Open Access Article
Open Access ArticleGreen synthesis of biocompatible silver nanoparticles using Trillium govanianum rhizome extract: comprehensive biological evaluation and in silico analysis†
Syed Ifrah
Manzoor
a,
Farhat
Jabeen
a,
Rajan
Patel
 b,
M. Moshahid
Alam Rizvi
c,
Khalid
Imtiyaz
c,
Maqsood Ahmad
Malik
b,
M. Moshahid
Alam Rizvi
c,
Khalid
Imtiyaz
c,
Maqsood Ahmad
Malik
 *d and
Tanveer A.
Dar
*d and
Tanveer A.
Dar
 *a
*a
aDepartment of Clinical Biochemistry, University of Kashmir, Srinagar-190006, Jammu and Kashmir, India. E-mail: tanveerali@kashmiruniversity.ac.in
bCentre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi-110025, India
cDepartment of Biosciences, Faculty of Life Sciences, Jamia Millia Islamia, New Delhi-110025, India
dDepartment of Chemistry, Faculty of Sciences, Jamia Millia Islamia, New Delhi-110025, India. E-mail: mamalik@jmi.ac.in
First published on 18th December 2024
Abstract
Green synthesis of silver nanoparticles (AgNPs), using medicinal plants, is widely practiced due to the diverse bioactive compounds present in these plants, which can act synergistically with the AgNPs. Notably, the rhizome of Trillium govanianum Wall Ex. Royle is rich in bioactive phytochemicals such as steroids, saponins, glycosides, and fatty acids. This study aims to synthesize and characterize AgNPs mediated by the Trillium govanianum rhizome (TGRAgNPs) and comprehensively investigate their biological activities. The results indicated that phytochemicals in the rhizome may function as reducing and capping agents in the synthesis of TGRAgNPs. Overall, this study presents a simple, rapid, cost-effective, and eco-friendly method for nanoparticle production. Subsequently, the TGRAgNPs were characterized using UV-vis spectroscopy, FTIR, XRD, DLS, zeta potential, SEM-EDX, and TEM analyses. GC-MS analysis of the extract identified several phytochemicals that might play crucial role in the synthesis of TGRAgNPs. Moreover, TGRAgNPs demonstrated significant antioxidant, anticancer (against HCT-116 cell lines), anti-inflammatory, and DNA protection activities. Additionally, hemolytic assay confirmed the non-toxic and hemo-compatible nature of TGRAgNPs at all tested concentrations. The findings suggest that the green synthesized TGRAgNPs have promising potential as biocompatible antioxidant, anti-inflammatory, and anticancer agents in biomedicine.
1. Introduction
Nanoscience is rapidly emerging as a pivotal field for producing various metal nanoparticles, which have diverse applications in biomedicine and diagnostics.1,2 Due to their unique physicochemical properties, these metal nanoparticles hold great promise in biotechnology, biomedicine, optoelectronics, diagnostics, therapy, and bio-sensing.3,4 Silver nanoparticles (AgNPs) have garnered significant attention for their exceptional biocompatibility, low toxicity, and multiple biological activities, including antimicrobial, anticancer, and antioxidant effects.5,6 Consequently, the synthesis of AgNPs has become a focal point for the scientific community. Several physicochemical methods, such as gamma-ray radiation, autoclaving, electrochemical techniques, and chemical reduction approaches, are being explored to achieve efficient yields of AgNPs.7 However, these methods are often expensive, time-consuming, energy-intensive, and require extreme experimental conditions like high temperatures, pressures, and sophisticated equipment. The adverse effects of toxic agents adsorbed and capped on nanoparticle surfaces remain a significant concern.8,9 Conversely, biological approaches using bio-resources such as plants and microorganisms are becoming increasingly popular for synthesizing AgNPs with biomedical applications. These methods are cost-effective, less time-consuming, have reduced toxicity, and are more sustainable compared to traditional physicochemical techniques.10–12 Furthermore, these biological agents are inherently biocompatible and biodegradable, allowing for simple scale-up to large-scale production.13 Generally, for a process to be suitable for its scale-up to industrial level, a number of criteria need to be satisfied such as – (i) the raw materials, products, by-products, and wastes must be environmentally friendly, (ii) the required raw material should be producible, industrially abundant, and available in large scale, (iii) the conditions required for the process should be well defined with an ability to be controlled automatically and (iv) the production process should be economically appropriate for investment. In light of these, green synthesized nanoparticles are emerging as potential candidates for large-scale nanoparticle production. Among the synthesizing agents, plant extracts are particularly advantageous for the green synthesis of AgNPs as the phytochemical constituents in the extracts prevent nanoparticle aggregation and efficiently reduce metal precursors, resulting in the rapid formation of stable nanoparticles.14–16 More importantly, the coated bioactive phytochemical constituents exhibit synergistic effects with the synthesized AgNPs. Due to this, traditional Indian medicinal herbs have attracted increased attention for their use in nanoparticle synthesis.17–21To date, several Indian medicinal herbs such as Ocimum tenuiflorum,22Lysinibacillus fusiformis,23Azadirachta indica,24ficus bengalensis,25ocimum sanctum,26Tridax procumbens27etc., have been employed in green fabrication of AgNPs with significant biocompatibility and multifarious biological activities such as antioxidant, anti-inflammatory, cytotoxicity and DNA protection. In addition to this, Solanum trilobatum, Indoneesiella echioides, Azadirachta indica and Achillea biebersteinii-mediated AgNPs exhibit potent biological activities, including antimicrobial, antioxidant, and cytotoxicity.28–31 Furthermore, P. fulges-mediated AgNPs showed effective cytotoxicity against various cell lines, while banana peel extract-mediated synthesized nanoparticles exhibit potent antibacterial properties.32Trillium govanianum Wall. ex D. Don, native to high-altitude ranges of the Himalayas, is also a high-value medicinal plant from the family Melanthiacea. Morphologically, Trillium govanianum is a small herb characterized by three ovate leaves and a distinct stalk. It is abundant in bioactive phytochemicals, including fatty acids, phenols, hydrocarbons, saponins, carbohydrates, and steroids, contributing to its diverse biological activities.33 Among different parts of the plant, rhizomes have garnered significant attention due to the rich content of potent bioactive compounds such as diosgenin and glycosides, which are widely utilized in preparing steroidal and sex hormones. So far, only a single study has reported on Trillium govanianum-mediated AgNPs with antimicrobial properties.34 The present investigation marks the first comprehensive exploration of a simple, sustainable, rapid and cost-effective method for the green synthesis of highly biocompatible AgNPs, utilizing aqueous extract from Trillium govanianum rhizomes. Additionally, we conducted a detailed evaluation of the green synthesized biocompatible AgNPs for various biological activities such as antioxidant, anti-inflammatory, cytotoxicity and DNA damage protection. Moreover, in silico analysis was carried out to identify possible target molecules and pathways implicated in the biological activities of the synthesized silver nanoparticles.
