 Open Access Article
Open Access ArticleSensor array based on silver nanoprisms for the determination of ethanol content and resolution of water–ethanol mixtures
Masoud Shariati-Rad *ab and
Mahya Hosseinpouria
*ab and
Mahya Hosseinpouria
aDepartment of Analytical Chemistry, Faculty of Chemistry, Razi University, Kermanshah, Iran. E-mail: mshariati_rad@yahoo.com; Fax: +98 833 4274559
bResearch Group of Design and Fabrication of Kit, Razi University, Kermanshah, Iran
First published on 10th July 2025
Abstract
Determination of the authenticity of a drink as well as the existence or amount of its ethanol content is very important. Herein, silver nanoprisms were successfully used for the discrimination of different water–ethanol mixtures. The nanoparticles were characterized via transmission electron microscopy (TEM) and UV-vis spectrophotometry. Moreover, using the color profile of the array, a reliable relationship was established for the determination of ethanol content in water–ethanol mixtures. Silver nanoprisms were prepared via a green procedure using extracts of walnut peel, onion and red berry in the presence and absence of sodium citrate. The color variation of the prepared silver nanoprisms was used for discrimination. The RGB color indices of the sensor array were organized in matrices and processed via principal component analysis (PCA) for discrimination and clustering.
1. Introduction
Ethanol is a colorless, clear, volatile, flammable liquid that mixes with water in any proportion and has a pungent odor. The detection and quantification of alcohols with high sensitivity, selectivity and accuracy are crucial in many different areas. Accurate and rapid measurement of ethanol is important in clinical and forensic analyses to analyze human body fluids, e.g., blood, serum, saliva, urine, breath and sweat. The food, beverage (wine, beer and spirits) and pulp industries require simple, fast and economic analytical methods to control fermentation processes and the quality of the obtained products.Ethanol content is one of the most important elements in food items and is used to ensure their authenticity; ethanol in higher doses can alter or damage the central neural system.1,2
The most common form of adulteration concerning ethyl alcohol is the addition of water. This is because ethanol can be easily mixed with water, and it is difficult to detect this adulteration visually.
Many analytical methods have been developed during for determination of ethanol and other aliphatic alcohols, such as methanol. These include the use of chemical methods, such as redox titrations, colorimetric methods, specific gravity and refractive index measurements, chromatographic and spectroscopic methods.3
In 2015, Zhang et al. reported a full evaporation headspace gas chromatographic method for simultaneously determining the ethanol and methanol content in wines.4 In 2005, Nordon et al. reported a non-invasive NIR and Raman spectrometric methods for determination of ethanol content.5 In 2019, Michałowska-Kaczmarczyk reported a redox titration method for determination of ethanol content in beverages. However, a prior separation of ethanol by distillation of the sample is necessary.6 In 2011, Pinyou and Youngvises reported a colorimetric method for determination of ethanol content based on the reaction of ethanol with ceric ion in acidic medium to produce a red colored product having maximum absorption at 415 nm.7 In 2009, Alemea and Costa reported a specific gravity measurement method for determination of ethanol content as a routine assay to evaluate the quality of gasoline with multivariate PLS calibration method.8
Although these methods are precise and reliable, they are complex and time consuming and require prior separation processes (distillation and pervaporation) and expensive instrumentation. Conversely, colorimetric methods are simple and inexpensive and can be used without complicated instruments. The current colorimetric methods are commonly based on sample imaging by cameras.
