 Open Access Article
Open Access ArticleAn overlooked cyclase plays a central role in the biosynthesis of indole diterpenes†
Rosannah C.
Cameron
 ,
Daniel
Berry
,
Daniel
Berry
 ,
Alistair T.
Richardson
,
Alistair T.
Richardson
 ,
Luke J.
Stevenson
,
Luke J.
Stevenson
 ,
Yonathan
Lukito
,
Yonathan
Lukito
 ,
Kelly A.
Styles
,
Kelly A.
Styles
 ,
Natasha S. L.
Nipper
,
Natasha S. L.
Nipper
 ,
Rose M.
McLellan
,
Rose M.
McLellan
 and
Emily J.
Parker
and
Emily J.
Parker
 *
*
Ferrier Research Institute, Victoria University of Wellington, Maurice Wilkins Centre for Molecular Biodiscovery, Wellington 6012, New Zealand. E-mail: emily.parker@vuw.ac.nz
First published on 25th April 2025
Abstract
Indole diterpenes (IDTs) are a large class of highly complex fungal natural products that possess a wide array of intriguing bioactivities. While IDTs are structurally diverse, the first four steps of IDT biosynthesis are highly conserved and result typically in the formation of a tetrahydropyran (THP)-ring containing structure, most commonly paspaline. The biosynthetic genes responsible for these steps are the most extensively studied of all IDT genes and collectively define the core biosynthetic pathway. Here we show that the fourth fundamental step, formation of the THP ring, is catalysed by a terpene cyclase encoded by an overlooked and uncharacterised fifth gene, idtA. All previously delineated biosynthetic routes have incorrectly attributed this step to the terpene cyclase IdtB, leading to imprecise pathway reconstructions and ignoring the fully evolved biosynthetic solution for core IDT generation. Moreover, while IdtA terpene cyclases are found in Eurotiomycetes fungi, in Sordariomycetes fungi this step is catalysed by the unrelated protein IdtS, demonstrating that two distinct solutions to this chemistry exist. All biosynthetic gene clusters known to specify production of THP-containing IDTs include an idtA or idtS gene. These findings reset the paradigm for core IDT biosynthesis and support accurate heterologous biosynthesis of these complex natural products.
Introduction
Indole diterpenes (IDTs) are a complex and structurally diverse class of filamentous fungal natural products that are comprised of indole connected to a variably cyclised diterpenoid moiety.1–3 Additional tailoring steps on the core scaffold then massively amplify the structural diversity and associated bioactivity of IDTs. Several hundred IDTs have been identified to date, the majority of which possess a hexacyclic paspaline-type IDT core, which includes a tetrahydropyran (THP) ring derived from the diterpenoid moiety (Fig. 1).2 These THP-containing IDTs include the archetypical IDT paxilline, as well as the terpendoles, lolitrems, penitrems, shearinines, janthitrems and aflatrems.4–13 There are only a few known examples that lack this THP ring, such as the nodulisporic acids (Fig. 1A).14 Here we demonstrate that, contrary to all previously published biosynthetic routes that proposed THP ring formation was catalysed by IdtB-type terpene cyclases, this chemistry is delivered by a discrete cluster-encoded cyclase.2,15–23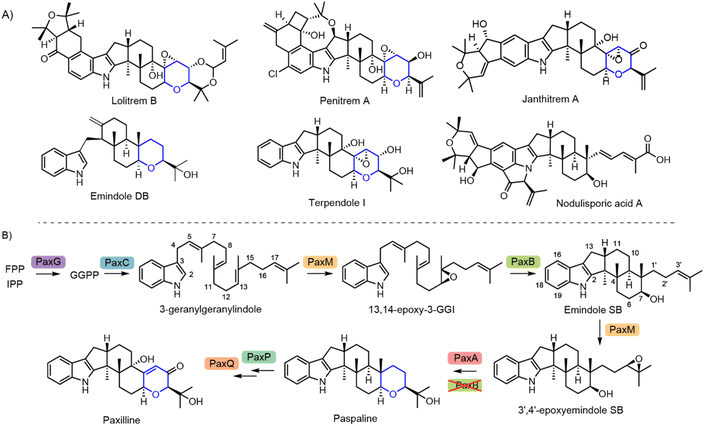 | ||
| Fig. 1 (A) Representative IDTs with the THP ring shown in blue. (B) The biosynthetic pathway for paxilline. | ||
The first IDT biosynthetic gene cluster (BGC), which specifies the production of the paspaline-type IDT paxilline was identified in 2001 from Penicillium paxilli.24 Through a series of gene deletion, complementation, and heterologous reconstruction experiments four genes from this PAX BGC (paxG, paxC, paxM and paxB) were identified as being sufficient to deliver paspaline (Fig. 1B).20,22,25 Paspaline biosynthesis is initiated with the formation of geranylgeranyl pyrophosphate (GGPP) catalysed by the cluster-encoded GGPP synthase, PaxG. An indole prenyl transferase (PaxC) then catalyses the first dedicated step of IDT biosynthesis, producing 3′-geranylgeranylindole (3′-GGI).