2. Experimental
2.1. Materials
Silver nitrate (AgNO3, 99.5% purity), 1,1-diphenyl-2-picrylhydrazyl (DPPH), (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT), L-Ascorbic acid (99% purity), and methanol were purchased from Sigma Aldrich, USA. Trillium govanianum rhizome leaves were collected from the higher reaches of the Kashmir Himalayas. All other chemicals were of analytical grade with highest purity and were used without any additional purification.2.2. Preparation and phytochemical analysis of T. govanianum rhizome extract
The aqueous extract of T. govanianum was prepared by dissolving 5 mg of the extract in 10 mL of distilled water. To remove the debris/impurities, the aqueous extract was filtered through standard filter paper followed by Whatman filter paper No. 1. The filtered solution, referred here as Trillium govanianum rhizome (TGR) extract, was used as a reducing and stabilizing agent for the nanoparticle synthesis. Subsequently, Gas chromatography – Mass spectrometry (GC-MS) analysis of the TGR extract was performed using Agilent 7000 D triple quadrupole GC/MSD. Helium was used as the carrier gas at a flow rate of 1 mL min−1. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v sample dilution in hexane was made, and 1.0 μL of sample was injected. The temperature of the column was first adjusted to 50 °C. It was then progressively raised to 150 °C at a 3 °C min−1 rate and eventually raised to 250 °C at 10 °C min−1. The electron emission and ionization energy were 100 μA and 70 eV, respectively. The inbuilt NIST library of the GC-MS instrument was used to establish the structure, name, and molecular structure of the phytochemicals of TGR extract.
100 v/v sample dilution in hexane was made, and 1.0 μL of sample was injected. The temperature of the column was first adjusted to 50 °C. It was then progressively raised to 150 °C at a 3 °C min−1 rate and eventually raised to 250 °C at 10 °C min−1. The electron emission and ionization energy were 100 μA and 70 eV, respectively. The inbuilt NIST library of the GC-MS instrument was used to establish the structure, name, and molecular structure of the phytochemicals of TGR extract.
2.3. Synthesis of Trillium govanianum-mediated AgNPs (TGRAgNPs)
For the preparation of TGRAgNPs, 50 mL of 0.1 M silver nitrate aqueous solution was taken in a 250 mL beaker with constant stirring at 30 °C. To this solution, 50 mL of TGR extract was added and stirred for 15 minutes. Initially, the formation of TGRAgNPs was observed by a colour change from yellowish to dark brown, followed by its confirmation through UV-visible spectra measurements at different time intervals and days. The prepared TGRAgNPs were collected as pellets after centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 minutes, followed by several washings with deionized water and ethanol to remove unreacted material or impurities. The final green synthesized TGRAgNPs were dried at 60 °C for 24 h and stored in a dark glass bottle at 4 °C for further characterization and activity studies.
000 rpm for 20 minutes, followed by several washings with deionized water and ethanol to remove unreacted material or impurities. The final green synthesized TGRAgNPs were dried at 60 °C for 24 h and stored in a dark glass bottle at 4 °C for further characterization and activity studies.
