pariente, Rev. Neurol., 2023, 179, 812–830 CrossRef CAS.
- A. R. Monteiro, D. J. Barbosa, F. Remião and R. Silva, Biochem. Pharmacol., 2023, 211, 115522 CrossRef CAS PubMed.
- Y. Liu, M. Nguyen, A. Robert and B. Meunier, Acc. Chem. Res., 2019, 52, 2026–2035 CrossRef CAS PubMed.
- H. Gerber, F. Wu, M. Dimitrov, G. M. G. Osuna and P. C. Fraering, J. Biol. Chem., 2017, 292, 3751–3767 CrossRef CAS PubMed.
- M. Kitazawa, H.-W. Hsu and R. Medeiros, Toxicol. Sci., 2016, 152, 194–204 CrossRef CAS.
- X. Zhang, N. Surguladze, B. Slagle-Webb, A. Cozzi and J. R. Connor, Glia, 2006, 54, 795–804 CrossRef CAS.
- D. Religa, D. Strozyk, R. A. Cherny, I. Volitakis, V. Haroutunian, B. Winblad, J. Naslund and A. I. Bush, Neurology, 2006, 67, 69–75 CrossRef CAS.
- M. G. Savelieff, G. Nam, J. Kang, H. J. Lee, M. Lee and M. H. Lim, Chem. Rev., 2018, 119, 1221–1322 CrossRef PubMed.
- A. Robert, Y. Liu, M. Nguyen and B. Meunier, Acc. Chem. Res., 2015, 48, 1332–1339 CrossRef CAS.
-
B. Dewangan, K. Kumar, A. Kumar, P. R. Bodhe, S. Beni and B. Sahu, in Natural Product-based Synthetic Drug Molecules in Alzheimer's Disease, Springer Nature Singapore, 2023, pp. 347–374 Search PubMed.
- S. V. R. Jonnalagadda, A. J. Gerace, K. Thai, J. Johnson, K. Tsimenidis, J. M. Jakubowski, C. Shen, K. J. Henderson, P. Tamamis and M. Gkikas, J. Phys. Chem. B, 2020, 124, 487–503 CrossRef CAS PubMed.
- M. Handa, S. N. Sanap, R. S. Bhatta, G. P. Patil, S. Ghose, D. P. Singh and R. Shukla, Biomater. Adv., 2023, 154, 213663 CrossRef CAS PubMed.
- T. Dalvi, B. Dewangan, R. Das, J. Rani, S. D. Shinde, N. Vhora, A. Jain and B. Sahu, Cent. Nerv. Syst. Agents Med. Chem., 2020, 20, 157–176 CrossRef CAS PubMed.
- S. Bhargava, R. Kulkarni, B. Dewangan, N. Kulkarni, C. Jiaswar, K. Kumar, A. Kumar, P. R. Bodhe, H. Kumar and B. Sahu, RSC Med. Chem., 2023, 14, 2192–2205 RSC.
- S. A. Funke and D. Willbold, Curr. Pharm. Des., 2012, 18, 755–767 CrossRef CAS PubMed.
- Y. Bouter, J. S. L. Noguerola, P. Tucholla, G. A. N. Crespi, M. W. Parker, J. Wiltfang, L. A. Miles and T. A. Bayer, Acta Neuropathol., 2015, 130, 713–729 CrossRef CAS PubMed.
- M. Gold, Alzheimer's & Dementia: Translational Research & Clinical Intervention, 2017, 3, 402–409 Search PubMed.
- S. Dattatray Shinde, S. Kumar Behera, N. Kulkarni, B. Dewangan and B. Sahu, Bioorg. Med. Chem., 2024, 97, 117538 CrossRef CAS.
- A. Agis-Torres, M. Söllhuber, M. Fernandez and J. M. Sanchez-Montero, Curr. Neuropharmacol., 2014, 2, 2–36 CrossRef PubMed.
- N. Kumar, V. Kumar, P. Anand, V. Kumar, A. Ranjan Dwivedi and V. Kumar, Bioorg. Med. Chem., 2022, 61, 116742 CrossRef CAS.
- B. Kumar, A. R. Dwivedi, T. Arora, K. Raj, V. Prashar, V. Kumar, S. Singh, J. Prakash and V. Kumar, ACS Chem. Neurosci., 2022, 13, 2122–2139 CrossRef CAS.