Construction of hexacyclic paspaline from this acyclic precursor then requires iterative cycles of activation and cyclisation. In the first of these, the third olefinic moiety of the diterpene tail is epoxidised by the flavin-dependent monooxygenase PaxM. A precise regio- and stereo-specific cyclisation cascade is then catalysed by the unusual integral membrane type II terpene cyclase PaxB, producing the first stable cyclised IDT, emindole SB.1,26,27 Generation of the THP ring of paspaline is then primed through a second PaxM-catalysed epoxidation on the terminal terpene olefin of emindole SB. A subsequent cyclisation generates the THP ring, a reaction which was previously also attributed PaxB, or the equivalent IdtBs in BGCs from other fungi.15–17,19,20,22,23,25,28,29 Two P450 monooxygenases, PaxP and PaxQ, then convert paspaline to paxilline.20–22 Three additional genes are also present in the paxilline BGC: paxD and paxO encode enzymes that further decorate the indole moiety of paxilline—although these modifications that are not generally observed in P. paxilli—and a third gene, paxA, encoding a predicted helical transmembrane protein of unknown function. Intriguingly, orthologues (idtAs) of paxA are also found in other BGCs known to specify THP-containing IDTs (Fig. 2).
 | ||
| Fig. 2 IDT biosynthetic gene clusters from the Eurotiomycetes fungi P. paxilli (PAX), Penicillium janthinellum (JAN), Penicillium crustosum (PTM), Aspergillus desertorum (DES) showing idtA homologues in pink. The BCGs from Sordariomycetes fungi Epichloe festucae (LTM) and Tolypocladium album (TER) contain an alternative gene, idtS (shown in yellow). Letters above the PAX cluster correspond to the gene name and function of the encoded enzyme – as shown in Fig. 1B. This figure was generated using clinker.30 . | ||
Results and discussion
To investigate the function of PaxA we created paxA deletion strains (ΔpaxA) in P. paxilli (Fig. S1 and ESI 1.2†). In contrast to wild type, ΔpaxA strains were attenuated for the production of paxilline (Fig. 3, S5 and ESI 1.2.3†) and accumulated an IDT not observed in wild type (Fig. 3A and S4†). This compound, with a molecular weight of 421 Da, was isolated and characterized as 3′,4′-epoxyemindole SB (ESI 1.3†), the proposed substrate for THP ring formation (Fig. 1B).20Reintroduction of paxA into ΔpaxA strains complemented this chemotype, attenuating the 3′,4′-epoxyemindole SB peak and restoring paxilline biosynthesis (Fig. 3A, S6, S7 and ESI 1.2.4†). In a parallel study, we used a biomimetic approach to construct synthetic BGCs containing paxG, paxC, paxM, paxB, and optionally paxA, which were then transformed into the ΔPAX locus of a P. paxilli strain in which the entire PAX BGC had been deleted. Consistent with our hypothesis that PaxA catalyses THP ring formation, the synthetic BGC containing paxA conferred the ability to synthesise paspaline without appreciable accumulation of 3′,4′-epoxyemindole SB. Whereas the strains transformed with the synthetic BGC lacking paxA accumulated the epoxidised intermediate (Fig. S8, S9 and ESI 1.2.6†).
