Open Access Article
Open Access ArticleChemodynamic covalent adaptable network-induced robust, self-healing, and degradable fluorescent elastomers for multicolor information encryption†
Changyang
Li
ab,
Xing
Su
*ab,
Chuanbao
Cao
 ab,
Xiaodong
Li
b and
Meishuai
Zou
ab,
Xiaodong
Li
b and
Meishuai
Zou
 *ab
*ab
aAdvanced Technology Research Institute (Jinan), Beijing Institute of Technology, Jinan, 250300, China. E-mail: sx1020@126.com; zoums@bit.edu.cn
bSchool of Materials Science and Engineering, Beijing Institute of Technology, No. 5 South Zhongguancun Street, Haidian District, Beijing, 100081, China
First published on 30th December 2024
Abstract
Elastomers are of great significance in developing smart materials for information encryption, and their unique self-healing and highly flexible properties provide innovative solutions to enhance security and anti-counterfeiting effectiveness. However, challenges remain in the multifunctional combination of mechanical properties, self-healing, degradability, and luminescence of these materials. Herein, a chemodynamic covalent adaptable network (CCAN)-induced robust, self-healing, and degradable fluorescent elastomer is proposed. Thanks to the CCANs, the resulting elastomer exhibits a tensile strength of 33.44 MPa (300 times higher than that of a linear elastomer) and an elongation at break of 1265%, and its mechanical properties can be restored to about 20 MPa after 72 h of healing at room temperature, and a self-healing efficiency of 94.67% can be realized for 24 h at 70 °C. Simultaneously, the dynamic chemical balance of keto and enol structural transitions of curcumin chain segments can be driven by CCANs, realizing multi-color (from yellow to violet) display and broad wavelength (300–500 nm) excitation, which in turn enables surface read-write and color rosette and QR code pattern printing. In addition, it can also achieve adaptive degradation under biological, alkaline, and hot water conditions. This work has guiding significance for developing the next generation of high-performance multifunctional elastomer materials, which have potential applications in the field of smart anti-counterfeiting materials and smart flexible optoelectronics.
1 Introduction
In the 21st century, with the rapid development of human society, the speed and convenience of information transmission have been greatly improved, but this is also accompanied by the great risk of information leakage.1–4 Therefore, information encryption has become particularly important. So far, various functional materials, such as fluorescent/phosphorescent/quantum dot materials,5–8 structural color materials,9,10 liquid crystal elastomers,11,12 wrinkled photonic elastomers,13etc., have been developed as information encryption materials. Among them, elastomers have attracted much attention due to their excellent deformation and load-bearing capacity. To date, a variety of fluorescent materials based on elastomers have been developed, such as stretchable and highly mechanically robust intrinsic fluorescent elastomers,14 damage-detectable fluorescent elastomers,15,16 and high-strength repairable fluorescent elastomers.3,6 Furthermore, a series of colored encrypted materials have been developed by relying on doping luminescent materials,5,17 changing excitation light,7,18 and applying external stimuli (e.g., mechanical,19,20 thermal,21 and electrical22). Multi-color encryption significantly increases the technical difficulty of counterfeiting through color combinations and variations. Compared to single-color anti-counterfeiting technologies, multi-color schemes require more complex chemical and optical matching, posing higher technical barriers to counterfeiters. Meanwhile, elastomer-based multi-color encryption materials can combine self-healing and multi-color luminescent properties, so that even if the surface of the material is damaged, the original anti-counterfeiting function can still be maintained after repair and the service life can be extended. Although attention has been focused on the mechanical properties or self-healing properties of fluorescent elastomers to improve their stability and durability, these devices tend to have relatively short service intervals, and their post-processing has become a non-negligible problem. If non-degradable, a large amount of polymer waste will accumulate, which will cause huge pollution pressure on the ecological environment and seriously affect the health of human beings and other organisms. Therefore, it is of great significance to develop multifunctional elastomers that are degradable, self-healing, high-strength, and luminescent.Covalent adaptive networks (CANs) are polymer networks with dynamically reversible covalent bonds.23–25 Such materials are characterized by a molecular structure that contains reversible bond exchanges under certain conditions (e.g., temperature, light, and pH) to drive the rearrangement of network backbone segments, endowing the material with robustness, self-healing, recyclability, shape memory, and other dynamic properties. Recently, many materials with high strength, self-healing, and recyclability have been developed based on covalent adaptive networks. Zhang et al.26 introduced dynamic thiocarbamate bonds into light-curable methacrylates to prepare reprocessable and self-healing 4D-printed polyurethanes with Young's modulus of 1.2 GPa and tensile strength of 61.9 MPa. Zhai et al.27 proposed a dynamic covalent and supramolecular design of a dynamic covalent ionomer based on lipoic acid with hierarchical dynamic bonding, in which lithium bonding contributes to ion dissociation and dynamic disulfide bond recombination, and the integration of lithium bonding and binary hydrogen bonding enhances its mechanical properties, self-healing ability, reprocessing, and recyclability. Zhang et al.28 also demonstrated a fast reprocessable and closed-loop recyclable covalent adaptive network of spiroborate-connected ions. Also, Wei et al.29 combined low molecular weight polylactic acid and lipoic acid to obtain a covalent adaptive network with strong mechanical properties and controlled degradability. Zhao et al.30 also introduced dynamic aggregation-induced luminescent molecules as both dynamic cross-linking points and fluorescent probes into the covalent adaptive liquid crystal network, which achieved simultaneous, fast, and non-destructive visualization of the cross-linking structure and driving properties of the materials, and endowed them with early warning of the driving limit.
Despite these advances, thresholds remain. These methods are less applicable for fluorescent elastomers, and there still exists a contradiction between the design concepts of mechanical and self-healing properties, which is difficult to realize to achieve a balance between the two.31,32 You et al.33 attempted to use highly dynamic four-armed crosslinking units with internal catalytic oxime-carbamate groups to obtain ionogels based on covalently adapted networks as a way to balance their self-healing ability and mechanical properties but were still limited to a small strength (tensile strength 4.55 MPa). If degradation as well as highly tunable fluorescence properties are considered simultaneously, the design of such multifunctional elastomers is a great challenge, which has been seldom reported till now.34
Based on the above-mentioned concerns, we designed a unique chemodynamic covalent adaptative network (CCAN)-induced robust, self-healing, degradable, and luminescent elastomer. Curcumin, a naturally degradable material, acts as a chain extender. And dynamically reversible B–O bonds act as a cross-linking agent. The curcumin segments in the elastomer possess special keto- and enol-type reciprocal isomers, which can be used to trigger a dynamic chemical balance for structural transitions in CCANs. The fabricated PICB1.0PU elastomer shows a tensile strength of 33.44 MPa with elongation at break of 1265.34%, its tensile strength can be restored to about 20 MPa with the assistance of an aqueous ethanol solution for 72 h at room temperature, and its repair efficiency is as high as 94.67% for 24 h at 70 °C. Furthermore, it shows reprocessability and multi-environment adaptive degradation performance. Remarkably, the dynamic chemical balance of the keto group and enol structure of curcumin at the surface interface can be triggered by CCANs, that is, adjusted by a simple ink pH, to achieve multicolor (from yellow to violet) display and broad wavelength (300–500 nm) excitation on the same carrier. Based on this, functions such as multicolor display and information encryption are easily realized on the same carrier and at 395 nm excitation wavelength. Moreover, ethanol can regulate the microenvironment of the surface interface and promote the isomerization and rearrangement of curcumin to achieve erasure. This work not only provides new ideas for the development of high-performance multifunctional elastomeric materials but also sheds light on the design and application of colorful information encryption elastomeric materials.