- A. Kaur, S. Mann, A. Kaur, N. Priyadarshi, B. Goyal, N. K. Singhal and D. Goyal, Bioorg. Chem., 2019, 87, 572–584 CrossRef CAS.
- W. Qu, M. P. Kung, C. Hou, S. Oya and H. F. Kung, J. Med. Chem., 2007, 50, 3380–3387 CrossRef CAS.
- K. Arunrungvichian, C. Boonyarat, V. V. Fokin, P. Taylor and O. Vajragupta, ACS Chem. Neurosci., 2015, 6, 1331–1340 CrossRef CAS PubMed.
- M. R. Jones, E. Mathieu, C. Dyrager, S. Faissner, Z. Vaillancourt, K. J. Korshavn, M. H. Lim, A. Ramamoorthy, V. Wee Yong, S. Tsutsui, P. K. Stys and T. Storr, Chem. Sci., 2017, 8, 5636–5643 RSC.
- J. Jiaranaikulwanitch, P. Govitrapong, V. V. Fokin and O. Vajragupta, Molecules, 2012, 17, 8312–8333 CrossRef CAS.
- T. Dalvi, B. Dewangan, G. Agarwal, D. Shinde Suchita, A. Jain, A. Srivastava and B. Sahu, Bioorg. Med. Chem. Lett., 2021, 42, 127999 CrossRef CAS PubMed.
- M. C. Rosales Hernández, L. G. Fragoso Morales, J. Correa Basurto, M. Olvera Valdez, E. V. García Báez, D. G. Román Vázquez, A. P. Anaya García and A. Cruz, Int. J. Mol. Sci., 2022, 23, 12945 CrossRef.
- L. Blaikie, G. Kay and P. Kong Thoo Lin, MedChemComm, 2019, 10, 2052–2072 RSC.
- J. R. Horsley, B. Jovcevski, K. L. Wegener, J. Yu, T. L. Pukala and A. D. Abell, Biochem. J., 2020, 477, 2039–2054 CrossRef CAS PubMed.
- T. Lieblein, R. Zangl, J. Martin, J. Hoffmann, M. Hutchison, T. Stark, E. Stirnal, T. Schrader, H. Schwalbe and N. Morgner, eLife, 2020, 9, 1–30 CrossRef PubMed.
- J. Li, R. Liu, K. S. Lam, L. W. Jin and Y. Duan, Biophys. J., 2011, 100, 1076–1082 CrossRef CAS.
- B. Török, A. Sood, S. Bag, A. Kulkarni, D. Borkin, E. Lawler, S. Dasgupta, S. Landge, M. Abid, W. Zhou, M. Foster, H. Levine and M. Török, ChemMedChem, 2012, 7, 910–919 CrossRef PubMed.
- S. K. Pachahara, N. Chaudhary, C. Subbalakshmi and R. Nagaraj, J. Pept. Sci., 2012, 18, 233–241 CrossRef CAS.
- T. M. Kamenecka, B. Lyda, M. R. Chang and P. R. Griffin, MedChemComm, 2013, 4, 764–776 RSC.
- M. Dabur, J. A. Loureiro and M. C. Pereira, Int. J. Mol. Sci., 2020, 21, 2989 CrossRef CAS PubMed.
- B. Gomes, J. A. Loureiro, M. A. N. Coelho, M. Do and C. Pereira, Curr. Pharm. Des., 2015, 21, 5725–5735 CrossRef CAS PubMed.
- Y. Li, D. Xu, A. Sun, S. L. Ho, C. Y. Poon, H. N. Chan, O. T. W. Ng, K. K. L. Yung, H. Yan, H. W. Li and M. S. Wong, Chem. Sci., 2017, 8, 8279–8284 RSC.
- J. Wang, D. B. Wang, L. L. Sui and T. Luan, Arabian J. Chem., 2024, 17, 105794 CrossRef CAS.
- J. Zhu, J. Mo, H. z. Lin, Y. Chen and H. p. Sun, Bioorg. Med. Chem., 2018, 26, 3065–3075 CrossRef CAS PubMed.