In the last decade, rapid advances in microelectronics have resulted in the ready availability of digital and video cameras, mobile phones, and scanners with continually enhanced specifications at a very modest cost. Thus, digital images obtained by commercial devices, such as digital cameras, webcams, smartphone cameras, scanners, and tablets, are currently implemented in many different scientific and technical fields. The most common way to extract the information from digital images is their decomposition to color indices such as red (R), green (G) and blue (B). The RGB system is an additive system, which uses the combination of the colors R, G, and B to form a wide variety of color tones.1 In colorimetric sensor arrays, such devices and procedures are commonly employed to extract signals, which are then analyzed by multivariate chemometrics methods. Colorimetric sensor arrays constructed from diverse chemo-responsive colorants have been successfully used to discriminate different volatile organic compounds in breath, which were related to lung cancer,9,10 and in beers.11 A series of common soft drinks were discriminated by a colorimetric sensor array composed of different dyes.12
Sensor arrays based on nanoparticles have been constructed for the detection of different volatile or organic species.13,14 A colorimetric solvatochromic molecule-based sensor was designed that can discriminate between some solvents and some water–tetrahydrofuran mixtures.15 For specific ethanol detection, a sensor array was constructed by dyes.16
Amongst the nanoparticles, silver nanoparticles (AgNPs) have been widely used to develop various sensors, including fluorimetric,17,18 UV-vis spectrophotometric and colorimetric,19,20 and electrochemical sensors.21,22 Herein, using silver nanoprisms (AgNPrs), a colorimetric sensor array is introduced for the analysis of mixtures of ethanol and water. This is the first time AgNPs have been used for the discrimination of different ethanol–water mixtures. Moreover, principal component analysis (PCA) and hierarchical cluster analysis (HCA) were used to analyze the color indices of the sensor array to cluster the samples.23–26
2. Experimental
2.1. Materials and solutions
Ethanol (99.9%, w/w), ammonia (25%, w/w), silver nitrate (99.0%) and sodium citrate (99.9%) were all obtained from Merck (Darmstadt, Germany). For preparation of all solutions, deionized water was employed.Ammonia (10%), silver nitrate solution (0.01 mol L−1), and sodium citrate solution (0.01 mol L−1) were used in the preparation of the silver nanoparticles (AgNPs). Different AgNPs were prepared with extracts of walnut peel, onion and red berry in the presence and absence of sodium citrate. Six different AgNPs were obtained.
Different mixtures of ethanol–water were prepared by mixing volumes (0.0–10.0 mL) of ethanol with doubly distilled water. The mixtures have total volumes of 10.0 mL and different percent ethanol contents (Table 1).
| Number (class) | Percent of ethanol (v/v) |
|---|---|
| 1 | 0.0 |
| 2 | 10.0 |
| 3 | 20.0 |
| 4 | 30.0 |
| 5 | 40.0 |
| 6 | 50.0 |
| 7 | 60.0 |
| 8 | 70.0 |
| 9 | 80.0 |
| 10 | 90.0 |
| 11 | 100.0 |
| 12 | Unknown |
2.2. Synthesis of AgNPs
Six different AgNPs were prepared using walnut peel extract without and with sodium citrate (A and B), onion extract without and with sodium citrate (C and D) and berry extract without and with sodium citrate (E and F).The method was that used by Jin et al.27 with minor modifications. To 300 mL boiling distilled water, separately, 1.5 mL of extracts of walnut peel, onion and red berry were added. After about 25 min, 4 mL of ammonia (10%, w/w) and 1.0 mL of sodium citrate (0.01 mol L−1) were added, followed by the addition of silver nitrate solution (5.0 mL, 20 mmol L−1) while stirring for 2 min. The yellow AgNPs formed after 50 min. Note that, for preparing AgNPs without citrate as a capping agent, sodium citrate was not added.