2.4. Characterization of TGRAgNPs
The bioreduction of silver ions to silver nanoparticles was characterized by monitoring its UV-vis spectra using Cary-100 UV-visible spectrophotometer at wavelength range of 200–800 nm. The spectra were taken at different time intervals to confirm the complete reduction of Ag+ to Ag0. To assess the stability of the nanoparticles, the spectra were taken over a period of 5 days and at different temperatures. The functional groups responsible for capping/stabilizing the AgNPs and the phytochemical constituents of the plant extract were identified by FTIR (Fourier transform infrared) spectroscopy. The FTIR spectra were collected using the Burker Tensor 37 instrument. All the spectroscopic measurements were taken in 450–4000 cm−1 range. Scanning electron microscopy (SEM) was performed to confirm the surface morphology of the nanoparticle. For this study, a thin layer of the TGRAgNPs was placed on a carbon-coated copper grid. The grid was kept under a mercury lamp for 5 minutes to allow the sample to completely dry and the images were recorded. Furthermore, the size, shape and the corresponding selected area electron diffraction (SAED) were analysed with a transmission electron microscope (TEM). SAED is a two-dimensional electron diffraction pattern used to study the lattice parameters and crystallinity of materials. The samples for TEM analysis were prepared by dissolving the TGRAgNPs in distilled water. The sample was sonicated for 5 minutes, placed on the copper grid, and dried. The images were recorded at different magnification powers. The X-ray diffraction (XRD) technique was employed to ascertain the crystal structure of TGRAgNPs by examining the location and intensities of diffraction peaks, commonly observed in highly crystalline substances. The diffraction pattern was captured within a range of diffraction angles spanning from 20° to 80°. The XRD investigations were performed with a D8 Advance, Bruker, Germany X-ray diffractometer with Cu Kα (1.542 Å) radiation source. Dynamic light scattering (DLS) approach was employed to assess the hydrodynamic particle size distribution, polydispersity index (PDI), and ζ-potential of the nanoparticles using a Zetasizer nano ZS equipment (Malvern Instruments, Malvern, U.K.). The light scattering angle was set at 90°, and the size measurements were performed at 25 °C. The electrophoretic cell was utilized to measure the surface charge, also known as the ζ-potential, of the TGRAgNPs in an electric field. The preparation of the sample mainly involved the dilution of the nanoparticles with MilliQ water and subjecting them to ultrasonication for 5 minutes, resulting in the formation of a uniformly disseminated suspension.2.5. Free radical scavenging activity
For assessing the antioxidant activity of the TGRAgNPs, the DPPH assay was performed as per the established method35 with slight modifications. Different concentrations of TGRAgNPs (0.05, 0.1, 0.15, 0.2, and 0.25 mg mL−1) were added to 1 mL of DPPH (0.02 mg mL−1), with vigorous shaking followed by incubation at room temperature for 30 minutes. Ascorbic acid was used as a positive control, and methanol as a blank. The absorbance was measured at 515 nm and analyzed using a Cary 300 UV-vis spectrophotometer. The percentage scavenging activity was calculated by the following equation: | (1) |
2.6. Hemo-compatibility assay
Compatibility of TGRAgNPs for red blood cells (RBCs) was observed using a haemolytic assay as per the already established protocol,36 with slight modifications. Briefly, fresh blood samples (5 mL) were collected from healthy volunteers at the Diagnostic Centre of the Department of Clinical Biochemistry, University of Kashmir, in EDTA tubes. The blood samples were centrifuged at 2000 rpm for 15 minutes at 4 °C to separate the cells from the serum. After separation from the serum, the precipitated RBCs were washed thrice in PBS buffer (pH 7.4). RBCS were dispersed in PBS to prepare RBC suspension (1%). Following this, 1 mL of RBC suspension in separate tubes was mixed with different concentrations of TGRAgNPs (0.05, 0.1, 0.2, 0.5, and 1 mg mL−1). Following incubation at 37 °C, the tubes were centrifuged at 800 g for 5 minutes. The supernatant formed was collected, and measured its absorbance at 545 nm. RBCs were treated with phosphate-buffered saline (PBS) and Triton-X as negative and positive controls, respectively. All the experiments were done in triplicates. The percentage of haemolysis was calculated by the following formula: | (2) |
2.7. Cytotoxic activity assay
Cytotoxic effect of TGRAgNPs was performed on the HCT-116 cell line under in vitro conditions, using standard MTT assay.37 The cell lines were obtained from the National Centre for Cell Science, Pune, India. In this assay, HCT-116 cells were plated in a 96-well plate at a density of 1 × 104 cells per well and allowed to attach overnight. After removing the supernatant, different concentrations of TGRAgNPs (0.05–0.5 mg mL−1) were added to each well, and the cells were allowed to grow at 37 °C with 5% CO2 for 24 h. Following this, MTT was added to each well, and plates were kept at 37 °C for 4 h. After 4 h, MTT was replaced with 200 μL DMSO, and cells were agitated for 5 minutes. Absorbance at 570 nm was measured using an iMark Microplate reader (Biorad, USA). Absorbance measured was used to calculate the percentage of cell cytotoxicity using the formula: | (3) |
2.8. Anti-inflammatory activity
In vitro anti-inflammatory activity of TGRAgNPs was carried out as per the already established protocol38 with some minor modifications. Briefly, the reaction mixture consisted of 2.8 mL of PBS, 0.2 mL of 5% Bovine Serum Albumin (BSA), and different volumes of the TGRAgNPs with a concentration range of 0.1–0.4 mg mL−1. The total volume was made up to 5 mL using PBS. The mixture was then incubated at 37 °C for 20 minutes and then at 70 °C for 10 minutes. After cooling, the absorbance of the mixture was monitored at 660 nm. The solution of PBS (2.8 mL) and BSA (0.2 mL) was used as control, and diclofenac sodium was taken as a standard. The following equation was used to calculate the inhibition percentage of protein denaturation.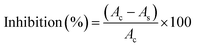 | (4) |