2 Results and discussion
2.1 Synthesis and characterization of elastomers
Adaptive chemistry combines both molecular and supramolecular chemical properties with reversible dynamic structures dominated by both non-covalent interactions and dynamic covalent bonds, which plays a unique role in molecular chain dissociation and association, favoring the functional combination of elastomers in terms of self-healing properties and degradability. The synthesis process of the CCAN-induced elastomer is illustrated in Fig. 1a. Bis-isocyanate terminal precursors are synthesized by polycondensation between polycaprolactone (PCL) polyols and isophorone diisocyanate (IPDI). Also, curcumin, a natural biodegradable biomass, is introduced into the polymer network as a chain extender to obtain linear polyurethane (PU) chains, denoted as PICPU elastomer. Subsequently, 1,4-benzenediboronic acid (BDA) is introduced into the polyurethane network to achieve dynamic reversible crosslinking of the linear PU backbone (Table S1†), denoted as PICBPU elastomer. Finally, PICBPU elastomer films are obtained by casting and drying. The molecular weight results of the different elastomers are presented in Fig. S1 and Table S2.† | ||
| Fig. 1 (a) Schematic illustration of the structures of the chemodynamic covalent adaptation network-induced elastomer. (b and c) FT-IR spectra. (d) Stress–strain curves of the different elastomers. | ||
Additionally, the surface elemental compositions of the PICBPU elastomer films are analyzed by energy dispersive X-ray spectroscopy (EDS), including C, N, O, B, etc. (Fig. S2†). In addition, the content of elemental B in the elastomer structure is determined by inductively coupled plasma optical emission spectrometer (ICP-OES) testing to be 0.35 wt%, which is close to the theoretical content (0.37 wt%). The structural compositions of PICBPU elastomers are determined by Fourier transform infrared spectroscopy (FT-IR), as shown in Fig. 1b and c. In Fig. 1b, the typical –NCO peak at 2243 cm−1 and its disappearance in the elastomer network35,36 indicate the complete consumption of the added monomers. The peaks at 1726 and 1233 cm−1 are attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O stretching vibrations in the carbamate bonds,37,38 while the bending vibration at 3367 cm−1 is attributed to the –NH groups in the carbamate bonds, proving the sequential incorporation of PCL polyols and curcumin into the main chain of the PICPU elastomer. Furthermore, the peak at 1628 cm−1 corresponds to the C
O and C–O stretching vibrations in the carbamate bonds,37,38 while the bending vibration at 3367 cm−1 is attributed to the –NH groups in the carbamate bonds, proving the sequential incorporation of PCL polyols and curcumin into the main chain of the PICPU elastomer. Furthermore, the peak at 1628 cm−1 corresponds to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak in the curcumin structures,39,40 whereas the attenuation and shift of the C
O peak in the curcumin structures,39,40 whereas the attenuation and shift of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrational peak at 1628 cm−1 is found after the addition of BDA, which is attributed to the formation of B–O bonds and B–O coordination bonds between the β-diketone structure in curcumin and BDA. The FT-IR results confirm the successful synthesis of the designed PICBPU elastomers.
O vibrational peak at 1628 cm−1 is found after the addition of BDA, which is attributed to the formation of B–O bonds and B–O coordination bonds between the β-diketone structure in curcumin and BDA. The FT-IR results confirm the successful synthesis of the designed PICBPU elastomers.
Moreover, the thermal properties of PICBPU elastomers are further tested by thermogravimetric analysis (TGA) and differential scanning calorimetry (DSC). As shown in Fig. S3,† the initial decomposition temperature of the elastomers is around 283 °C, indicating its good thermal stability. PCL is a crystallizable chain segment that exhibits a distinct melting peak at 40 °C in PICPU elastomers (Fig. S4†). With the addition of BDA, dynamic reversible B–O bonds are formed between the segments, breaking the orderly arrangement of the PCL segments and inhibiting their crystallization, as evidenced by the absence of a distinct melting peak at 40 °C in the DSC curves. With the increase of dynamic bonds, the glass transition temperature (Tg) gradually decreases, with Tg of PICB1.0PU around −40 °C, which favors the migration of chain segments and the reorganization of dynamic bonding at room temperature, which in turn affects its self-healing properties. Generally, the Tg of polymers increases with the increase of cross-linking density because the increase of cross-linking points restricts polymer molecular chain movement. However, elastomers based on B–O bonds as a reversible CCAN greatly reduce the limiting effect of the cross-linking point on the ability of molecular chain movement. In addition, materials with higher crystallinity usually have higher Tg because the crystalline structure provides stronger intermolecular interactions and rigidity, limiting the chain segment motion. Whereas the introduction of dynamic B–O bonds effectively reduces the formation of crystalline regions, as shown in the DSC curves (Fig. S4†), the degree of crystallinity of PCL is effectively reduced with the increase of B–O bonds. This means that the motility of the chain segments increases and thus the migration of chain segments can occur at lower temperatures, leading to a decrease in Tg. Overall, the B–O bonds were found to be effective in reducing the crystallinity of the elastomers due to their role as dynamic reversible cross-linking points, leading to a decrease in Tg. The dynamic mechanical properties of the different elastomers are also analyzed using a dynamic thermomechanical analyzer (DMA). The curves of storage modulus (E′), loss modulus (E′′), and loss factor (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ) at different temperatures are shown in Fig. S5.† According to the curves of storage modulus (Fig. S5a†) and DSC (Fig. S4†), the Tg of the four elastomers is between −40 and −30 °C, which is lower than room temperature, indicating that the molecular chains are in a relatively active state at room temperature with the ability to adjust the conformation and movement. This is conducive to activating the dynamic bonds in the hard chain phase, allowing the material to balance its self-healing ability and mechanical strength. Additionally, the results of the loss factor curve (Fig. S5c†) show that the loss factor of the elastomer presents a high level and a wide effective temperature range.
δ) at different temperatures are shown in Fig. S5.† According to the curves of storage modulus (Fig. S5a†) and DSC (Fig. S4†), the Tg of the four elastomers is between −40 and −30 °C, which is lower than room temperature, indicating that the molecular chains are in a relatively active state at room temperature with the ability to adjust the conformation and movement. This is conducive to activating the dynamic bonds in the hard chain phase, allowing the material to balance its self-healing ability and mechanical strength. Additionally, the results of the loss factor curve (Fig. S5c†) show that the loss factor of the elastomer presents a high level and a wide effective temperature range.