2.3. Instruments and software
Transmission electron microscopy (TEM) was performed using a Zeiss EM900 transmission electron microscope. UV-vis spectra of AgNPs in the presence of different ethanol–water mixtures were recorded on an Agilent 8453 UV-vis spectrophotometer with a diode array detector equipped with 1 cm path-length quartz cells. Digital images of the colorimetric sensor were recorded with the camera (50 megapixel) of a SAMSUNG Galaxy smartphone. To extract red (R), green (G) and blue (B) color indices as the analytical signal and for extraction of the profile of the constructed sensor array, ImageJ version 1.54h was employed. PCA toolbox for MATLAB version 1.5 was used to perform principal component analysis (PCA) and hierarchical cluster analysis (HCA).282.4. Principal component analysis (PCA) and hierarchical cluster analysis (HCA)
Principal component analysis (PCA) is a statistical method used for dimension reduction of data, feature extraction, and data visualization. It transforms a dataset of possibly correlated variables into a set of linearly uncorrelated variables called principal components (PCs), ordered by their ability to explain variance in the data. In PCA, orthogonal directions (principal components) in the data that maximize variance are identified. The first PC captures the most variance, the second PC (orthogonal to the first) captures the next most, and so on. This allows dimensionality reduction while preserving as much information as possible.29Hierarchical cluster analysis (HCA) is an unsupervised machine learning method used to build a hierarchy of clusters, enabling the exploration of data grouping at multiple scales. Unlike flat clustering methods (e.g., k-means), HCA does not require predefining the number of clusters and provides a tree-like structure (dendrogram) to visualize relationships between data points.30,31 In HCA, similar data points are grouped into nested clusters based on pairwise distances.
3. Results and discussion
3.1. Characterization of the synthesized AgNPs
The AgNPs prepared and the codes used for them in this work are reported in Table 2.| Coded silver nanoprisms (AgNPrs) | Condition for preparation of silver nanoparticle |
|---|---|
| A | With walnut peel extract without citrate |
| B | With walnut peel extract with citrate |
| C | With onion extract without citrate |
| D | With onion extract with citrate |
| E | With red berry extract without citrate |
| F | With red berry extract with citrate |
To investigate the morphology and size of the synthesized AgNPs, TEM analysis was carried out. The TEM micrographs are shown in Fig. 1. Analysis of the TEM micrographs showed a distribution with a mean particle size of 22.1 ± 4.9 nm. It is evident from the images that the shape of the synthesized AgNPs is as prism and its cross section is hexagonal. So, from this point, the synthesized AgNPs are referred to as Ag nanoprisms (AgNPrs).
 | ||
| Fig. 1 TEM micrographs of the AgNPs synthesized in the presence of citrate using walnut peel extract. | ||
In Fig. 2, the UV-vis spectra of the prepared AgNPs are shown. It can be seen that their maximum absorption peaks are located at 410–420 nm. This peak is a characteristic surface plasmon absorption band that indicates the presence of nano-silver in the solution.32
The differences in the full width at half maximum (FWHM) and maximum absorption wavelengths of the prepared AgNPs, as can be observed in Fig. 2, can be related to the different sizes of the AgNPs.33–36 This is the result of the different reducing agents used to prepare them.33–36
3.2. Construction of the sensor array
In order to introduce a method based on a sensor array for the determination of percent of ethanol in mixtures with water, six different AgNPrs were used (Table 2). The AgNPrs were prepared based on the procedure explained in Section 2.2.27The sensor array was constructed in such a way that to each row of the 72 well plate, 100 μL of one of the AgNPrs was added. Then, to each column of the plate, 50 μL of one of the ethanol–water mixtures was added and mixed well with the AgNPrs. After 5 min, an image of the plate was taken and used for analysis. This procedure was repeated three times. The images of the sensor array can be seen in Fig. 3. Because of the use of 6 AgNPrs and 12 mixtures, in total, 72 wells of the plate were filled.
 | ||
| Fig. 3 Image of three replicates of the constructed sensor array. Each row in the array corresponds to AgNPrs reported in Table 2 and each column corresponds to the mixtures reported in Table 1. | ||
Eleven different mixtures of water and ethanol were prepared (Table 1). To each mixture, a class was assigned. In addition, a mixture was used as an unknown mixture. Therefore, in total, 12 different classes can be analyzed.