2.9. DNA damage protection assay
The DNA damage protection potential of TGRAgNPs was evaluated by DNA nick assay as described earlier39 with minor modifications. In this, a mixture of 20 μL of calf thymus DNA and 1 μL of different concentrations of TGRAgNPs (0.1–0.9 mg mL−1) were added to 3 μL of Fenton's reagent (30 mM H2O2, 100 mM ascorbic acid and 20 mM ferric nitrate). The reaction mixture was then incubated at 37 °C for 20 minutes. Post incubation, 3 μL of orange G dye (6×) was added to the mixture. Following this, the reaction mixture was loaded on 1% agarose gel, and electrophoresis was run for 10 minutes at 155 V, followed by ethidium bromide staining. Observed bands were visualized using an imager gel documentation system (Benchtop UV Transilluminator).2.10. Experimental and statistical analysis
All the experiments were conducted in triplicate and One-Way ANOVA with Bonferroni t-test was applied to analyse inter-group differences and the results obtained were presented either as mean value ± standard deviation or the corresponding percentage error, wherever applicable, with respective p values as indicated in the respective result sections. The spectra and profiles displayed are indicative of a minimum of three separate measurements for each concentration.3. Network pharmacology-based analysis
3.1. Physicochemical parameters of palmitic acid
The SMILES notation for palmitic acid was provided as input to SwissADME, a publicly accessible online tool (https://www.swissadme.ch/), designed for rapid and robust prediction of physicochemical characteristics and pharmacokinetic parameters, including ADME (Absorption, Distribution, Metabolism, and Excretion) properties. SwissADME employs state-of-the-art computational methodologies to efficiently compute drug-likeness (DL), oral bioavailability (OB), Caco-2 permeability, lipophilicity, and other relevant features for naturally occurring small molecules. The evaluation of the compound's drug-likeness was determined in accordance with Lipinski's rules and other guidelines.403.2. Identification of palmitic acid target genes
In this, the saturated data file (SDF), containing the chemical structure of palmitic acid, was acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). Subsequently was imported to two separate web-based server databases, PharmMapper (https://lilab.ecust.edu.cn/pharmmapper/)41 and SwissTarget42 for identification of its related target genes. The list of potential target genes was then retrieved as a CSV file, and the gene names were standardized to gene symbols by consulting the UniProt database (https://www.uniprot.org).3.3. Protein–protein interaction (PPI) network construction
The PPI network is a pictorial representation of the highly specific protein–protein edge interaction for the identification of key genes and potential biomarkers involved in several biological processes. For this, the Network Analyst free web tool, STRING, was used to construct the PPI network based on the physical connections between the palmitic acid gene targets.43,44 A minimum cutoff confidence score of 0.70 was used to develop the network, and the output data was analysed using cytoscape 3.8.2.3.4. Gene ontology (GO) and pathway enrichment analysis
For this, functional analysis was carried out to evaluate the biological activities of the palmitic acid gene targets through GO and pathway analysis by using Shiny GO.45 Generally, GO is a powerful computational tool for performing enrichment analysis on gene sets wherein the GO terms and pathways are primarily grouped into three biological functions i.e., biological process (BP), Cellular component (CC) and Molecular function (MF).46 The pathway-related data was extracted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. KEGG pathway is a collection of pathway maps used to understand the high-level functioning of biological processes, including metabolism, human diseases and drug development.4. Results and discussion
Developing reliable and eco-friendly approaches for generating biologically active nanoparticles is an emerging area of research in nanotechnology. Generally, green synthesis employs environmental friendly natural materials, instead of chemicals, for nanoparticle synthesis. Among these, plant-based extracts have been widely utilized to synthesize different NPs with defined size, shape, and composition. Indeed, these extracts contain several classes of phytochemicals such as polyphenols, alkaloids, saponins, etc. that carry out reduction of metal.47 Moreover, plant synthesized NPs, as compared to chemically synthesized ones, exhibit multifarious biological activities with least side effects. In light of this, the present study employed Trillium govanianum rhizome aqueous extract for green synthesis of silver nanoparticles, termed Trillium govanianum-silver nanoparticles (TGRAgNPs), followed by their characterization and evaluation of several biological activities.4.1. Synthesis of Ag NPs and GC-MS analysis of TGR extract
Generally, the synthesis of nanoparticles is ascertained by the visual colour change from pale yellow to brown. In light of this, 50 mL of 0.1 M AgNO3 solution (Ag+) was incubated with 50 mL of TGR extract (5 mg mL−1), gradually changing the solution colour from pale yellow to dark brown within 15 minutes. Colour change, with time-dependent darkness of the brown colour, implies the reduction of Ag+ to Ag0 and resulting in the synthesis of TGRAgNPs. Post few hours, no change in colour intensity was observed, indicating complete reduction of silver ions in the reaction mixture. It is reported that the appearance of dark brown colour is due to the surface plasmon resonance (SPR) of AgNPs, regarded as an optical property of AgNPs.48Fig. 1 illustrates the schematic representation of the synthesis of TGRAgNPs as well as the observed change in solution colour. | ||