- N. Steiner, R. Balez, N. Karunaweera, J. M. Lind, G. Münch and L. Ooi, Neurochem. Int., 2016, 95, 46–54 CrossRef CAS PubMed.
- S. A. Back, R. Khan, X. Gan, P. A. Rosenberg and J. J. Volpe, J. Neurosci. Methods, 1999, 91, 47–54 CrossRef CAS PubMed.
- C. Xue, T. Y. Lin, D. Chang and Z. Guo, R. Soc. Open Sci., 2017, 4, 160696 CrossRef.
- M. Biancalana and S. Koide, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 804, 1405–1412 CrossRef.
- K. Rajasekhar, M. Chakrabarti and T. Govindaraju, Chem. Commun., 2015, 51, 13434–13450 RSC.
- J. Mayes, C. Tinker-Mill, O. Kolosov, H. Zhang, B. J. Tabner and D. Allsop, J. Biol. Chem., 2014, 289, 12052–12062 CrossRef CAS PubMed.
- L. Hong, T. M. Carducci, W. D. Bush, C. G. Dudzik, G. L. Millhauser and J. D. Simon, J. Phys. Chem. B, 2010, 114, 11261–11271 CrossRef CAS.
- S. Mann, A. Kaur, A. Kaur, N. Priyadarshi, B. Goyal, N. K. Singhal and D. Goyal, ACS Chem. Neurosci., 2023, 14, 1631–1645 CrossRef CAS PubMed.
- J. H. Jang and Y. J. Surh, Biochem. Biophys. Res. Commun., 2005, 331, 1421–1428 CrossRef CAS PubMed.
- U. Keil, A. Bonert, C. A. Marques, I. Scherping, J. Weyermann, J. B. Strosznajder, F. Müller-Spahn, C. Haass, C. Czech, L. Pradier, W. E. Müller and A. Eckert, J. Biol. Chem., 2004, 279, 50310–50320 CrossRef CAS PubMed.
- M. H. Tran, K. Yamada, A. Olariu, M. Mizuno, X. H. Ren and T. Nabeshima, FASEB J., 2001, 15, 1407–1409 CrossRef CAS.
- A. Elmann, A. Telerman, R. Ofir, Y. Kashman and O. Lazarov, J. Nat. Med., 2018, 72, 626–631 CrossRef CAS.
- X. J. Han, Y. Y. Hu, Z. J. Yang, L. P. Jiang, S. L. Shi, Y. R. Li, M. Y. Guo, H. L. Wu and Y. Y. Wan, Mol. Med. Rep., 2017, 16, 4521–4528 CrossRef CAS.
- W. Zhang, J. Xu, J. Dong, Z. Huang and L. Cao, Neurosci. Bull., 2023, 39, 873–876 CrossRef CAS PubMed.
- L. Zheng, A. Cedazo-Minguez, M. Hallbeck, F. Jerhammar, J. Marcusson and A. Terman, Transl. Neurodegener., 2012, 1, 19 CrossRef CAS.
- Y. Kim, T. Y. Ha, M. S. Lee and K. A. Chang, Int. J. Biol. Sci., 2025, 21, 1014 CrossRef CAS PubMed.
- M. Y. Cha, S. H. Han, S. M. Son, H. S. Hong, Y. J. Choi, J. Byun and I. Mook-Jung, PLoS One, 2012, 7, e34929 CrossRef CAS PubMed.
- J. X. Chen and S. Du Yan, J. Alzheimer's Dis., 2007, 12, 177–184 CAS.
- X. Wang, B. Su, G. Perry, M. A. Smith and X. Zhu, Free Radical Biol. Med., 2007, 43, 1569–1573 CrossRef CAS PubMed.
- C.-C. Chou, R. Vest, M. A. Prado, J. Wilson-Grady, J. A. Paulo, Y. Shibuya, P. Moran-Losada, T.-T. Lee, J. Luo, S. P. Gygi, J. W. Kelly, D. Finley, M. Wernig, T. Wyss-Coray and J. Frydman, Nat. Cell Biol., 2025, 27, 619–632 CrossRef CAS PubMed.
- C. Caspersen, N. Wang, J. Yao, A. Sosunov, X. Chen, J. W. Lustbader, H. W. Xu, D. Stern, G. McKhann and S. Du Yan, FASEB J., 2005, 19, 2040–2041 CrossRef CAS PubMed.