The color change of the AgNPrs in the presence of different ethanol–water mixtures is clearly seen (Fig. 3). The color of the AgNPrs is the result of the surface plasmon phenomenon.37 This phenomenon is affected by the medium and the size and the cap of the corresponding AgNPrs.37 Any physicochemical variations in Ag nanostructure can alter its surface plasmon; simply, it is the base of a chemical sensor.38 The polarities of water and ethanol are different. Moreover, ethanol possesses an organic moiety which interacts with AgNPs in a different manner. The capping of AgNPrs with citrate can change their color and interaction with the medium.
Ethanol molecules can adsorb onto AgNP surfaces via hydrogen bonding between the hydroxyl (–OH) group of ethanol and oxygen-containing surface groups (e.g., citrate or oxide layers).39 In ethanol–water mixtures, the stability of AgNPs can be modulated by altering the solvent's polarity. Ethanol reduces the polarity of aqueous solutions, destabilizing AgNPs by compressing the electrical double layer, which promotes aggregation. In these conditions, the presence of capping agents like citrate can result in different aggregation behaviors of AgNPs with and without citrate.40
3.3. Profile of the colors
To obtain an overview of variation of the color with the ethanol content of the mixtures, using ImageJ version 1.54h, the overall image of the plate with filled wells was selected and its profile was plotted. The profile plots the variation in gray value of the selected part with pixels in the horizontal direction. The resulting profile can be seen in Fig. 4a. The maxima of the curve belong to the boundaries between the wells. The important parts of the plot are the minima. As can be seen, there exists a gradual, relatively linear change in the intensities in the minima with the ethanol contents of the mixtures. In order to build a relation between the ethanol content and the calculated gray values, the values in the minima were selected as signal and were plotted against the ethanol contents of the samples. By excluding the sample with 0.0% ethanol content, an excellent logarithmic relation can be obtained (Fig. 4c). The coefficient of determination for the linear semi-log relation is 0.9946 and the F-statistic is 930. These statistics prove an excellent linear relationship with high prediction ability. Using this calibration, the ethanol content of the unknown sample was estimated to be 67.7%. The true ethanol content of the unknown sample is 75.0%, which shows a 9.7% difference.3.4. Principal component analysis
In order to quantify the signal, which is initially in the form of color, different segments of the images (Fig. 3) corresponding to each well were cropped and transferred to ImageJ version 1.54h. Each mixture response is represented as the red (R), green (G) and blue (B) values of each of the 6 AgNPs, i.e., an 18 dimensional row vector. These vectors for 3 replicates of 12 analyzed samples (Table 2) can be arranged row-wise to produce a matrix with 36 × 18 dimensions.The matrix constructed was used for analysis by PCA. PCA is a chemometrics method which abstracts the initial data and lowers its dimensions to more limited new dimensions (variables) named scores. In fact, scores are the coordinates in the new axes. It must be mentioned that, during this transfer, the information in the original data is retained.
The scores calculated by PCA for the analyzed data can be seen in Fig. 5a. The first score (PC1) explained 99.77% of the variation in the analyzed data. In this analysis, the data were not preprocessed. As can be seen from Fig. 5a, a distinct clustering of the mixtures was obtained for most of the mixtures. However, overlaps of class 7 with class 10 and class 9 with class 11 can be observed. Class 12, which is the unknown mixture is located between classes 10, 11, 6 and 7. This result demonstrates that the unknown mixture probably contains 70–80% ethanol. In reality, the unknown mixture is a 75% ethanol sample. Therefore, the analysis based on the sensor array is confirmed to work satisfactorily in its expected function.
 | ||
| Fig. 5 Score plots obtained by analysis of the RGB data from the image of the sensor array (Fig. 3) via PCA without preprocessing (a), after mean centering (b) and after auto-scaling (c). | ||
Analysis of the data was also performed by preprocessing. By mean centering and subsequently applying PCA, the scores shown in Fig. 5b resulted. The figure shows that different mixtures are distinctly clustered. In this case, the unknown sample is located between classes 6, 7, 9, 10 and 11. Therefore, its ethanol content can be estimated to be in the range of 70–80%.