| Fig. 1 Schematic illustration of Trillium govanianum rhizome extract mediated green synthesis of TGRAgNPs. | ||
The phytochemical components present in the methanolic rhizome extract of T. govanianum were further identified by GC-MS analysis. After careful analysis of the GC-MS chromatogram (Fig. S1, ESI†), the chemical profile of the rhizome extract and percent amount of the individual components obtained are summarized in Table S1 (ESI†). As can be seen from the results, GC-MS analysis of the TGR extract indicated presence of different classes of phytochemicals such as fatty acid esters, alkaloids, phenols, saponins, carbohydrates, and steroids (Table S1, ESI†). As can be seen from the results, 28 major compounds were identified in the methanolic extract of T. govanianum rhizome with corresponding significant peaks observed in the GC-MS chromatogram (Fig. S1, ESI†). Altogether, the dominant phytochemicals mainly included n-hexadecanoic acid (100%), 9,12-octadecanoic acid methyl ester (67.5), palmitoleic acid (45.7), 9,12-octadecadienoic acid (29%), alpha tocopherol-beta-D-mannoside (35.3%), hexadecanoic acid, 2-hydroxy-1-ethyl ester (20.4%), 9-octadecadienoic acid, methyl ester (20.15%), 11,14,17-eicosatrienoic acid (18%), diosgenin (17%), methyl hexadec-9-enoate (13%) and triacontane (10%). Interestingly, hexadecanoic acid (palmitic acid) and 9,12-Octadecadienoic acid (linoleic acid), observed as major constituents of our extract, have been reported to exhibit anticancer activity.33,49 Moreover, diosgenin is reported to exhibit hypoglycaemic, hypolipidemic, antioxidant, anti-inflammatory, and antiproliferative activities while as tocopherol and astaxanthin are reported to have antioxidant properties.50,51 As observed in our results, similar other studies have also reported the presence of n-hexadecanoic acid methyl ester, n-hexadecanoic acid, 9,12-octadecadienoic acid (Z, Z) and octadecanoic acid in the methanolic rhizome extract of trillium govanianum.52
It is a well-established fact that these phytochemicals contribute towards the reduction of Ag+ by donating the electrons, transforming them into a large number of silver nuclei or zero-valent (Ag0) atoms (Fig. 2).53 The bioreduction process involves three main stages: activation, nucleation and growth. The reduction of Ag+ ions occur in the activation stage which is followed by the nucleation of the reduced Ag atoms. During the growth stage, the spontaneous coalescence of small AgNPs into large-sized AgNPs takes place. In this final stage, where the AgNPs attain their ultimate shape, there is an increase in their thermodynamic stability, a process known as Ostwald ripening. It has been shown that phytochemicals act as stabilizing agents by creating a protective phytochemical cage around the nanoparticles. Through this process, these phytochemicals prevent further aggregation through steric hindrance, resulting in the formation of a stable and uniformly shaped AgNPs.54
Interestingly, earlier studies on Trillium govanianum rhizome also indicated the presence of secondary metabolites such as fatty acid esters, glycosides, saponins, steroids, and carbohydrates in the rhizome.55 In light of the above, these secondary metabolites might have mediated the reduction of silver, thereby leading to the synthesis of nanoparticles. As represented in Fig. 1, the obtained Ag0 once formed nucleates into small clusters, which in turn create the nanoparticles. As observed in GC-MS analysis, the aqueous TGR extract is rich in diverse phytochemicals with an ability to effectively chelate metal ions and convert them into nanoparticles. The ability to produce NPs is ascribed to their abundance of essential functional groups, including numerous hydroxyl and carboxylic acid groups, as depicted in Fig. 1. As a result, the abundant phytochemicals, such as palmitic acid in the extract, facilitated the easy conversion of Ag+ to Ag0 through bioreduction (Fig. 2). Additionally, the organic functional groups present in the extract can bind to the surface of silver nanoparticles, preventing their aggregation and oxidation.
4.2. Characterization of TGRAgNPs
 | ||
| Fig. 3 (a) UV-visible spectra of TGR extract and TGR extract synthesised AgNPs (Inset: optical images of TGRAgNPs at different time intervals), (b) FTIR spectra and (c) XRD diffractogram of TGRAgNPs. | ||
In addition, the stability of the TGRAgNPs was monitored at different temperatures, and the ageing effect was studied for 5 days (Fig. S2a and b, ESI†). As can be seen from the results, TGRAgNPs showed a characteristic SPR λmax peak at 426 nm with a slight increase in absorption intensity without any shift in the peak wavelength over the time period of 5 days (Fig. S2a, ESI†). In fact, the obtained suspension maintained a high degree of stability even after extended periods of storage and ageing. The existence of AgNPs during the first stages of the reaction indicated that even a short reaction time is sufficient for the formation of the nanoparticles. Furthermore, the thermal stability of the synthesized TGRAgNPs was monitored by heating the reaction mixture at different temperatures i.e., 5, 30, 40, and 60 °C (Fig. S1b, ESI†). At 60 °C, the maximum intensity of SPR λmax peak was still observed at 426 nm with a temperature-dependent increase in absorption, which in turn demonstrates the temperature-induced increased rate of AgNP synthesis. Overall, the results concluded that while increasing the reaction temperature, the absorbance intensity also increased without any shift in the peak position.