- P. H. Reddy and M. F. Beal, Trends Mol. Med., 2008, 14, 45–53 CrossRef CAS PubMed.
- Y. He, H. Hara and G. Núñez, Trends Biochem. Sci., 2016, 41, 1012–1021 CrossRef CAS PubMed.
- E. S. Jung, K. Suh, J. Han, H. Kim, H. S. Kang, W. S. Choi and I. Mook-Jung, Aging Cell, 2022, 21, e13623 CrossRef CAS PubMed.
- Y. Liu, Y. Dai, Q. Li, C. Chen, H. Chen, Y. Song, F. Hua and Z. Zhang, Neurosci. Lett., 2020, 736, 135279 CrossRef CAS.
- A. Nakanishi, N. Kaneko, H. Takeda, T. Sawasaki, S. Morikawa, W. Zhou, M. Kurata, T. Yamamoto, S. M. F. Akbar, T. Zako and J. Masumoto, Inflammation Regener., 2018, 38, 27 CrossRef CAS PubMed.
- D. Doens and P. L. Fernández, J. Neuroinflammation, 2014, 11, 48 CrossRef PubMed.
- M. T. Heneka, R. M. McManus and E. Latz, Nat. Rev. Neurosci., 2018, 19, 610–621 CrossRef CAS PubMed.
- S. S. Shaftel, W. S. T. Griffin and K. M. Kerry, J. Neuroinflammation, 2008, 5, 7 CrossRef PubMed.
- R. Cacabelos, X. A. Alvarez, L. Fernandez-Novoa, A. Franco, R. Mangues, A. Pellicer and T. Nishimura, Methods Find. Exp. Clin. Pharmacol., 1994, 16, 141–151 CAS.
- L. Xie, C. Lockhart, S. R. Bowers, D. K. Klimov and M. S. Jafri, Biomolecules, 2025, 15, 89 CrossRef CAS PubMed.
- S. Tomaselli, V. Esposito, P. Vangone, N. A. J. Van Nuland, A. M. J. J. Bonvin, R. Guerrini, T. Tancredi, P. A. Temussi and D. Picone, ChemBioChem, 2006, 7, 257–267 CrossRef CAS PubMed.
- C. Nerelius, A. Sandegren, H. Sargsyan, R. Raunak, H. Leijonmarck, U. Chatterjee, A. Fisahn, S. Imarisio, D. A. Lomas, D. C. Crowther, R. Strö Mberg and J. Johansson, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 9191–9196 CrossRef CAS.
- A. Kaur and B. Goyal, J. Mol. Graphics Modell., 2023, 124, 108558 CrossRef CAS PubMed.
- Y. Jiang, X. Jiang, X. Shi, F. Yang, Y. Cao, X. Qin, Z. Hou, M. Xie, N. Liu, Q. Fang, F. Yin, W. Han and Z. Li, iScience, 2019, 17, 87–100 CrossRef CAS PubMed.
- E. Khalili Samani, M. R. Mofid and M. Malakoutikhah, J. Pept. Sci., 2020, 26, e3227 CrossRef CAS PubMed.
- D. Gheidari, M. Mehrdad and Z. Karimelahi, Sci. Rep., 2024, 14, 26431 CrossRef CAS PubMed.
- N. M. Abdelazeem, W. M. Aboulthana, A. S. Hassan, A. A. Almehizia, A. M. Naglah and H. M. Alkahtani, Saudi Pharm. J., 2024, 32, 102025 CrossRef CAS PubMed.
- B. Muthuraj, S. Layek, S. N. Balaji, V. Trivedi and P. K. Iyer, ACS Chem. Neurosci., 2015, 6, 1880–1891 CrossRef CAS PubMed.
- S. Choudhury and A. K. Dasmahapatra, Mol. Diversity, 2025, 29, 2445–2461 CrossRef CAS PubMed.
- S. S. Pawar and S. H. Rohane, Asian J. Res. Chem., 2021, 14, 1–3 CrossRef.