By auto-scaling the data as a preprocessing method and subsequent PCA analysis, the scores shown in Fig. 5c were calculated. Different mixtures are clustered distinctly. However, an overlap between classes 5 and 6 can be seen. Here, the unknown mixture can be observed between classes 5, 6, 7, 9 and 11. Therefore, an ethanol content of 65–70% can be attributed to this mixture.
In PCA, for calculation of the scores, coefficients are required. These coefficients are named loadings. For each original variable, a loading is calculated. In the current work, the variables are the R, G and B indices of different AgNPrs. In fact, the magnitude of each loading reflects the significance of the corresponding variable. Therefore, using loading plots, it is possible to observe which original variable is important. In order to eliminate the effect of the magnitude of the variables on the loadings, the loadings for the case where the data have been preprocessed by auto-scaling were studied (Fig. 6).
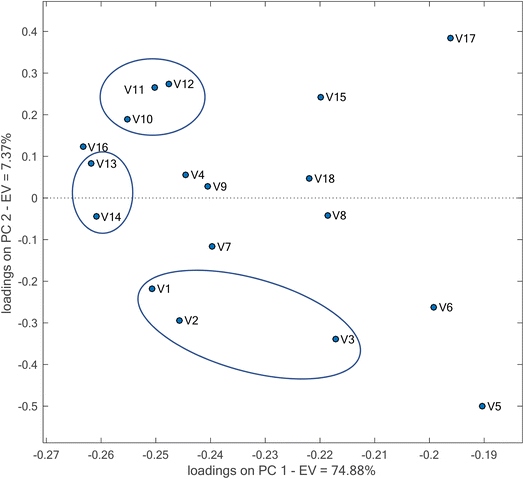 | ||
| Fig. 6 Loading plot obtained by PCA for the constructed sensor array after preprocessing of the data with auto-scaling. | ||
Because of the higher variability of the data explained by PC1, detection of the more significant variables was focused on PC1. A variable is significant and effective in clustering when it has a higher loading. It can be seen that AgNPrs with codes A (v1, v2, v3), D (v10, v11, v12) and E (v13, v14) have a large role in the clustering. These AgNPrs are those prepared respectively by walnut skin without citrate, onion extract with citrate and berry extract with citrate. Therefore, the capping of AgNPs with citrate with organic characteristics facilitates the interaction of ethanol with it.
Another clustering method, HCA, was also used. In this case, the data were auto-scaled, and clustering was performed based on the Euclidean distance calculated for complete data. In Fig. 7, the resulting dendrogram is shown. The dendrogram shows that, in most cases, the clustering was performed correctly. However, one of the replicates in each of classes 5 (sample 13) and 6 (sample 18) has been displaced. The unknown mixture with replicate numbers 34, 35 and 36 can be seen in a major cluster with classes 9 (samples 25, 26 and 27) and 11 (samples 31, 32 and 33). Therefore, an ethanol content of about 90% can be predicted for it. Altogether, for the unknown sample, an ethanol content in the range of 70–90% is predicted by the applied clustering methods.
 | ||
| Fig. 7 Dendrogram obtained by HCA processing of the data of the constructed sensor array. For HCA, data were auto-scaled and Euclidean distances were calculated for complete points. | ||
The method introduced can be used to predict the ethanol content of an water–ethanol mixture. The method, needing no instrument, can successfully be used for this purpose.