Considering the introduction of CCANs in elastomers and the formation of dynamic crosslinking points between elastomer frameworks, these elastomers may exhibit excellent mechanical properties. Accordingly, to assess the mechanical strength and toughness of the elastomers, the stress–strain curves of the elastomer are measured. The results are shown in Fig. 1d and Table S3.† The tensile strength and fracture elongation of the curcumin-extended PICPU elastomer are 0.10 MPa and 2161.42 ± 3.76%, respectively. In contrast, the mechanical properties of the PICPU elastomer are substantially improved after BDA cross-linking. The tensile strengths of PICB0.25PU, PICB0.5PU, and PICB1.0PU elastomers are 9.35 ± 0.96, 16.07 ± 0.95, and 33.44 ± 1.49 MPa, respectively, while the fracture elongations are 1983.62 ± 95.69%, 1795.29 ± 142.21%, and 1265.34 ± 43.75%, respectively. Furthermore, with the continuous increase of BDA content, there is little change in the tensile strength and fracture elongation of the elastomer, as shown in Fig. S6.† Overall, the tensile strength of PICBPU elastomers is significantly improved, with the tensile strength of PICB1.0PU elastomer being over 300 times that of the ordinary curcumin-extended PU, which is attributed to the formation of CCANs. Also, we prepared PCL-IPDI and PCL-IPDI-BDA polymer networks to serve as a control. From Fig. S7,† it can be found that the tensile strength of the PCL-IPDI sample was around 0.2 MPa, whereas it increased to around 2 MPa with the addition of BDA, which suggests that the B–OH groups in BDA may form hydrogen bonds with the carbamate groups, which in turn improves the mechanical properties of the polymers. To this end, we investigated the hydrogen bonding interactions of BDA by FT-IR spectroscopy. The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching region was deconvoluted into two subpeaks belonging to free C
O stretching region was deconvoluted into two subpeaks belonging to free C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and hydrogen-bonded C
O and hydrogen-bonded C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively. The percentage of hydrogen-bonded C
O, respectively. The percentage of hydrogen-bonded C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in PCL-IPDI was calculated to be 26.19%, whereas the percentage of hydrogen-bonded C
O in PCL-IPDI was calculated to be 26.19%, whereas the percentage of hydrogen-bonded C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in PCL-IPDI-BDA was 28.06%, which is a slight increase in the percentage of hydrogen bonding, suggesting that BDA forms hydrogen bonds in the PCL-IPDI network and improves its mechanical properties. However, this small difference (∼1.9%) is in contrast to the significant improvement in mechanical properties observed with the addition of BDA, mainly because there are other interactions such as B–N coordination bonding to provide additional cross-linking sites,41,42 as shown in Fig. S7.† Although the formation of partial hydrogen bonds and B–N coordination bonds by BDA in PCL-IPDI-BDA results in cross-linking points and improves its mechanical properties, it is still much lower than the properties achieved by the system containing curcumin units (tensile strength of 33.44 MPa and elongation at break of 1265.34%). This is due to the fact that curcumin not only acts as a chain extender in the main chain, but can also provide reactive groups for BDA, forming dynamic cross-linking points with B–O bonds by esterification of the enolized structure of curcumin and the B–OH groups in BDA, resulting in the formation of CCANs, which are much stronger than the network formed by physical bonds alone. In addition, curcumin is an acid/alkali-sensitive and photosensitive biomaterial, and the B–O bond is a dynamic chemical bond sensitive to water. Therefore, the photostability and humidity stability of the elastomers are also investigated as shown in Fig. S8,† as well as the effect of pH adjustment on the mechanical properties during the synthesis process as shown in Fig. S9.† Overall, the prepared elastomers show weaker humidity stability and good photostability, and the pH modulation of the system has less effect on the formation of the CCAN in the structures, which may be due to the volatilization of the acid and the reaction of the alkali with CO2 during the later curing process.
O in PCL-IPDI-BDA was 28.06%, which is a slight increase in the percentage of hydrogen bonding, suggesting that BDA forms hydrogen bonds in the PCL-IPDI network and improves its mechanical properties. However, this small difference (∼1.9%) is in contrast to the significant improvement in mechanical properties observed with the addition of BDA, mainly because there are other interactions such as B–N coordination bonding to provide additional cross-linking sites,41,42 as shown in Fig. S7.† Although the formation of partial hydrogen bonds and B–N coordination bonds by BDA in PCL-IPDI-BDA results in cross-linking points and improves its mechanical properties, it is still much lower than the properties achieved by the system containing curcumin units (tensile strength of 33.44 MPa and elongation at break of 1265.34%). This is due to the fact that curcumin not only acts as a chain extender in the main chain, but can also provide reactive groups for BDA, forming dynamic cross-linking points with B–O bonds by esterification of the enolized structure of curcumin and the B–OH groups in BDA, resulting in the formation of CCANs, which are much stronger than the network formed by physical bonds alone. In addition, curcumin is an acid/alkali-sensitive and photosensitive biomaterial, and the B–O bond is a dynamic chemical bond sensitive to water. Therefore, the photostability and humidity stability of the elastomers are also investigated as shown in Fig. S8,† as well as the effect of pH adjustment on the mechanical properties during the synthesis process as shown in Fig. S9.† Overall, the prepared elastomers show weaker humidity stability and good photostability, and the pH modulation of the system has less effect on the formation of the CCAN in the structures, which may be due to the volatilization of the acid and the reaction of the alkali with CO2 during the later curing process.
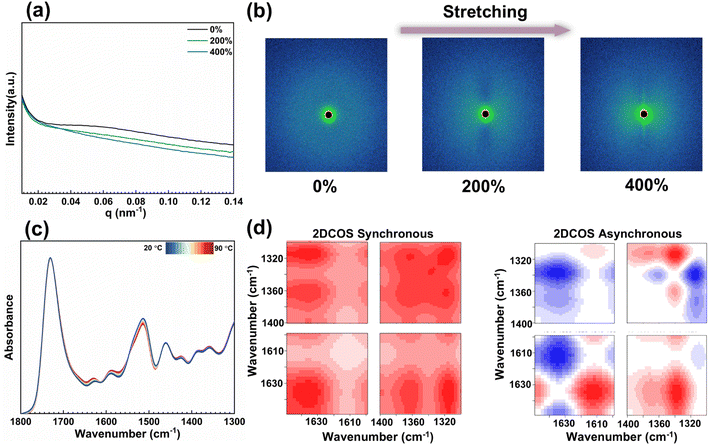
The reasons why the mechanical properties of this elastomer can be significantly improved are as follows. Curcumin molecular chain segments are constantly in dynamic chemical balance in terms of enol–keto conformation. When they are in the enol conformation, the generated hydroxyl groups are supposed to undergo a dynamic borate esterification reaction with the boronic acid groups in BDA, forming a rigid chemically cross-linked hard chain segment. These CCANs can retain a considerable number of rigid chemical cross-links when the elastomer is subjected to external forces, providing high strength. They can also be dissociated in reaction to large amounts of stress. These significantly avoid stress concentration and enhance the toughness of the material. To elucidate the underlying mechanisms of the mechanical behavior of PICB1.0PU elastomers, 2D-SAXS is performed to analyze the structural changes during the dynamic phase of the tensile process (Fig. 2b). In the static state, scattering halos are found in the samples, indicating a clear microphase separation in the PICB1.0PU elastomer. When the elastomer is stretched up to 200%, the 2D-SAXS scattering pattern turns into a butterfly shape, indicating deformation of the hard phases along the stretching direction. As the PICB1.0PU elastomer is continuously stretched to 400% strain, the long axis of the elliptical 2D-SAXS scattering pattern gradually becomes longer, indicating that the hard phases gradually decompose with the stretching of the PICB1.0PU elastomer. To facilitate a more comprehensive assessment of the dynamic microstructural changes, the 1D integration curves of the SAXS patterns are compared (Fig. 2a). As the strain increases from 0% to 400%, the average distance between the hard phases perpendicular to the tensile direction decreases while the strength decreases, indicating a high orientation of the hard phases along the stretching direction. This indicates that CCANs can achieve effective dissociation under external forces to dynamically adapt to external mechanical loads. We also collect temperature-dependent FT-IR spectra (20–90 °C) to further understand the dynamic interaction mechanism of PICB1.0PU elastomers. These spectra can track the motion of different functional groups, thus providing insights into molecular interactions (Fig. 2c). The spectrum bands located at 1650–1600 and 1400–1300 cm−1 belong to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups in curcumin and the B–O bonds, respectively, thus allowing the tracking of dynamic bonding changes. According to the variable temperature FT-IR spectra of PICB1.0PU elastomers, the C
O groups in curcumin and the B–O bonds, respectively, thus allowing the tracking of dynamic bonding changes. According to the variable temperature FT-IR spectra of PICB1.0PU elastomers, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching bands are red-shifted, and B–O bonds are blue-shifted on heating, indicating dissociation of C