- J. Huang and A. D. Mackerell, J. Comput. Chem., 2013, 34, 2135–2145 CrossRef CAS PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov and A. D. Mackerell, J. Comput. Chem., 2010, 31, 671–690 CrossRef CAS PubMed.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- A. Salimi, H. Li and J. Y. Lee, Int. J. Biol. Macromol., 2021, 184, 887–897 CrossRef CAS PubMed.
- J. H. Zhao, H. L. Liu, Y. F. Liu, H. Y. Lin, H. W. Fang, Y. Ho and W. B. Tsai, J. Biomol. Struct. Dyn., 2009, 26, 481–490 CrossRef CAS PubMed.
- F. Maghsoodi, T. D. Martin and E. Y. Chi, ACS Omega, 2023, 8, 10148–10159 CrossRef CAS PubMed.
- H. Saito, H. Nagao, K. Nishikawa and K. Kinugawa, J. Chem. Phys., 2003, 119, 953–963 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- G. Wei, N. Mousseau and P. Derreumaux, Prion, 2007, 1, 3 CrossRef PubMed.
- A. Tiwari, R. Singh, S. Kumar, A. Sunkaria and A. Jain, ACS Chem. Neurosci., 2025, 16, 500–512 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2026 |
Click here to see how this site uses Cookies. View our privacy policy here.  ,
Parijat
Swain
,
Srimanta
Patra
,
Praveen Reddy
Bodhe
,
Neeraj
Kulkarni
,
Parijat
Swain
,
Srimanta
Patra
,
Praveen Reddy
Bodhe
,
Neeraj
Kulkarni
 and
Bichismita
Sahu
and
Bichismita
Sahu
 *
*
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, respectively). The samples were treated with ThT and fluorescence at 450 nm excitation and 485 nm emission wavelengths was recorded. The percent inhibition of aggregation was calculated in comparison to the control. According to preliminary screening, out of 13 molecules (Fig. S8), compounds 1a, 2a, 2f, and 2r showed anti-aggregation properties. These selected compounds were tested further in triplicate and compared with the standard amyloid β aggregation inhibitor curcumin, Fig. 2-A2. In the presence of compound 2f, the Cu2+ induced Aβ peptide aggregation was significantly reduced (IC50 value 16.8 ± 1.8 μM) (SI S139). Furthermore, a time-dependent ThT fluorescence assay was conducted to assess the effect of compound 2f on the growth kinetics of Aβ aggregation from monomeric to aggregated species.55 The fibrillation of Aβ exhibited a characteristic sigmoidal growth curve, encompassing an initial nucleation phase, a subsequent rapid growth phase, and a final saturation phase, reached after approximately 20 hours of incubation. In the absence of compound 2f, a substantial increase in ThT fluorescence intensity was observed for the untreated Aβ peptide, indicating significant aggregation. In contrast, the presence of 2f significantly suppressed the aggregation of Aβ, as evidenced by the reduced ThT fluorescence. Fig. 2-A3 shows that 2f inhibits Aβ42 aggregation with an efficiency of 52.73%, drastically reducing the growth phase after 4 h of incubation. Hence, compound 2f was further used for subsequent cellular studies.
2 ratio, respectively). The samples were treated with ThT and fluorescence at 450 nm excitation and 485 nm emission wavelengths was recorded. The percent inhibition of aggregation was calculated in comparison to the control. According to preliminary screening, out of 13 molecules (Fig. S8), compounds 1a, 2a, 2f, and 2r showed anti-aggregation properties. These selected compounds were tested further in triplicate and compared with the standard amyloid β aggregation inhibitor curcumin, Fig. 2-A2. In the presence of compound 2f, the Cu2+ induced Aβ peptide aggregation was significantly reduced (IC50 value 16.8 ± 1.8 μM) (SI S139). Furthermore, a time-dependent ThT fluorescence assay was conducted to assess the effect of compound 2f on the growth kinetics of Aβ aggregation from monomeric to aggregated species.55 The fibrillation of Aβ exhibited a characteristic sigmoidal growth curve, encompassing an initial nucleation phase, a subsequent rapid growth phase, and a final saturation phase, reached after approximately 20 hours of incubation. In the absence of compound 2f, a substantial increase in ThT fluorescence intensity was observed for the untreated Aβ peptide, indicating significant aggregation. In contrast, the presence of 2f significantly suppressed the aggregation of Aβ, as evidenced by the reduced ThT fluorescence. Fig. 2-A3 shows that 2f inhibits Aβ42 aggregation with an efficiency of 52.73%, drastically reducing the growth phase after 4 h of incubation. Hence, compound 2f was further used for subsequent cellular studies.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and 1
3 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio with CuSO4 and ZnSO4 at a concentration of 50 μM for 30 minutes at room temperature in MeOH. Hyperchromic shift was observed for interaction of 2f with Cu2+, whereas hypochromic shift was observed for the interaction with Zn2+. A small change in absorbance observed for Fe3+ and 2f interaction suggesting limited interaction between 2f and Fe3+.