4. Conclusions
Using AgNPrs in a sensor array, it was possible to differentiate between ethanol–water mixtures. Differentiating and detecting fraud in such mixtures and many foodstuffs is of great importance. Due to the success of the designed sensor array in the current work, it is recommended to be applied for such a procedure. The method can be utilized as an alternative to sophisticated instrumental methods.Data availability
Data described in the manuscript are available on request.Conflicts of interest
There are no conflicts to declare.References
- F. C. Böck, G. A. Helfer, A. B. da Costa, M. B. Dessuy and M. F. Ferrão, Rapid determination of ethanol in sugarcane spirit using partial least squares regression embedded in smartphone, Food Anal. Methods, 2018, 11, 1951–1957 CrossRef.
- L. Curbani, J. M. L. N. Gelinski and E. M. Borges, Determination of ethanol in beers using a flatbed scanner and automated digital image analysis, Food Anal. Methods, 2020, 13, 249–259 CrossRef.
- A. M. Azevedo, D. M. F. Prazeres, J. M. Cabral and L. P. Fonseca, Ethanol biosensors based on alcohol oxidase, Biosens. Bioelectron., 2005, 21, 235–247 CrossRef CAS.
- C.-Y. Zhang, N.-B. Lin, X.-S. Chai and D. G. Barnes, A rapid method for simultaneously determining ethanol and methanol content in wines by full evaporation headspace gas chromatography, Food Chem., 2015, 183, 169–172 CrossRef CAS PubMed.
- A. Nordon, A. Mills, R. T. Burn, F. M. Cusick and D. Littlejohn, Comparison of non-invasive NIR and Raman spectrometries for determination of alcohol content of spirits, Anal. Chim. Acta, 2005, 548(1–2), 148–158 CrossRef CAS.
- A. M. Michałowska-Kaczmarczyk and T. Michałowski, Stoichiometric approach to redox back titrations in ethanol analyses, Ann. Adv. Chem., 2019, 3, 1–6 Search PubMed.
- P. Pinyou, N. Youngvises and J. Jakmunee, Flow injection colorimetric method using acidic ceric nitrate as reagent for determination of ethanol, Talanta, 2011, 84(3), 745–751 CrossRef CAS PubMed.
- H. G. Alemea, L. M. Costa and P. G. Barbeira, Determination of ethanol and specific gravity in gasoline by distillation curves and multivariate analysis, Talanta, 2009, 78(4–5), 1422–1428 CrossRef.
- P. G. Mazzone, X. F. Wang, Y. Xu, T. Mekhail, M. C. Beukemann, J. Na, J. W. Kemling, K. S. Suslick and M. Sasidhar, Exhaled breath analysis with a colorimetric sensor array for the identification and characterization of lung cancer, J. Thorac. Oncol., 2011, 7(1), 137–142 CrossRef.
- X. Zhong, D. Li, W. Du, M. Yan, Y. Wang, D. Huo and C. Hou, Rapid recognition of volatile organic compounds with colorimetric sensor arrays for lung cancer screening, Anal. Bioanal. Chem., 2018, 410, 3671–3681 CrossRef CAS.
- K. S. Suslick, N. A. Rakow and A. Sen, Colorimetric sensor arrays for molecular recognition, Tetrahedron, 2004, 60(49), 11133–11138 CrossRef CAS.
- C. Zhang and K. S. Suslick, A colorimetric sensor array for organics in water, J. Am. Chem. Soc., 2005, 127(33), 11548–11549 CrossRef CAS PubMed.
- T. Lin, Y. Wu, Z. Li, Z. Song, L. Guo and F. Fu, Visual monitoring of food spoilage based on hydrolysis-induced silver metallization of Au nanorods, Anal. Chem., 2016, 88(22), 11022–11027 CrossRef CAS PubMed.
- H. Xia, J. Hu, J. Tang, K. Xu, X. Hou and P. Wu, A RGB-type quantum dot-based sensor array for sensitive visual detection of trace formaldehyde in air, Sci. Rep., 2016, 6(1), 36794 CrossRef CAS.