4.3. FTIR analysis
Identification of the secondary metabolites, most probably responsible for the reduction and capping of TGRAgNPs, was accomplished by using FTIR spectroscopy. The FTIR spectra of the TGR extract exhibited prominent absorption peaks at 3290, 2924, 2822, 1614, 1516, 1376, and 994 cm−1, as depicted in Fig. 3b. The phytochemical examination of the TGR extract indicated the presence of flavonoids, alkaloids, steroids, rosins, saponins, and proteins. The FTIR spectrum of TGRAgNPs exhibited prominent absorption peaks at 3305, 2932, 1605, 1359, and 1002 cm−1, indicating the presence of phytoconstituents that function as capping agents (Fig. 3b). Comparative analysis of the FTIR peaks of the TGRAgNPs and TGR extract identified a complete absence of the peak 1516 cm−1 in the nanoparticles, implying that the fatty acids with –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional groups are responsible for the stabilization and fabrication of nanoparticles. Furthermore, significant changes in other peak positions were observed in the FTIR spectra of the TGR extract, corresponding to TGRAgNPs, which can be attributed to the reduction, capping, and stabilization of the synthesized TGRAgNPs. The signal at 3290 cm−1 shifted to 3305 cm−1, indicating the participation of phenolic compounds in the TGR extract in stretching O–H bonds. The peak observed at 2932 cm−1 is a result of the stretching of the C–H bonds in the methylene or aliphatic group and is also a distinctive feature of triterpenoid saponins. The band observed at 1614 cm−1 shifted to a higher wavenumber of 1605 cm−1, indicating the presence of alkenyl or aromatic stretching. The band observed at 1359 cm−1 indicates the existence of the –C–O stretching, characteristic of phenol or tertiary alcohols. Similarly, the band at 1002 cm−1 corresponds to the O–H stretching of the phenol group. Most of the peaks correspond to the phenolic groups of the polyphenols, triterpenoids, alkaloids, steroids, and tannins adequately present in the extract. They might be, in turn, responsible for the formation of TGRAgNPs. Similar to our results, earlier studies of FTIR characterization of AgNPs have indicated that the peak at 3340 cm−1 is due to O–H bonds in OH functional groups, C–H stretching of alkanes resulted in peaks at 2925 cm−1, peak at 1736 cm−1 corresponds to carbonyl groups and a distinct peak at 1636 cm−1 may be related to C
O functional groups are responsible for the stabilization and fabrication of nanoparticles. Furthermore, significant changes in other peak positions were observed in the FTIR spectra of the TGR extract, corresponding to TGRAgNPs, which can be attributed to the reduction, capping, and stabilization of the synthesized TGRAgNPs. The signal at 3290 cm−1 shifted to 3305 cm−1, indicating the participation of phenolic compounds in the TGR extract in stretching O–H bonds. The peak observed at 2932 cm−1 is a result of the stretching of the C–H bonds in the methylene or aliphatic group and is also a distinctive feature of triterpenoid saponins. The band observed at 1614 cm−1 shifted to a higher wavenumber of 1605 cm−1, indicating the presence of alkenyl or aromatic stretching. The band observed at 1359 cm−1 indicates the existence of the –C–O stretching, characteristic of phenol or tertiary alcohols. Similarly, the band at 1002 cm−1 corresponds to the O–H stretching of the phenol group. Most of the peaks correspond to the phenolic groups of the polyphenols, triterpenoids, alkaloids, steroids, and tannins adequately present in the extract. They might be, in turn, responsible for the formation of TGRAgNPs. Similar to our results, earlier studies of FTIR characterization of AgNPs have indicated that the peak at 3340 cm−1 is due to O–H bonds in OH functional groups, C–H stretching of alkanes resulted in peaks at 2925 cm−1, peak at 1736 cm−1 corresponds to carbonyl groups and a distinct peak at 1636 cm−1 may be related to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching.57,58 On the other hand, an obvious peak at 1376 cm−1 could be related to the stretching vibrations of alcohols, ethers, esters, carboxylic acids, and amino groups, while the peak at 994 cm−1 might be attributed to C–H bending vibrations.57 In addition to this, it has been shown that the FTIR spectrum of green synthesized AgNPs exhibited absorption peaks at 3305 cm−1, 2932 cm−1, 1605 cm−1, 1359 cm−1 and 1002 cm−1 corresponding to O–H, C–H, alkene (C
O stretching.57,58 On the other hand, an obvious peak at 1376 cm−1 could be related to the stretching vibrations of alcohols, ethers, esters, carboxylic acids, and amino groups, while the peak at 994 cm−1 might be attributed to C–H bending vibrations.57 In addition to this, it has been shown that the FTIR spectrum of green synthesized AgNPs exhibited absorption peaks at 3305 cm−1, 2932 cm−1, 1605 cm−1, 1359 cm−1 and 1002 cm−1 corresponding to O–H, C–H, alkene (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), bending (C–H) and C–O bonds, respectively.58 Altogether, FTIR findings for the TGRAgNPs are quite consistent with the phytochemical analysis of the TGR extract.
C), bending (C–H) and C–O bonds, respectively.58 Altogether, FTIR findings for the TGRAgNPs are quite consistent with the phytochemical analysis of the TGR extract.
4.4. XRD analysis of TGRAgNPs
The biosynthesized TGRAgNPs were analysed by XRD for their crystalline nature and phase purity, as shown in Fig. 3c. The XRD pattern of TGRAgNPs exhibited four prominent diffraction peaks at 2θ = 37.79, 44.77, 64.89, and 76.70° indexed as (111), (200), (220), and (311) (JCPDS file No. 04-0783).59 The high intense peak in the XRD spectrum demonstrates the polycrystalline phase composition of TGRAgNPs, confirming the production of face-centered cubic (fcc) crystalline TGRAgNPs. The diffraction peaks of (111), (200), (220), and (311) planes were also observed in the green synthesis of AgNPs.60 XRD patterns confirmed the structure of TGRAgNPs as a cubic structure and confirmed the purity of the as-prepared AgNPs using TGR extract. Moreover, several reducing agents in the TGR extract are responsible for stabilizing TGRAgNPs, thus providing them with a crystalline structure quite well studied in various biosynthesized nanoparticles. The average crystallite size of TGRAgNPs was calculated using the Debye–Scherrer formula61 using eqn (5). From this, the calculated average crystallite size of the TGRAgNPs was found to be ∼32 nm. Overall, the results demonstrated that TGRAgNPs exist as small and monodispersed nanoparticles.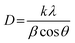 | (5) |