O stretching bands are red-shifted, and B–O bonds are blue-shifted on heating, indicating dissociation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups and B–O bonds, respectively. To provide higher resolution for fine molecular motions, two-dimensional correlation spectroscopy (2D COS) analysis is carried out, which is sensitive to a variety of fine dynamic interactions and helps to provide comprehensive information at the molecular level. 2D COS is generated from all variable-temperature IR spectra, both synchronous and asynchronous (Fig. 2d). According to Noda's judging rule,43,44 the order of precedence in the heating process can be determined as (→ indicates prior or earlier; see Table S4† for operational details to support the information):1340 cm−1 → 1361 cm−1 → 1612 cm−1 → 1314 cm−1 → 1635 cm−1, i.e., ν(B–O) (ligand bonding) → ν(B–O) (chemical bonding) → ν(C
O groups and B–O bonds, respectively. To provide higher resolution for fine molecular motions, two-dimensional correlation spectroscopy (2D COS) analysis is carried out, which is sensitive to a variety of fine dynamic interactions and helps to provide comprehensive information at the molecular level. 2D COS is generated from all variable-temperature IR spectra, both synchronous and asynchronous (Fig. 2d). According to Noda's judging rule,43,44 the order of precedence in the heating process can be determined as (→ indicates prior or earlier; see Table S4† for operational details to support the information):1340 cm−1 → 1361 cm−1 → 1612 cm−1 → 1314 cm−1 → 1635 cm−1, i.e., ν(B–O) (ligand bonding) → ν(B–O) (chemical bonding) → ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (bonded) → ν(B–O) (free) → ν(C
O) (bonded) → ν(B–O) (free) → ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (free). Note that the two new peaks at 1635 and 1612 cm−1 identified by the 2D COS asynchronous spectra are split from the original peak at 1628 cm−1 in the 1D spectra and are attributed to free C
O) (free). Note that the two new peaks at 1635 and 1612 cm−1 identified by the 2D COS asynchronous spectra are split from the original peak at 1628 cm−1 in the 1D spectra and are attributed to free C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and bonded C
O and bonded C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively. The results indicate that the B–O bonds first respond to temperature perturbation, which in turn leads to the dissociation of CCANs. Overall, the CCANs in this work are driven by the intrinsic dynamic chemical balance of curcumin, which can dynamically adapt to the external mechanical loads. Resultantly, it is of great significance for the improvement of the mechanical properties of elastomers. PICB1.0PU elastomer is chosen as the subject for further study because of its superior properties.
O bonds, respectively. The results indicate that the B–O bonds first respond to temperature perturbation, which in turn leads to the dissociation of CCANs. Overall, the CCANs in this work are driven by the intrinsic dynamic chemical balance of curcumin, which can dynamically adapt to the external mechanical loads. Resultantly, it is of great significance for the improvement of the mechanical properties of elastomers. PICB1.0PU elastomer is chosen as the subject for further study because of its superior properties.
2.2 Self-healing and reprocessing properties
With the assistance of CCANs, the fabricated elastomers are inherently reversible, with the ability to dissociate, reorganize, and rearrange between different structures to achieve damage recovery, improve their damage tolerance, extend the service life of the material, and reduce the safety hazards associated with damaged materials. Therefore, the effect of self-healing temperature and conditions on the self-healing ability of PICB1.0PU elastomer is studied by testing cut dumbbell-shaped samples, as illustrated in Fig. 3a. Firstly, the self-healing performance of PICB1.0PU elastomer at different healing times at room temperature is evaluated. As shown in Fig. 3b and c, the tensile strength of the fractured PICB1.0PU elastomer increases with increasing healing time. When healed for 72 h at room temperature, the tensile strength can recover to 15.33 MPa, with a self-healing efficiency of 45.83%. Additionally, B–O bonds are sensitive to water, and under the action of water, the exchange efficiency of B–O bonds can be greatly improved,36,45 which is expected to further enhance the self-healing efficiency of PICB1.0PU elastomer at room temperature. However, PICB1.0PU elastomer is composed of a large amount of hydrophobic PCL and curcumin, and its surface exhibits certain hydrophobicity (Fig. S10a†),46–48 which is unfavorable for water wetting the surface, making it difficult to achieve the water-assisted effect. Therefore, it is necessary to increase the wettability of water with the surface to enhance the activity and exchange efficiency of B–O bonds at the fracture interface. Therefore, a mixed solution of ethanol and water is chosen to effectively wet the interface (Fig. S10b†). As shown in Fig. 3d and e, when the fractured PICB1.0PU elastomer interface is treated with a mixed solution of ethanol and water for 5 min and then healed for 72 h at room temperature, the tensile strength can recover to 20.08 MPa, with a self-healing efficiency of 60.04%. It can be seen that after solvent treatment, the tensile strength and self-healing efficiency of the fractured PICB1.0PU elastomer are greatly improved, proving that the activity and exchange efficiency of B–O bonds at the fracture interface are effectively enhanced with the assistance of solvent. However, due to the limited number of B–O bonds between molecular chains, there is little change in self-healing efficiency when the healing time is increased to 96 h (Fig. S11†). Additionally, macroscopic photos of the healed PICB1.0PU elastomer are shown in Fig. 3g, which show that it does not fracture after bending, twisting, and stretching, further showing that the fractured specimen is effectively repaired. Fig. 3h illustrates the self-healing mechanisms of PICB1.0PU elastomers at room temperature, mainly including the recombination and exchange of hydrogen bonds and B–O bonds. Current self-healing elastomers require not only the introduction of dynamic bonding but also a certain degree of flexibility of the polymer chains. Otherwise, the lack of polymer chain interfacial mobility at the contact interface often causes few interfacial dynamic bonds, which makes it difficult to realize self-healing behavior under mild conditions. In contrast, as for the hard segments in PICB1.0PU, both curcumin and BDA have rigid conjugated benzene ring structures, which can be chemically crosslinked by esterification. As for the soft chain segments, the polyester molecules with ester bonds are not as flexible as polyether. Accordingly, the overall design of PICB1.0PU seems to favor high strength and toughness instead of self-healing capability. However, the CCANs allow the fracture interface of the elastomeric material to realize spontaneous dynamic bonding/debonding in the absence of external stimuli such as force, light, heat, etc. This creates the conditions for this structurally rigid elastomer to exhibit self-healing ability under mild conditions.The CCANs in this work can trigger additional dynamic chemical mechanisms at higher temperatures, as well as significantly enhancing polymer chain mobility, leading to a significant increase in self-healing efficiency. Compared to conventional urethane bonds, phenol–carbamate bonds have a lower dissociation temperature,49,50 which is expected to further enhance the elastomer's self-healing performance. To explore the dissociation temperature of phenol–carbamate bonds in PICB1.0PU elastomers, in situ infrared spectroscopy is conducted at different temperatures, as shown in Fig. 4a. It is observed that when the temperature reached 70 °C, the –NCO groups were detected at 2272 cm−1, indicating the dissociation of phenol–carbamate bonds. Therefore, the self-healing efficiency of fractured PICB1.0PU samples healed at 70 °C at different times is investigated. From Fig. 4b and c, it can be seen that the tensile strength of fractured PICB1.0PU elastomers increases with increasing healing time, with a self-healing efficiency reaching as high as 94.67% after only 24 h of healing. The FTIR spectrum after healing is the same as that of the original elastomer (Fig. S12†), which also proves the recovery of its broken bonds. When the temperature is below 70 °C, the self-healing efficiency of samples healed for 24 h is only around 60% (Fig. S11†), further demonstrating the significant role of dissociation and recombination of phenol–carbamate bonds in enhancing self-healing performance. In summary, the fabricated PICB1.0PU elastomers exhibit outstanding self-healing performance, which can meet the requirements for damage repair in different usage environments.