To interrogate directly the catalytic role of PaxA we set up a series of feeding experiments using ΔPAX strains into which paxM alone, paxM + paxA, or paxM + paxB had been reintroduced. These strains were fed by the addition of purified emindole SB. Whereas strains expressing paxM + paxA efficiently converted this to paspaline, the strains expressing paxM alone or paxM + paxB accumulated 3′,4′-epoxyemindole SB (Fig. 4, S15 and ESI 1.4†). Combined, these results demonstrate that PaxA catalyses the conversion of 3′,4′-epoxyemindole SB to paspaline in vivo. The terpene cyclase PaxB, which was previously assigned this role, does not contribute to THP ring formation.20,22
The structural model for PaxA reveals that this protein is a helical integral membrane protein reminiscent of the terpene cyclase PaxB, but with no clear structural or sequence relationship (Fig. 5 and S16–S18†). Interestingly however, PaxA bears similarity to other meroterpenoid cyclases that specifically catalyse the formation of THP or tetrahydrofuranyl (THF) rings.31–33 Notably in Acremonium egyptiacum, which generates meroterpenoids ascofuranone and ascochlorin, both IdtA- and IdtB-like terpene cyclases are encoded.31 However, rather than operating sequentially as they do in paspaline biosynthesis, in A. egyptiacum they operate to bifurcate the pathway to the two alternative meroterpenoid outcomes. Our modelling of 3′,4′-epoxyemindole SB into PaxA reveals a likely active site nestled between helices at the C-terminal end of the helical bundle (Fig. 5A). While the putative binding site is lined by hydrophobic residues as expected to support the required conformation for reaction, an aspartate residue is located appropriately to promote THP formation through a favoured 6-exo-tet cyclisation (Fig. 5B and C). Accordingly, we found that substituting Asp279 for Ala (D279A) also generated a 3′,4′-epoxyemindole SB accumulating chemotype (Fig. 5D, S19 and ESI 1.5†). An equivalent Asp residue has been shown to be important for the ascofuranone pathway THF-forming biosynthetic cyclase AscI.31 We note that Cys66, which is conserved across all confirmed IdtA cyclases (Fig. S18†), resides ∼4 Å from the hydroxyl group of 3′,4′-epoxyemindole SB and may play a role in proton loss in THP formation.
 | ||
| Fig. 5 Structural model of PaxA (generated by AlphaFold34), with substrate 3′,4′-epoxyemindole SB (cyan). (A) PaxA is a helical integral membrane protein, 3′,4′-epoxyemindole SB shown as spheres (B) Close up of predicted binding site showing Asp279. (C) Acid catalysed THP formation. (D) EIC traces showing IDT production at m/z 422 for two Asp to Ala substitution mutants (D279A). Maximum ion intensity counts are shown on the y-axis. | ||
It was notable that some paspaline formation was detected in cultures of ΔpaxA strains (Fig. 3, S4 and ESI 1.2.3†), in the paxGCMB heterologous reconstruction strains that lacked paxA (Fig. S8 and ESI 1.2.6†), and in feeding studies where PaxM was present in the absence of PaxA (Fig. 4 and S15†). These observations indicate that spontaneous conversion of 3′,4′-epoxyemindole SB to paspaline is occurring either in vivo or during chemical extraction. The stereochemical outcome in this reaction is set by the prior PaxM-catalysed epoxidation step, and 6-exo-tet THP formation is the expected reaction outcome. Indeed, we have observed this conversion directly in vitro, with isolated samples of 3′,4′-epoxyemindole SB slowly converting to paspaline, and as expected, we show that this reaction is accelerated by the addition of weak acid (Fig. S20 and ESI 1.6†). This observed PaxA-independent formation of paspaline (and correspondingly paxilline in strains containing paxP and paxQ) explains why previous investigations have overlooked the role of PaxA.15–23 Similarly, the equivalent non-enzymatic formation of the THF ring for the epoxy intermediate in ascofuranone biosynthesis was also observed.31