4.5. Particle size analysis
The dynamic light scattering measurements were performed to determine the hydrodynamic diameter, particle size distribution (PSD), and polydispersity index (PDI) of TGRAgNPs. The particle size distribution and zeta potential of the TGRAgNPs is shown in Fig. S3(a and b) (ESI†). The average hydrodynamic size of TGRAgNPs was 131.9 nm, which is larger than the size observed in the TEM analysis. The phytochemicals present in the TGR act as capping agents, resulting in the increase of the hydrodynamic diameter of TGRAgNPs. The PDI value of TGRAgNPs was found to be 0.148, indicating that they were relatively polydispersed.Zeta potential, regarded as an indicator of surface charge potential, is an important parameter for understanding the stability of nanoparticles in aqueous suspensions. Any charge on the surface of nanoparticles provides stability and prevents aggregation of nanoparticles by repulsion between like-charged particles. In view of this, the Zeta potential value of TGRAgNPs was found to be −38.0 mV (Fig. S3(b), ESI†). A high absolute value of zeta potential specifies a high electrical charge on the surface of the nanoparticles, which in turn might lead to strong repellent forces among the particles, thus preventing their agglomeration.62 Moreover, the strong negative charge of TGRAgNPs demonstrates negative charges all across the nanoparticles surface, indicating their great colloidal dispersivity and long-term stability. Literature studies have reported that nanoparticles with a zeta potential value higher than +30 mV or lower than −30 mV are considered quite stable in the dispersion medium.63 Results obtained here indicate the successful formation of the nanoparticles with long-term stability. The negative potential value might be due to the absorption of capping biomolecules present in the TGR extract. Overall, the results are quite consistent with some earlier reports, such as F. religiosa leaf extract mediated synthesis of AgNPs.64
4.6. Morphological analysis
Scanning electron microscopy (SEM) analysis was used to determine the surface morphology of TGRAgNPs, and energy dispersive X-ray analysis (EDX) was used to investigate the elemental composition of the as-prepared TGRAgNPs. The nanoparticles were primarily observed to be uniformly dispersed in polymorphic forms, with a small percentage being slightly aggregated, ellipsoidal, and irregularly granulated (Fig. 4a and b). Similar kinds of nanoparticles were observed during the Taraxacum officinale mediated synthesis of AgNPs.65 The size of TGRAgNPs from SEM micrographs was found to be 30–50 nm. From the EDX spectrum of TGRAgNPs shown in Fig. 4c, a high-intensity peak of Ag metal at 3 keV indicates that silver is the major constituent (80.30%) in the prepared sample. Other constituents like carbon (16.70%) and oxygen (3%) could be due to the presence of phytochemical compounds of TGR extract on the surface of the TGRAgNPs, validating phytochemicals as the capping and reducing agents.Furthermore, the surface morphology, size and particle size distribution were confirmed from TEM analysis of the TGRAgNPs at different nanoscale magnifications (Fig. 5a and b). The TEM images at different nanoscale magnifications revealed that TGRAgNPs are slightly agglomerated and nearly spherical to irregular shape. The particle size distribution histogram, (Fig. 5c), obtained using ImageJ software, represents the size distribution of the particles which in turn confirms the polydispersity of TGRAgNPs. Moreover, the results revealed that the determined particle size varied between 15–55 nm (Fig. 5c). The average particle size of TGRAgNPs was found to be ∼38 nm, nearly similar to the crystallite size and SEM observation of these nanoparticles. As can be seen in Fig. 5e, the high-resolution TEM images showed characteristics of lattice fringes with a lattice fringe width of 0.242 nm, corresponding to Ag (111) crystal planes.66 In addition, a typical selected-area diffraction (SAED) pattern demonstrates the continuous ring patterns without additional diffraction spots and rings of secondary phases, revealing their crystalline structure. TGRAgNPs exhibit four bright rings, indexed as (111), (200), (220), and (311) lattice planes of the crystalline silver face-centered cubic (fcc) structure, respectively (Fig. 5f).67 In addition to this, the results observed in the SAED pattern (Fig. 5f) are in good agreement with the XRD results of TGRAgNPs, wherein the most intense peak observed at 37.79° 2θ is related to Ag (111) crystal planes.68
 | ||
| Fig. 5 TEM micrographs at (a) 100 nm (b) 50 nm nanoscale magnifications, (c) size distribution histogram, (d) HR-TEM (e) HR-TEM with zoom-in showing lattice fringes and (f) SAED patterns of TGRAgNPs. | ||
4.7. Antioxidant activity