Based on the CCANs, compared to traditional irreversible crosslinking points, the elastomer chains can dissociate and restructure between segments. Thus, the recyclability of the elastomer is investigated, with the schematic process shown in Fig. 4d. The results indicate that the prepared elastomer, assisted by DMF solvent at 70 °C, can dissolve and form films again, and after undergoing three cycles of dissolution and film formation, its tensile strength and elongation at break are similar to those of the original samples, showing that CCANs can realize multistage dynamic chemical dissociation for important implications in the field of recycling and reprocessing.
2.3 Fluorescence properties
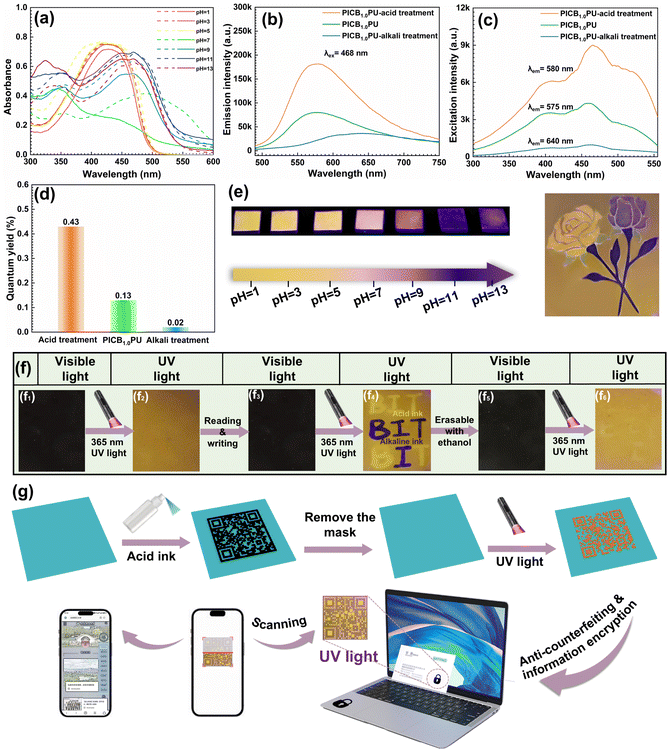
It has been reported that curcumin molecules possess high electron delocalization and π–π conjugated structures, which can be used as fluorescent dyes based on aggregation-induced mechanisms.51,52 In addition, curcumin exhibits pH sensitivity, which can be modulated by regulating the environmental pH to trigger ketone and enol transitions in the curcumin structures (Fig. S13†),53–55 as a way to achieve different fluorescent colors (Fig. S14†). Here, curcumin is introduced into the PU backbone as a chain-expanding component, which is located in the hard phase concentrated area and can further increase its entanglement density through B–O bond cross-linking (Fig. S15†). This satisfies its aggregation-induced luminescence conditions, which can potentially lead to the generation of fluorescent properties and acid/alkali-induced color changes on the elastomer surface. To investigate this, the UV-Vis absorption spectra of curcumin ethanol solution and curcumin- and BDA-liganded ethanol solutions are first compared (Fig. 5a). From Fig. S14,† it can be observed that the curcumin ethanol solution exhibits strong absorption peaks in the range of 300–500 nm, indicating that it can produce fluorescence under UV irradiation. Moreover, under different pH conditions, there is a noticeable shift in the excitation wavelength, with alkaline conditions showing a significant red shift compared to acidic conditions, attributed to the sensitivity of curcumin to acid/alkaline conditions resulting in structural isomerization. After curcumin coordinates with BDA, there is a significant red shift in both the maximum excitation wavelength and the excitation wavelength under acidic conditions (Fig. 5a), as well as a weak blue shift under alkaline conditions, without losing its fluorescence properties, showing a wide range of excitation wavelengths. Moreover, different color changes can be observed under different pH conditions, as shown in Fig. S16.† On this basis, the fluorescence properties of acid-treated PICB1.0PU, PICB1.0PU, and alkali-treated PICB1.0PU elastomers are investigated, and their emission spectra, excitation spectra, and quantum yields are recorded. As shown in Fig. 5b, it can be found that the maximum emission wavelengths of acid-treated PICB1.0PU, original PICB1.0PU, and alkali-treated PICB1.0PU are 580 nm, 575 nm, and 640 nm, respectively, and the maximum emission intensities gradually decrease. In the excitation spectra, it can be found that the maximum excitation intensity of the three PU samples decreases gradually at the excitation wavelength of 468 nm (Fig. 5c). In addition, the quantum yields of acid-treated PICB1.0PU, original PICB1.0PU, and alkali-treated PICB1.0PU are 0.43%, 0.13%, and 0.02%, respectively, and their quantum yields gradually decrease (Fig. 5d). Compared with the original PICB1.0PU, the enol structures of curcumin in acid-treated PICB1.0PU samples are converted to ketone structures, while the enolized structures in alkali-treated PICB1.0PU samples are further increased. Analyzing the above results suggests that the ketone structures coordinated with BDA are more conducive to electron transfer and produce stronger fluorescence properties compared to the enol structures. Furthermore, the macroscopic views of PICB1.0PU under visible and UV irradiation are observed, showing reddish-black color under visible light (Fig. 5f1) and yellow color under UV irradiation (Fig. 5f2), which suggests that fluorescent properties are produced. Moreover, changes in the fluorescence properties of the PICB1.0PU samples after immersion in ethanol solutions of different pH values are observed (Fig. 5e). It can be found that under UV irradiation, the PICB1.0PU samples show different colors, which transition from bright yellow to dark blue and then to purple as the pH increases. Also, through the masking method and modularity, based on the different pH-based induction, a colorful rose pattern demonstration is achieved in the same carrier at 395 nm excitation wavelength in the right image of Fig. 5d. Analysis of the FT-IR spectra of the original and acid/alkaline treated elastomer samples (Fig. S17†) reveals changes in the intensity and peak shift of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O absorption peak at 1628 cm−1 after acid or alkaline treatment. The surface C
O absorption peak at 1628 cm−1 after acid or alkaline treatment. The surface C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O absorption peak area increases significantly under acid induction, indicating that the proportion of its ketone structure increases, while the C
O absorption peak area increases significantly under acid induction, indicating that the proportion of its ketone structure increases, while the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O absorption peak area significantly decreases under alkali induction, indicating that the proportion of its enol structure increases. This indicates that the curcumin structure on the elastomer surface undergoes structural isomerization induced by different pH, with the ratio of its ketone and enol structures reaching a dynamic chemical balance. Overall, the CCAN network can be triggered by acid/alkali-induced structural changes to achieve a multicolor fluorescence display. In addition, in order to further understand the effect of this structural change of acid and alkaline surface treatment on the mechanical properties of elastomers, we test the mechanical properties of the elastomers after immersion in different pH solutions, as shown in Fig. S18.† The results also show that the surface treatment has little effect on the elastomer, which may be due to the fact that this pH-adjusted structural change in curcumin is limited to the surface only, as illustrated in the inset in Fig. S18.†
O absorption peak area significantly decreases under alkali induction, indicating that the proportion of its enol structure increases. This indicates that the curcumin structure on the elastomer surface undergoes structural isomerization induced by different pH, with the ratio of its ketone and enol structures reaching a dynamic chemical balance. Overall, the CCAN network can be triggered by acid/alkali-induced structural changes to achieve a multicolor fluorescence display. In addition, in order to further understand the effect of this structural change of acid and alkaline surface treatment on the mechanical properties of elastomers, we test the mechanical properties of the elastomers after immersion in different pH solutions, as shown in Fig. S18.† The results also show that the surface treatment has little effect on the elastomer, which may be due to the fact that this pH-adjusted structural change in curcumin is limited to the surface only, as illustrated in the inset in Fig. S18.†
Furthermore, we conduct reading and writing tests on the surface of the PICB1.0PU elastomer using ethanol solutions with pH of 3 and 11 as acidic ink and alkaline ink, respectively. As shown in Fig. 5e, when writing the English letters “BIT” on the surface with acidic ink and alkaline ink, it can be observed that under visible light, they appear red-black (Fig. 5f3), while under UV irradiation, they appear with a yellow background, displaying the letters “BIT” in bright yellow and navy blue (Fig. 5f4), respectively, thus achieving their readability and information encryption functions. Notably, the surface-drawn patterns can be removed with anhydrous ethanol, as shown in Fig. 5f5 and f6, achieving erasable and rewritable functions on the elastomer surface. Specific ketone- and enol-type structural balances in curcumin on the elastomer surface can be exquisitely regulated by acidic and alkaline inks. The obtained dynamic chemical balance can be maintained for a long time even after the inks were evaporated. Interestingly, when the ethanol solution is used to treat the surface, it drastically destroys the obtained dynamic chemical balance states, and thus the corresponding chemical history is eliminated. After the evaporation of ethanol, the keto- and enol-chemical balance in the curcumin structure can be restored to the original state, realizing facile information erasure. Additionally, as a proof-of-concept for anti-counterfeiting labels, QR code anti-counterfeiting labels are created using the masking method by spraying acidic ink, as shown in Fig. 5g. Under UV irradiation, the surface presents a visible QR code pattern that is scannable. These observations demonstrate the enormous potential of PICBPU elastomers in data storage and encryption applications.