As noted, we identified orthologues (idtAs) of paxA in BGCs from Eurotiomycetes fungi that are known to biosynthesise similar THP-containing IDTs (Fig. 2, S16 and S17†). Accordingly, we tested the function of JanA and PtmA, encoded within the JAN and PTM clusters that specify the paspaline-derived janthitremane and penitrem IDTs, respectively. Introduction of the janA or ptmA genes into one of our ΔpaxA strains eliminated 3′,4′-epoxyemindole SB accumulation and restored paxilline production (Fig. 3B, S21–24 and ESI 1.7†), demonstrating that JanA and PtmA are indeed functional orthologues of PaxA. Additionally, we showed that desA, which is a paxA homologue located within the emindole DA-specifying DES cluster from Aspergillus desertorum, also complemented these chemotypes when added to a ΔpaxA strain (Fig. 3, S25, S26 and ESI 1.7†).15 This is notable as while emindole DB, like paspaline, contains a THP ring (Fig. 1A), this product likely arises from the epoxidation of emindole DA and its subsequent cyclisation catalysed by DesA, implying a wider repertoire of chemistry for the IdtA cyclases solely than the formation of paspaline.15 This finding implies that some substrate promiscuity is demonstrated by DesA.
Our attention then turned to the IDT BGCs from Sordariomycete fungi, which do not include paxA homologues (Fig. 2). For some this was to be anticipated; the nodulisporic acids do not contain a THP ring, being derived directly from emindole SB rather than via paspaline, so a paxA homologue is not required.23,35,36 However, several of these BGCs are known to specify paspaline-derived IDTs, yet do not contain an idtA gene. Notably, these include the LTM cluster from Epichloë festucae and the TER cluster from Tolypocladium album, which are respectively responsible for delivering the lolitrems and the terpendoles.17,37 We noted that these clusters include the homologous uncharacterised genes ltmS and terS, which encode proteins with a predicted helical transmembrane topology that is similar to PaxA despite lacking any significant sequence homology (Fig. S27 and ESI 1.8†). Introduction of the ltmS gene into a paxA deletion strain restored efficient conversion of 3′,4′-epoxyemindole SB to paspaline and production of paxilline, demonstrating that LtmS is functionally equivalent to PaxA (Fig. 3, S28, S29 and ESI 1.8†). Consistent with this observation and supporting an important role for LtmS in lolitrem biosynthesis, E. festucae ΔltmS strains grown inside their grass hosts accumulated 3′,4′-epoxyemindole SB and showed a reduction in the production of lolitrem B (Fig. S30–33 and ESI 1.8.3†).
Collectively these results indicate clearly that these unassigned idtA and idtS genes encoded within IDT BGCs specify cyclases with key roles in IDT biosynthesis. While the previously proposed biosynthetic pathways for the paspaline and related IDTs included a dual role for the terpene cyclase IdtB in constructing both emindole SB and the full paspaline skeleton, our work here shows that this long-held paradigm is incorrect; the distinct cluster-encoded IdtA and IdtS cyclases deliver the THP functionality.15–17,19,20,22,23,25,28,29 Moreover, through IdtA and IdtS, Eurotiomycetes and Sordariomycetes fungi have evolved two seemingly independent solutions for delivering this chemistry, reflecting the importance of this catalysis for efficient IDT biosynthesis.