- N. He, C. Zhang, X. Qi, S. Zhao, Y. Tao, G. Yang, T. H. Lee, X. Wang, Q. Cai, D. Li and M. Lu, Draft genome sequence of the mulberry tree Morus notabilis, Nat. Commun., 2013, 4(1), 2445 CrossRef.
- H. Lin, Z. X. Man, B. B. Guan, Q. S. Chen, H. Jin and Z. L. Xue, In situ quantification of volatile ethanol in complex components based on colorimetric sensor array, Anal. Methods, 2017, 9(40), 5873–5879 RSC.
- W. Peng, Y. Qin, W. Li, M. Chen, D. Zhou, H. Li, J. Cui, J. Chang, S. Xie, X. Gong and B. Tang, Nonenzyme cascaded amplification biosensor based on effective aggregation luminescence caused by disintegration of silver nanoparticles, ACS Sens., 2020, 5(7), 1912–1920 CrossRef CAS.
- Y. Li, S. Chen, D. Lin, Z. Chen and P. Qiu, A dual-mode nanoprobe for the determination of parathion methyl based on graphene quantum dots modified silver nanoparticles, Anal. Bioanal. Chem., 2020, 412, 5583–5591 CrossRef CAS.
- A. Rossi, M. Zannotti, M. Cuccioloni, M. Minicucci, L. Petetta, M. Angeletti and R. Giovannetti, Silver nanoparticle-based sensor for the selective detection of nickel ions, Nanomaterials, 2021, 11(7), 1733 CrossRef CAS PubMed.
- S. Karakuş, G. Baytemir and N. Taşaltın, Digital colorimetric and non-enzymatic biosensor with nanoarchitectonics of Lepidium meyenii-silver nanoparticles and cotton fabric: real-time monitoring of milk freshness, Appl. Phys. A:Mater. Sci. Process., 2022, 128(5), 390 CrossRef.
- Q. K. Vu, T. H. Nguyen, A. T. Le, N. P. Vu, X. D. Ngo, T. K. Nguyen, T. T. Nguyen, C. Van Pham, T. L. Nguyen, T. L. T. Dang and M. Tonezze, Enhancing electron transfer and stability of screen-printed carbon electrodes modified with AgNP-reduced graphene oxide nanocomposite, J. Electron. Mater., 2022, 51(3), 1004–1012 CrossRef CAS.
- J. Liu, R. Siavash Moakhar, A. Sudalaiyadum Perumal, H. N. Roman, S. Mahshid and S. Wachsmann-Hogiu, An AgNP-deposited commercial electrochemistry test strip as a platform for urea detection, Sci. Rep., 2020, 10(1), 9527 CrossRef CAS.
- M. A. Farag, M. M. Elmassry and S. H. El-Ahmady, The characterization of flavored hookahs aroma profile and in response to heating as analyzed via headspace solid-phase microextraction (SPME) and chemometrics, Sci. Rep., 2018, 8(1), 17028 CrossRef PubMed.
- A. Serag, A. Zayed, A. Mediani and M. A. Farag, Integrated comparative metabolite profiling via NMR and GC–MS analyses for tongkat ali (Eurycoma longifolia) fingerprinting and quality control analysis, Sci. Rep., 2023, 13(1), 2533 CrossRef CAS PubMed.
- N. Hegazi, A. R. Khattab, H. H. Saad, B. Abib and M. A. Farag, A multiplex metabolomic approach for quality control of Spirulina supplement and its allied microalgae (Amphora & Chlorella) assisted by chemometrics and molecular networking, Sci. Rep., 2024, 14(1), 2809 CrossRef CAS PubMed.
- Y. Wang, D. Huo, H. Wu, J. Li, Q. Zhang, B. Deng, J. Zhou, M. Yang and C. Hou, A visual sensor array based on an indicator displacement assay for the detection of carboxylic acids, Microchim. Acta, 2019, 186, 1–12 CrossRef.