5 ratio with CuSO4 and ZnSO4 at a concentration of 50 μM for 30 minutes at room temperature in MeOH. Hyperchromic shift was observed for interaction of 2f with Cu2+, whereas hypochromic shift was observed for the interaction with Zn2+. A small change in absorbance observed for Fe3+ and 2f interaction suggesting limited interaction between 2f and Fe3+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded into each 96 well plate and cells were allowed to adhere. After 24 h, the cells were then treated with 50 μM of the synthesized compound (dissolved in DMSO and diluted in DMEM media) for the next 24 h. Thereafter, the entire medium was removed and 10% volume of Alamar blue reagent was then added (from stock solution of 1 mg ml−1) and incubated for an additional 4 h at 37 °C. The fluorescence measurements were performed at an excitation wavelength of 560 nm and emission wavelength of 590 nm using a Varioskan Lux multiplate reader (Thermo Fischer). For the positive control, cells were treated with 1.66% v/v DMSO, cultured in DMEM media only, and considered as 100% viable, and the cell viability was calculated based on this.
000 cells were seeded into each 96 well plate and cells were allowed to adhere. After 24 h, the cells were then treated with 50 μM of the synthesized compound (dissolved in DMSO and diluted in DMEM media) for the next 24 h. Thereafter, the entire medium was removed and 10% volume of Alamar blue reagent was then added (from stock solution of 1 mg ml−1) and incubated for an additional 4 h at 37 °C. The fluorescence measurements were performed at an excitation wavelength of 560 nm and emission wavelength of 590 nm using a Varioskan Lux multiplate reader (Thermo Fischer). For the positive control, cells were treated with 1.66% v/v DMSO, cultured in DMEM media only, and considered as 100% viable, and the cell viability was calculated based on this.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) cells were cultured in chamber slides followed by treatment with Aβ42 + Cu2+ and 2f (24 h). Post treatment, the cells were fixed with 4% PFA (15 min at RT). Then, the cells were blocked with immunofluorescence blocking buffer (5% BSA in PBS and 0.3% Triton-X). Further, the cells were incubated with primary antibodies: NLRP3 (MA5-52041, Sigma-Aldrich; 1
000) cells were cultured in chamber slides followed by treatment with Aβ42 + Cu2+ and 2f (24 h). Post treatment, the cells were fixed with 4% PFA (15 min at RT). Then, the cells were blocked with immunofluorescence blocking buffer (5% BSA in PBS and 0.3% Triton-X). Further, the cells were incubated with primary antibodies: NLRP3 (MA5-52041, Sigma-Aldrich; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) and IL-1β (P420B, Sigma-Aldrich; 1
250) and IL-1β (P420B, Sigma-Aldrich; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) at 4 °C overnight followed by secondary antibody anti-rabbit Alexa Fluor 488 (1
250) at 4 °C overnight followed by secondary antibody anti-rabbit Alexa Fluor 488 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, A11008, Abcam) and goat anti-rat Alexa Fluor 647 (A-21434, Thermo Fisher Scientific; 1
500, A11008, Abcam) and goat anti-rat Alexa Fluor 647 (A-21434, Thermo Fisher Scientific; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) incubation (4–6 h at RT). Next, the cells were washed with PBS 3 times. DAPI was used as a counterstain before visualizing in a confocal microscope (SP8 Leica Light Microscope).
500) incubation (4–6 h at RT). Next, the cells were washed with PBS 3 times. DAPI was used as a counterstain before visualizing in a confocal microscope (SP8 Leica Light Microscope).