2.4 Degradation performances
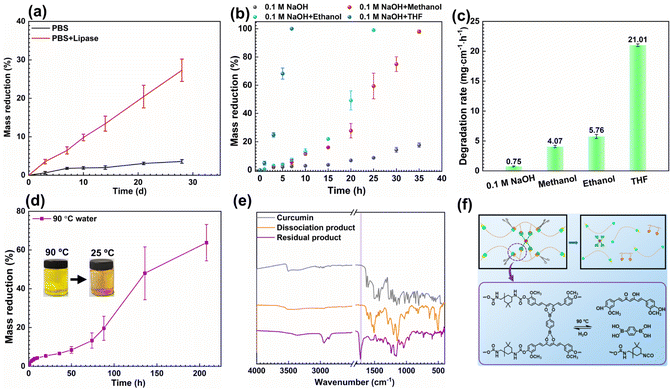
Elastomers have abundant applications in the field of smart materials due to their excellent strength and ductility, and the consequent accumulation of waste and environmental pollution, thus making the issue of their degradability particularly important. The prepared CCAN-induced elastomers are rich in reversible dynamic structures that facilitate the dissociation of molecular chains and are expected to achieve the collapse of the polymer network and thus polymer fragmentation with the help of hydrolyzable polyester and degradable curcumin. Here, based on the structural characteristics of PICB1.0PU elastomers, their degradability properties are tested and evaluated.As is well known, polyesters can undergo enzymatic chain cleavage under natural and biological conditions, ultimately completing degradation and metabolism.56,57 Therefore, we evaluate the degradability of PICB1.0PU elastomers in phosphate-buffered saline (PBS) with or without lipase. As shown in Fig. 6a, the change in mass loss of the PICB1.0PU elastomer over time is displayed. It can be observed that with increasing immersion time, the mass loss gradually increases, reaching approximately 30 wt% after 30 days in PBS solution with lipase. This indicates that under the action of lipase, the PCL polyol soft chain segments of the PICB1.0PU elastomer undergo decomposition. Furthermore, by analyzing the FT-IR spectra of the PICB1.0PU elastomer before and after degradation (Fig. S19†), it is observed that the absorption peaks of the C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the PCL segments at approximately 1233 cm−1 and 1726 cm−1, respectively, decrease or disappear, and the characteristic peaks around 3200 cm−1 become broader.58,59 This indicates that the ester bonds in the PU backbone were cleaved, suggesting that its degradation is mainly due to the cleavage of molecular chain segments.
O of the PCL segments at approximately 1233 cm−1 and 1726 cm−1, respectively, decrease or disappear, and the characteristic peaks around 3200 cm−1 become broader.58,59 This indicates that the ester bonds in the PU backbone were cleaved, suggesting that its degradation is mainly due to the cleavage of molecular chain segments.
Next, curcumin is a naturally biodegradable biomaterial that can undergo chemical degradation under different conditions, such as light exposure, alkaline environments, and auto-oxidation.55,60 Therefore, we also tested the degradation of curcumin in 0.1 M sodium hydroxide by UV-Vis spectrophotometric analysis as shown in Fig. S20.† It can be noticed that the intensity of the absorption spectrum of curcumin under alkaline conditions decreased dramatically compared to the initial one, which indicates that curcumin decomposed under alkaline conditions. Based on this, when curcumin is used as a chain extender for PU elastomers, PICB1.0PU elastomers are expected to chemically degrade under alkaline conditions. To investigate this, the degradation of PICB1.0PU elastomers in 0.1 M NaOH solution (organic solvent/H2O = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) at different times is explored, as shown in Fig. 6b. It can be observed that the PICB1.0PU elastomer can be completely degraded within 7 h in THF solution, followed by ethanol and methanol, and lastly in pure NaOH solution with a mass loss of up to 30 wt% within 35 h. By calculation, the degradation rates of the elastomers in THF, ethanol, methanol, and pure NaOH solution are determined to be 21.01, 5.76, 4.07, and 0.75 mg cm−1 h−1, respectively (Fig. 6c). Through analysis of the FT-IR spectra of the elastomers before and after degradation (Fig. S21†), it is observed that the characteristic peak of the C
1, v/v) at different times is explored, as shown in Fig. 6b. It can be observed that the PICB1.0PU elastomer can be completely degraded within 7 h in THF solution, followed by ethanol and methanol, and lastly in pure NaOH solution with a mass loss of up to 30 wt% within 35 h. By calculation, the degradation rates of the elastomers in THF, ethanol, methanol, and pure NaOH solution are determined to be 21.01, 5.76, 4.07, and 0.75 mg cm−1 h−1, respectively (Fig. 6c). Through analysis of the FT-IR spectra of the elastomers before and after degradation (Fig. S21†), it is observed that the characteristic peak of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 1628 cm−1 in the curcumin component of the backbone disappears. This indicates that the curcumin component undergoes decomposition under alkaline conditions, leading to the breakage and cleavage of the PU chain segments.
O at 1628 cm−1 in the curcumin component of the backbone disappears. This indicates that the curcumin component undergoes decomposition under alkaline conditions, leading to the breakage and cleavage of the PU chain segments.
Finally, it was mentioned earlier that the phenol-carbamate bonds can undergo dissociation at 70 °C, curcumin is insoluble in water, and the isocyanate groups are sensitive to water. Based on this, the chain extender curcumin is expected to dissociate when placed in an aqueous solution above 70 °C. Therefore, the degradation of PICB1.0PU elastomers in hot water at 90 °C is investigated, as shown in Fig. 6d. It can be observed that the PICB1.0PU elastomer dissociates in hot water at 90 °C for 200 h, with a mass loss of up to 60 wt%. From the analysis of the FT-IR spectra before and after the degradation of the elastomers (Fig. 6e), it can be found that the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O intensity of curcumin in the elastomer is reduced at 1628 cm−1, implying that the curcumin component is dissociated from the backbone. Additionally, analysis of the gel permeation chromatography (GPC) curve of the elastomer after degradation (Fig. S22†) reveals a molecular weight reduction to 8457 g mol−1, nearly 8 times lower than the original, confirming the successful detachment of curcumin from the backbone. Furthermore, the yellow color of the hot water after elastomer dissociation in Fig. 6d also confirms this. Moreover, when the 90 °C hot water containing degradation products is cooled to room temperature, the solution color becomes lighter, and a red precipitate forms at the bottom of the liquid. Analysis of the degradation products in aqueous solution by FTIR and NMR characterization confirmed that the dissociated products are curcumin components (Fig. 6e and S23†). This implies that there is a dynamic chemical-structural balance in the curcumin structure in the CCANs, which can undergo dynamic dissociation, offering the possibility of recycling and reusing small molecules such as curcumin and BDA. The degradation mechanism of PICB1.0PU elastomers in hot water is illustrated in Fig. 6f, primarily stemming from the dissociation and association of phenol–carbamate bonds, the hydrophobicity of curcumin, the water solubility of BDA, and the sensitivity of isocyanate groups to water.