Data availability
The data supporting this article have been included as part of the ESI† available at [https://doi.org/10.1039/d5sc02009c].Author contributions
RCC & EJP conceived the study; RCC, EJP, DB, ATR, LJS, YL & RMM designed the experiments; NSLN provided key materials; RCC, DB, ATR, LJS, YL & KAS collected and analysed the data. RCC, ATR & EJP wrote the manuscript. EJP and RCC acquired the funding. All authors corrected the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the New Zealand Ministry of Business, Innovation, and Employment (Grant # RTVU1809). It was also supported by a Returning Carer's Research Fund grant (Grant # 400654) awarded to Rosannah Cameron by Victoria University of Wellington. Additionally, we would like to acknowledge Scott Cameron for his help with R-scripts used in this work and Miriam Hunt for technical assistance.References
- T. Ozaki, A. Minami and H. Oikawa, Biosynthesis of indole diterpenes: a reconstitution approach in a heterologous host, Nat. Prod. Rep., 2023, 40, 202–213 RSC.
- J. Niu, J. Qi, P. Wang, C. Liu and J. M. Gao, The chemical structures and biological activities of indole diterpenoids, Nat. Prod. Bioprospect., 2023, 13, 3 CrossRef CAS PubMed.
- M. Jiang, Z. Wu, L. Liu and S. Chen, The chemistry and biology of fungal meroterpenoids (2009–2019), Org. Biomol. Chem., 2021, 19, 1644–1704 RSC.
- R. T. Gallagher, J. Clardy and B. J. Wilson, Aflatrem, a tremorgenic toxin from Aspergillus flavus, Tetrahedron Lett., 1980, 21, 239–242 CrossRef CAS.
- J. P. Springer, J. Clardy, J. M. Wells, R. J. Cole and J. W. Kirksey, The structure of paxilline, a tremorgenic metabolite of Penicillium paxilli bainier, Tetrahedron Lett., 1975, 16, 2531–2534 CrossRef.
- H. Tomoda, N. Tabata, D. J. Yang, H. Takayanagi and S. Omura, Terpendoles, novel ACAT inhibitors produced by Albophoma yamanashiensis, J. Antibiot., 1995, 48, 793–804 CrossRef CAS PubMed.
- W. L. Imlach, S. C. Finch, J. Dunlop and J. E. Dalziel, Structural determinants of lolitrems for inhibition of BK large conductance Ca2+-activated K+ channels, Eur. J. Pharmacol., 2009, 605, 36–45 CrossRef CAS PubMed.
- T. Yamaguchi, K. Nozawa, T. Hosoe, S. Nakajima and K.-i. Kawai, Indoloditerpenes related to tremorgenic mycotoxins, penitrems, from Penicillium crustosum, Phytochemistry, 1993, 32, 1177–1181 CrossRef CAS.
- A. E. de Jesus, P. S. Steyn, F. R. van Heerden and R. Vleggaar, Structure elucidation of the janthitrems, novel tremorgenic mycotoxins from Penicillium janthinellum, J. Chem. Soc., Perkin Trans. 1, 1984, 697–701, 10.1039/P19840000697.
- A. L. Wilkins, C. O. Miles, R. M. Ede, R. T. Gallagher and S. C. Munday, Structure elucidation of janthitrem B, a tremorgenic metabolite of Penicillium janthinellum, and relative configuration of the A and B rings of janthitrems B, E, and F, J. Agric. Food Chem., 1992, 40, 1307–1309 CrossRef CAS.
- J. Penn, R. Swift, L. J. Wigley, P. G. Mantle, J. N. Bilton and R. N. Sheppard, Janthitrems B and C, two principal indole-diterpenoids produced by Penicillium janthinellum, Phytochemistry, 1993, 32, 1431–1434 CrossRef CAS.
- R. T. Gallagher, J. Finer, J. Clardy, A. Leutwiler, F. Weibel, W. Acklin and D. Arigoni, Paspalinine, a tremorgenic metabolite from Claviceps paspali Stevens et Hall, Tetrahedron Lett., 1980, 21, 235–238 CrossRef CAS.
- G. N. Belofsky, J. B. Gloer, D. T. Wicklow and P. F. Dowd, Antiinsectan alkaloids: Shearinines A–C and a new paxilline derivative from the ascostromata of Eupenicillium shearii, Tetrahedron, 1995, 51, 3959–3968 CrossRef CAS.