- J. C. Jin, Z. Q. Xu, P. Dong, L. Lai, J. Y. Lan, F. L. Jiang and Y. Liu, One-step synthesis of silver nanoparticles using carbon dots as reducing and stabilizing agents and their antibacterial mechanisms, Carbon, 2015, 94, 129–141 CrossRef CAS.
- D. Ballabio, A MATLAB toolbox for Principal Component Analysis and unsupervised exploration of data structure, Chemom. Intell. Lab. Syst., 2015, 149, 1–9 CrossRef CAS.
- C. M. Bishop and N. M. Nasrabadi, Pattern Recognition And Machine Learning, Springer, 2006, vol. 4, 4, p. 738 Search PubMed.
- T. Hastie, R. Tibshirani and J. Friedman, The Elements Of Statistical Learning, 2009 Search PubMed.
- L. Kaufman and P. J. Rousseeuw, Finding Groups in Data: an Introduction to Cluster Analysis, John Wiley & Sons, 2009 Search PubMed.
- P. S. Kumar, S. Abhilash, K. Manzoor, S. V. Nair, H. Tamura and R. Jayakumar, Preparation and characterization of novel β-chitin/nanosilver composite scaffolds for wound dressing applications, Carbohydr. Polym., 2010, 80(3), 761–767 CrossRef CAS.
- K. Varghese Alex, P. Tamil Pavai, R. Rugmini, M. Shiva Prasad, K. Kamakshi and K. C. Sekhar, Green synthesized Ag nanoparticles for bio-sensing and photocatalytic applications, ACS Omega, 2020, 5(22), 13123–13129 CrossRef CAS PubMed.
- D. Paramelle, A. Sadovoy, S. Gorelik, P. Free, J. Hobley and D. G. Fernig, A rapid method to estimate the concentration of citrate capped silver nanoparticles from UV-visible light spectra, Analyst, 2014, 139(19), 4855–4861 RSC.
- T. E. Agustina, W. Handayani and C. Imawan, June. The UV-VIS spectrum analysis from silver nanoparticles synthesized using Diospyros maritima blume, Leaves extract, in 3rd KOBI Congress, International and National Conferences (KOBICINC 2020), Atlantis Press, 2021, pp. 411–419 Search PubMed.
- J. F. B. Rodrigues, E. P. S. Junior, K. S. Oliveira, M. R. R. Wellen, S. S. Simões and M. V. L. Fook, Multivariate Model Based on UV-Vis Spectroscopy and Regression in Partial Least Squares for Determination of Diameter and Polydispersity of Silver Nanoparticles in Colloidal Suspensions, J. Nanomater., 2020, 1279862 CAS.
- M. Zayats, A. B. Kharitonov, S. P. Pogorelova, O. Lioubashevski, E. Katz and I. Willner, Probing photoelectrochemical processes in au− cds nanoparticle arrays by surface plasmon resonance: application for the detection of acetylcholine esterase inhibitors, J. Am. Chem. Soc., 2003, 125(51), 16006–16014 CrossRef CAS.
- A. Amirjani and D. F. Haghshenas, Ag nanostructures as the surface plasmon resonance (SPR)- based sensors: a mechanistic study with an emphasis on heavy metallic ions detection, Sens. Actuators, B, 2018, 273, 1768–1779 CrossRef CAS.
- H. Zhu, E. Prince, P. Narayanan, K. Liu, Z. Nie and E. Kumacheva, Colloidal stability of nanoparticles stabilized with mixed ligands in solvents with varying polarity, Chem. Commun., 2020, 56(58), 8131–8134 RSC.
- M. Yoosefian and N. Etminan, The role of solvent polarity in the electronic properties, stability and reactivity trend of a tryptophane/Pd doped SWCNT novel nanobiosensor from polar protic to non-polar solvents, RSC Adv., 2016, 6(69), 64818–64825 RSC.
| This journal is © The Royal Society of Chemistry 2025 |