O intensity of curcumin in the elastomer is reduced at 1628 cm−1, implying that the curcumin component is dissociated from the backbone. Additionally, analysis of the gel permeation chromatography (GPC) curve of the elastomer after degradation (Fig. S22†) reveals a molecular weight reduction to 8457 g mol−1, nearly 8 times lower than the original, confirming the successful detachment of curcumin from the backbone. Furthermore, the yellow color of the hot water after elastomer dissociation in Fig. 6d also confirms this. Moreover, when the 90 °C hot water containing degradation products is cooled to room temperature, the solution color becomes lighter, and a red precipitate forms at the bottom of the liquid. Analysis of the degradation products in aqueous solution by FTIR and NMR characterization confirmed that the dissociated products are curcumin components (Fig. 6e and S23†). This implies that there is a dynamic chemical-structural balance in the curcumin structure in the CCANs, which can undergo dynamic dissociation, offering the possibility of recycling and reusing small molecules such as curcumin and BDA. The degradation mechanism of PICB1.0PU elastomers in hot water is illustrated in Fig. 6f, primarily stemming from the dissociation and association of phenol–carbamate bonds, the hydrophobicity of curcumin, the water solubility of BDA, and the sensitivity of isocyanate groups to water.
Overall, elastomers based on CCANs are capable of degradation in different environments and rates due to the hydrolyzability of polyester, chemical degradability of curcumin, and thermal dissociation of phenol–carbamate bonds with excellent environmental selectivity and adaptability, and are expected to have potential applications in multiple fields.
3 Conclusions
In summary, a high-strength, self-healing, degradable, and reprocessable multicolor fluorescent elastomer is successfully developed based on CCANs. The resulting elastomer shows a tensile strength 300 times higher than that of an ordinary linear elastomer and exhibits good toughness, room temperature self-healing, and reprocessability. Meanwhile, the self-healing efficiency of the obtained elastomers is as high as 94.67% at 70 °C for 24 h with the assistance of phenol–carbamate bonds. Based on the dynamic chemical balance of keto and enol structural transitions triggered by CCANs, a wide color band display can be achieved by simple ink pH adjustment, surface reading and writing, and pattern printing can be easily realized. Upon the hydrolyzability of polyester, chemical degradability of curcumin, thermal dissociation of phenol–carbamate bonds, and dynamic reversible dissociation of CCANs in the elastomers, different degradation environments (enzyme degradation, alkaline degradation, hot water degradation) and degradation rates can be realized with good environmental selectivity and adaptability. This work provides valuable guidance for the development of self-healing, degradable, and high-strength multifunctional materials for the field of smart anti-counterfeiting materials and smart flexible optoelectronics.Data availability
Data will be made available on request from the authors.Author contributions
C. Y. L. conducted the experiments and analyzed the data. X. S. and C. B. C. participated in the discussion; X. D. L. and M. S. Z. conceived the project and prepared the manuscript. All authors have approved the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the Natural Science Foundation of Shandong Province (ZR2024QE214), National Key Research and Development Program of China (Grant No. 2022YFB3806103), National Natural Science Foundation of China (Grant No. 12102010), and China Postdoctoral Science Foundation (2024M754107).References
- J. Bai, L. Z. Zhang, Z. X. Shi and X. S. Jiang, Adv. Funct. Mater., 2023, 33, 2212556 CrossRef.
- J. J. Gao, M. Tian, Y. R. He, H. J. Yi and J. B. Guo, Adv. Funct. Mater., 2021, 32, 2107145 CrossRef.
- Y. P. Li, Y. Jin, W. H. Zeng, H. Y. Jin, X. Shang and R. Zhou, ACS Appl. Mater. Interfaces, 2023, 15, 35469–35482 CrossRef PubMed.
- H. Ding, et al. , Chem. Sci., 2023, 14, 4633–4640 RSC.
- J. Wei, et al. , Nat. Commun., 2023, 14, 4839 CrossRef PubMed.
- J. Huang, W. J. Yao, X. J. Cui, L. B. Si, D. Yang, X. Y. Liu and W. T. Liu, Chem. Eng. Sci., 2024, 293, 120030 CrossRef.
- L. Song, X. Qiao, J. Sun, N. Yi, M. Wang, Z. Zhao, R. Xie, W. Chen and Y. Xia, Carbohydr. Polym., 2023, 304, 120500 CrossRef CAS.
- L. Zhou, K. Li, Y. Chang, Y. Yao, Y. Peng, M. Li and R. He, Chem. Sci., 2024, 15, 10046–10055 RSC.
- P. Shi, E. Miwa, J. He, M. Sakai, T. Seki and Y. Takeoka, ACS Appl. Mater. Interfaces, 2021, 13, 55591–55599 CrossRef CAS PubMed.
- X. Li, J. Li, W. Wei, F. Yang, M. Wu, Q. Wu, T. Xie and Y. Chen, Macromolecules, 2021, 54, 1557–1563 CrossRef CAS.
- Q. Y. Fan, Y. T. Tang, H. N. Sun, D. K. Guo, J. W. Ma and J. B. Guo, Adv. Mater., 2024, 2401315 CrossRef CAS PubMed.
- H. Q. Wang, Y. Q. Tang, Z. Y. Huang, F. Z. Wang, P. F. Qiu, X. F. Zhang, C. Li and Q. Li, Angew. Chem., Int. Ed., 2023, 62, e202313728 CrossRef CAS PubMed.
- R. C. Lin, Y. Qi, D. H. Kou, W. Ma and S. F. Zhang, Adv. Funct. Mater., 2022, 32, 2207691 CrossRef CAS.
- J. W. Li, Z. R. Zheng, Y. N. Ma, Z. X. Dong, M. H. Li and J. Hu, Small, 2024, 2402130 CrossRef CAS.
- H. T. Wu, H. Wang, M. Luo, Z. Y. Yuan, Y. W. Chen, B. Q. Jin, W. Q. Wu, B. J. Ye, H. J. Zhang and J. R. Wu, Mater. Horiz., 2024, 11, 1548–1559 RSC.
- X. X. Chen, et al. , ACS Appl. Mater. Interfaces, 2020, 12, 30847–30855 CrossRef PubMed.
- J. Z. Wei, H. R. Li, J. Yang, C. M. Yue and Y. G. Wang, ACS Appl. Polym. Mater., 2023, 5, 5716–5726 CrossRef.
- H. T. Deng, H. Wang, Y. Tian, Z. Lin, J. X. Cui and J. Chen, Mater. Horiz., 2023, 10, 5256–5262 RSC.
- X. Wang, J. Xu, Y. Zhang, T. Wang, Q. Wang, S. Li, Z. Yang and X. Zhang, Nat. Commun., 2023, 14, 4712 CrossRef.