- J. G. Ondeyka, G. L. Helms, O. D. Hensens, M. A. Goetz, D. L. Zink, A. Tsipouras, W. L. Shoop, L. Slayton, A. W. Dombrowski, J. D. Polishook, D. A. Ostlind, N. N. Tsou, R. G. Ball and S. B. Singh, Nodulisporic acid A, a novel and potent insecticide from a Nodulisporium sp. isolation, structure determination, and chemical transformations, J. Am. Chem. Soc., 1997, 119, 8809–8816 CrossRef CAS.
- R. Bundela, R. C. Cameron, A. J. Singh, R. M. McLellan, A. T. Richardson, D. Berry, M. J. Nicholson and E. J. Parker, Generation of alternate indole diterpene architectures in two species of Aspergilli, J. Am. Chem. Soc., 2023, 145, 2754–2758 CrossRef CAS PubMed.
- R. M. McLellan, R. C. Cameron, M. J. Nicholson and E. J. Parker, Aminoacylation of indole diterpenes by cluster-specific monomodular NRPS-like enzymes, Org. Lett., 2022, 24, 2332–2337 CrossRef CAS PubMed.
- Y. Jiang, T. Ozaki, M. Harada, T. Miyasaka, H. Sato, K. Miyamoto, J. Kanazawa, C. Liu, J. I. Maruyama, M. Adachi, A. Nakazaki, T. Nishikawa, M. Uchiyama, A. Minami and H. Oikawa, Biosynthesis of indole diterpene lolitrems: Radical-induced cyclization of an epoxyalcohol affording a characteristic lolitremane skeleton, Angew. Chem., Int. Ed., 2020, 59, 17996–18002 CrossRef CAS PubMed.
- M.-C. Tang, H.-C. Lin, D. Li, Y. Zou, J. Li, W. Xu, R. A. Cacho, M. E. Hillenmeyer, N. K. Garg and Y. Tang, Discovery of unclustered fungal indole diterpene biosynthetic pathways through combinatorial pathway reassembly in engineered yeast, J. Am. Chem. Soc., 2015, 137, 13724–13727 CrossRef CAS PubMed.
- C. Liu, K. Tagami, A. Minami, T. Matsumoto, J. C. Frisvad, H. Suzuki, J. Ishikawa, K. Gomi and H. Oikawa, Reconstitution of biosynthetic machinery for the synthesis of the highly elaborated indole diterpene penitrem, Angew. Chem., Int. Ed., 2015, 54, 5748–5752 CrossRef CAS PubMed.
- K. Tagami, C. Liu, A. Minami, M. Noike, T. Isaka, S. Fueki, Y. Shichijo, H. Toshima, K. Gomi, T. Dairi and H. Oikawa, Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae, J. Am. Chem. Soc., 2013, 135, 1260–1263 CrossRef CAS PubMed.
- S. Saikia, E. J. Parker, A. Koulman and B. Scott, Defining paxilline biosynthesis in Penicillium paxilli: functional characterization of two cytochrome P450 monooxygenases, J. Biol. Chem., 2007, 282, 16829–16837 CrossRef CAS PubMed.
- S. Saikia, E. J. Parker, A. Koulman and B. Scott, Four gene products are required for the fungal synthesis of the indole-diterpene, paspaline, FEBS Lett., 2006, 580, 1625–1630 CrossRef CAS PubMed.
- T. R. Hibbard, R. M. McLellan, L. J. Stevenson, A. T. Richardson, M. J. Nicholson and E. J. Parker, Functional crosstalk between discrete indole terpenoid gene clusters in Tolypocladium album, Org. Lett., 2023, 25, 7470–7475 CrossRef CAS PubMed.
- C. Young, L. McMillan, E. Telfer and B. Scott, Molecular cloning and genetic analysis of an indole-diterpene gene cluster from Penicillium paxilli, Mol. Microbiol., 2001, 39, 754–764 CrossRef CAS PubMed.