- L. D. C. de Castro, J. Lub, O. N. Oliveira and A. P. H. J. Schenning, Angew. Chem., Int. Ed., 2024, e202413559 Search PubMed.
- K. R. Schlafmann, M. S. Alahmed, K. L. Lewis and T. J. White, Adv. Funct. Mater., 2023, 33, 2305818 CrossRef.
- J. Li, J. Li, H. Li, C. Wang, M. Sheng, L. Zhang and S. Fu, ACS Appl. Mater. Interfaces, 2021, 13, 27200–27208 CrossRef PubMed.
- J. M. Lehn, Chem. Soc. Rev., 2007, 36, 151–160 RSC.
- J. M. Lehn, Adv. Polym. Sci., 2013, 261, 155–172 CrossRef.
- J. M. Lehn, Angew Chem. Int. Ed. Engl., 2015, 54, 3276–3289 CrossRef PubMed.
- C. Cui, L. An, Z. Zhang, M. Ji, K. Chen, Y. Yang, Q. Su, F. Wang, Y. Cheng and Y. Zhang, Adv. Funct. Mater., 2022, 32, 2203720 CrossRef.
- C. Dang, Y. Shao, S. Ding, H. Qi and W. Zhai, Adv. Mater., 2024, 36, 2406967 CrossRef CAS PubMed.
- H. Chen, Y. Hu, C. Luo, Z. Lei, S. Huang, J. Wu, Y. Jin, K. Yu and W. Zhang, J. Am. Chem. Soc., 2023, 145, 9112–9117 CrossRef CAS.
- X. Wei, X. Zhang, T. Chen, J. Huang, T. Li, X. Zhang, S. Wang and W. Dong, ACS Macro Lett., 2024, 13, 1112–1118 CrossRef CAS.
- Z. C. Jiang, Y. Y. Xiao, J. B. Hou, X. S. Chen, N. Yang, H. Zeng and Y. Zhao, Angew. Chem., Int. Ed., 2022, 61, e202211959 CrossRef CAS.
- T. Ye, J. Tan, T. Wu, F. Zhang, S. Chen and C. Wang, Sci. China:Chem., 2024 DOI:10.1007/s11426-024-2367-8.
- W. Yao, Q. Tian, J. Shi, C. Luo and W. Wu, Adv. Funct. Mater., 2021, 31, 2100211 CrossRef CAS.
- H. Huang, W. Sun, L. Sun, L. Zhang, Y. Wang, Y. Zhang, S. Gu, Z. You and M. Zhu, Proc. Natl. Acad. Sci. U.S.A., 2024, 121, e2404726121 CrossRef CAS.
- N. Roy, B. Bruchmann and J. M. Lehn, Chem. Soc. Rev., 2015, 44, 3786–3807 RSC.
- C. H. Gao, L. Y. Sun, S. K. Wang, S. T. Cao, H. Z. Liu, P. C. Chen and Y. Q. Wang, J. Appl. Polym. Sci., 2023, 140, e54710 CrossRef CAS.
- H. Y. Fei, G. M. Juan, N. Yu, X. H. Fei, Z. Hao and Z. Yue, Chem. Eng. J., 2023, 468, 143742 CrossRef.
- P. Schara, A. M. Cristadoro, R. P. Sijbesma and Ž. Tomović, Macromolecules, 2023, 56, 8866–8877 CrossRef.
- F. Zeng, et al. , Macromolecules, 2022, 55, 8741–8748 CrossRef.
- H. L. Xu, S. W. Zhao, A. Q. Yuan, Y. L. Zhao, X. Wu, Z. Wei, J. Lei and L. Jiang, Small, 2023, 19, 2300626 CrossRef PubMed.
- Q. H. Zhang, et al. , Adv. Mater., 2018, 30, 1801435 CrossRef.
- Y. Fu, S. Chen, X. Chen, H. Zheng, Y. Ling, Z. Liu, M. Wang and L. Lan, Adv. Funct. Mater., 2024, 2314561 CrossRef.
- K. Song, W. Ye, X. Gao, H. Fang, Y. Zhang, Q. Zhang, X. Li, S. Yang, H. Wei and Y. Ding, Mater. Horiz., 2021, 8, 216–223 RSC.
- H. Xiang, X. X. Li, B. H. Wu, S. T. Sun and P. Y. Wu, Adv. Mater., 2023, 35, 2209581 CrossRef.
- S. T. Sun, H. Tang, P. Y. Wu and X. H. Wan, Phys. Chem. Chem. Phys., 2009, 11, 9861–9870 RSC.
- Z. Z. Han, Z. P. Chen, Z. Yue, Z. J. Lin and L. C. Hui, Adv. Funct. Mater., 2022, 32, 2201959 CrossRef.
- X. X. Zhang, J. Zhao, L. Ma, X. J. Shi and L. L. Li, J. Mater. Chem. A, 2019, 7, 24532–24542 RSC.
- Y. Q. Zheng, Z. J. Bao, Y. X. Fan and X. Y. Wang, J. Mol. Liq., 2023, 370, 121065 CrossRef.
- A. C. d. Silva, P. D. d. F. Santos, J. T. d. P. Silva, F. V. Leimann, L. Bracht and O. H. Gonçalves, Trends Food Sci. Technol., 2018, 72, 74–82 CrossRef.
- Y. J. Jia, L. Z. Zhang, M. L. Qin, Y. Li, S. J. Gu, Q. B. Guan and Z. W. You, Chem. Eng. J., 2022, 430, 133081 CrossRef.
- Y. J. Jia, Q. B. Guan, L. Z. Zhang, R. E. Neisiany, N. N. Yan, Y. Li and Z. W. You, Sci. China Mater., 2022, 65, 2553–2564 CrossRef.
- Y. M. Guo, C. Yang, Y. J. Zhang and T. Tao, Spectrochim. Acta, Part A, 2022, 265, 120359 CrossRef PubMed.
- Y. Liu, C. Zhang, H. Pan, L. Li, Y. J. Yu and B. M. Liu, Asian J. Pharm. Sci., 2021, 16, 419–431 CrossRef PubMed.
- S. Mondal, S. Ghosh and S. P. Moulik, J. Photochem. Photobiol., B, 2016, 158, 212–218 CrossRef PubMed.
- F. Mohammed, F. Rashid-Doubell, S. Cassidy and F. Henari, Spectrochim. Acta, Part A, 2017, 183, 439–450 CrossRef.
- B. J. Zheng and D. J. McClements, Molecules, 2020, 25, 2791 CrossRef PubMed.
- C. C. Xu and Y. Hong, Bioact. Mater., 2022, 15, 250–271 Search PubMed.
- D. Zhao, X. Li, Q. Li, C. Yue, Y. Wang and H. Li, Chem. Sci., 2024, 15, 13306–13312 RSC.
- F. L. Li, Z. F. Xu, H. Hu, Z. Y. Kong, C. Chen, Y. Tian, W. W. Zhang, W. Bin Ying, R. Y. Zhang and J. Zhu, Chem. Eng. J., 2021, 410, 128363 CrossRef.
- R. Guo, Q. Zhang, Y. S. Wu, H. B. Chen, Y. H. Liu, J. Wang, X. Duan, Q. Chen, Z. Ge and Y. Zhang, Adv. Mater., 2023, 35, 2212130 CrossRef PubMed.
- W. Y. Jan, P. MinHsiung, C. AnnLii, L. L. In, H. Y. Soon, H. C. Yao and L. J. Kun, J. Pharm. Biomed. Anal., 1997, 15, 1867–1876 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: All experimental procedures, methods, GPC, EDS, ICP-OES, TGA, DSC, DMA, 2D-SAXS, and other characterizations. See DOI: https://doi.org/10.1039/d4sc06855f |
| This journal is © The Royal Society of Chemistry 2025 |