- B. Scott, C. A. Young, S. Saikia, L. K. McMillan, B. J. Monahan, A. Koulman, J. Astin, C. J. Eaton, A. Bryant, R. E. Wrenn, S. C. Finch, B. A. Tapper, E. J. Parker and G. B. Jameson, Deletion and gene expression analyses define the paxilline biosynthetic gene cluster in Penicillium paxilli, Toxins, 2013, 5, 1422–1446 CrossRef CAS PubMed.
- L. Barra and I. Abe, Chemistry of fungal meroterpenoid cyclases, Nat. Prod. Rep., 2021, 38, 566–585 RSC.
- M. Baunach, J. Franke and C. Hertweck, Terpenoid biosynthesis off the beaten track: unconventional cyclases and their impact on biomimetic synthesis, Angew. Chem., Int. Ed., 2015, 54, 2604–2626 CrossRef CAS PubMed.
- M. J. Nicholson, C. J. Eaton, C. Starkel, B. A. Tapper, M. P. Cox and B. Scott, Molecular cloning and functional analysis of gene clusters for the biosynthesis of indole-diterpenes in Penicillium crustosum and P. janthinellum, Toxins, 2015, 7, 2701–2722 CrossRef CAS PubMed.
- E. J. Ludlow, S. Vassiliadis, P. N. Ekanayake, I. K. Hettiarachchige, P. Reddy, T. I. Sawbridge, S. J. Rochfort, G. C. Spangenberg and K. M. Guthridge, Analysis of the indole diterpene gene cluster for biosynthesis of the epoxy-janthitrems in Epichloe endophytes, Microorganisms, 2019, 7, 560–582 CrossRef CAS PubMed.
- C. L. M. Gilchrist and Y.-H. Chooi, clinker & clustermap.js: automatic generation of gene cluster comparison figures, Bioinformatics, 2020, 37(16), 2473–2475 CrossRef PubMed.
- Y. Araki, T. Awakawa, M. Matsuzaki, R. Cho, Y. Matsuda, S. Hoshino, Y. Shinohara, M. Yamamoto, Y. Kido, D. K. Inaoka, K. Nagamune, K. Ito, I. Abe and K. Kita, Complete biosynthetic pathways of ascofuranone and ascochlorin in Acremonium egyptiacum, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 8269–8274 CrossRef CAS PubMed.
- X. M. Mao, Z. J. Zhan, M. N. Grayson, M. C. Tang, W. Xu, Y. Q. Li, W. B. Yin, H. C. Lin, Y. H. Chooi, K. N. Houk and Y. Tang, Efficient biosynthesis of fungal polyketides containing the dioxabicyclo-octane ring system, J. Am. Chem. Soc., 2015, 137, 11904–11907 CrossRef CAS PubMed.
- T. S. Lin, Y. M. Chiang and C. C. Wang, Biosynthetic pathway of the reduced polyketide product citreoviridin in Aspergillus terreus var. aureus revealed by heterologous expression in Aspergillus nidulans, Org. Lett., 2016, 18, 1366–1369 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- K. C. Van de Bittner, M. J. Nicholson, L. Y. Bustamante, S. A. Kessans, A. Ram, C. J. van Dolleweerd, B. Scott and E. J. Parker, Heterologous biosynthesis of nodulisporic acid F, J. Am. Chem. Soc., 2018, 140, 582–585 CrossRef CAS PubMed.
- A. T. Richardson, R. C. Cameron, L. J. Stevenson, A. J. Singh, Y. Lukito, D. Berry, M. J. Nicholson and E. J. Parker, Biosynthesis of nodulisporic acids: A multifunctional monooxygenase delivers a complex and highly branched array, Angew. Chem., Int. Ed., 2022, 61, e202213364 CrossRef CAS PubMed.
- T. Motoyama, T. Hayashi, H. Hirota, M. Ueki and H. Osada, Terpendole E, a kinesin Eg5 inhibitor, is a key biosynthetic intermediate of indole-diterpenes in the producing fungus Chaunopycnis alba, Chem. Biol., 2012, 19, 1611–1619 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d5sc02009c |
| This journal is © The Royal Society of Chemistry 2025 |