Based on our GC-MS analysis and earlier studies, TGR contains a range of phytochemicals, including fatty acids, steroids, carbohydrates, alkaloids, phenols, etc. Moreover, several authors have also reported the antioxidant activity of the rhizome of Trillium govanianum.69 In light of this, during the green synthesis of nanoparticles by TGRAgNPs, it is quite expected that the synthesized TGRAgNPs might be coated with the bioactive constituents of the extract. Previous literature reports have also shown possible capping of bioactive constituents during the green synthesis of nanoparticles.70 To evaluate this, DPPH assay was performed where DPPH is regarded as a lipophilic radical whose colour changes from purple to yellow on accepting hydrogen and electrons from the donor.71 In this, different concentrations of TGRAgNPs (0.05–0.25 mg mL−1) and TGR extract (0.05–0.25 mg mL−1), used as a control, were titrated into DPPH solution for the purpose of free radical scavenging. Ascorbic acid (0.3–8.3 μg mL−1) was used as a standard. The standard showed the radical scavenging activity of 81.5% at a concentration of 8.3 μg mL−1. As shown in Fig. 6a, TGRAgNPs revealed a significant dose-dependent free radical scavenging activity with the highest scavenging of 85.7% ± 1.4% at 0.25 mg mL−1 and the lowest of 29.8 ± 1.25% at 0.05 mg mL−1. On the other hand, the highest and lowest scavenging activities of TGR extract were, respectively, found to be 65.2 ± 0.7% and 19.5 ± 1% at 0.25 mg mL−1 and 0.05 mg mL−1. Similar studies with green synthesized silver nanoparticles also exhibited potential radical scavenging activity with concentration dependent inhibition.72 Moreover, the observed free radical scavenging activities of TGRAgNPs were quite comparable to the already reported green synthesized silver nanoparticles.73 It has been shown that scavenging ability of Chenopodium murale leaf extract-mediated silver nanoparticles was dose dependent with a highest and lowest activities of 65% and 13%, respectively.74 Similarly, A. tribuloides root extract-mediated AgNPs exhibited 64% activity while aqueous extract of E. scaber mediated showed 85% activity.75,76 In comparison to these studies, the TGRAgNPS exhibited much significant antioxidant activity and thus can serve as a potent antioxidant agent with a highest scavenging activity of ∼86%. Interestingly, a number of studies have shown that green synthesized nanoparticles show better radical scavenging potential as compared to chemically synthesized ones,77,78 probably due to the presence of a bioactive capping agents on their surfaces. Overall, results infer that TGRAgNPs exhibited potential antioxidant activity possibly due to the presence of the coated phytochemicals, which might include palmitic acid, diosgenin, astaxanthin and alpha tocopherol, observed during the GC MS analysis of the TGR extract (Table S1, ESI†).4.8. Hemo-compatibility of TGRAgNPs
With the increased clinical application of AgNPs, their biosafety is vital. Hence, assessment of hemo-compatibility of the synthesized TGRAgNPs is necessary in light of blood-contacting medical appliances. Generally, characteristics of AgNPs, such as size and surface area, interfere with their hemolytic activity. Normally, hemolysis of RBCs is due to their compromised membranes, resulting in hemolytic leakage. TGRAgNPs were elucidated for hemocompatibility by monitoring cytotoxicity on RBCs through haemoglobin (Hb) release. For that, suspended RBCs were titrated with different concentrations (0.05, 0.1, 0.2, 0.3, 0.4 and 0.5 mg mL−1) of TGRAgNPs. Results indicated a maximum hemolysis of 6 ± 0.2% at the highest concentration (0.5 mg mL−1) of TGRAgNPs with a minimum of 0.95 ± 0.1% at its lowest concentration (0.05 mg mL−1). TGR extract was used as a control, wherein the extract exhibited the highest haemolysis of 1.76 ± 0.3% at 0.5 mg mL−1 with a hemolysis of 0.09 ± 0.006% at its lowest concentration. PBS buffer (negative control) and TritonX-100 (positive control) exhibited 0.05% and 95% hemolysis, respectively. As shown in Fig. 6b, a hemolytic activity of 6.1% was observed in 0.5 mg mL−1, which is slightly higher than the acceptable value (i.e. 5%) for the clinical applications of the nanoparticles as per World Health Organization (WHO) guidelines. Except this, all other concentrations of the TGRAgNPs are well below the recommended value of WHO. Overall, the results infer that the synthesized TGRAgNPs are largely compatible with no significant hemolytic effect on RBCs. A number of studies have also observed the biocompatibility of green synthesized nanoparticles, such as cannabis sativa root extract mediated silver nanoparticles exhibited ∼7% haemolysis, and AgNPs synthesized from Annona reticulata leaves extract showed ∼14% haemolysis.79,80 Moreover, a number of polymeric nanoparticles have been shown to exhibit >10% hemolysis. Interestingly, as compared to these reports, the TGRAgNPs exhibit significant biocompatibility with promising biomedical applications. More importantly, green-synthesized nanoparticles are generally more biocompatible than chemically synthesized nanoparticles.81 So far, the actual mechanism for AgNPs-induced hemolysis is still not clear. However, it has been shown that on coming in contact with the body fluids, metallic silver undergoes ionization, leading to the release of Ag+, as per the response of the nanoparticle surface area, which in turn induces hemolysis of cells like RBCs, at least under in vitro conditions.82 In biological systems, neutralizing the ionized silver ions by ions like chloride, sulfide, etc., makes them unavailable for hemolytic activity. Moreover, the interaction of nanoparticles with other cellular components, such as proteins and antioxidants, might result in their cell lysis activity. In fact, literature studies report that there is always a release of Ag+ ions on the interaction of AgNPs with the RBC membrane.834.8. Cytotoxicity activity
The biological role of AgNPs has gained great interest in diagnosing and bioremediation of cancers. In this study, preliminary screening of the biosynthesized TGRAgNPs for cytotoxicity against the HCT-116 cell line was carried out through MTT Assay. 5-Fluorouracil (5FU), used as a standard, exhibited a cytotoxicity of 92.8 ± 2.8% at 4 μg mL−1. As shown in Fig. 7a, a concentration-dependent increase in cytotoxicity was observed with 84.4 ± 4.1% at 0.5mg mL−1 of TGRAgNPs. An IC50 value of 0.17 mg mL−1 was observed for TGRAgNPs-induced cytotoxicity. TGR Extract was used as a control. The highest cytotoxicity for extract was found to be 78 ± 4.4% at 0.5 mg mL−1. A number of other studies have also shown cytotoxicity of green synthesized nanoparticles. C.nudiflora mediated silver nanoparticles showed potent cytotoxicity against HCT-116 colon cancer cells with an IC50 value of 100 μg mL−1.84S. Jambolanum AgNPs caused a significant decrease in cell viability of the MCF-7 cells when compared to the plant extract.85 The AgNPs synthesized using Chaetomorpha linum exhibited dose-dependent toxicity, while Putranjiva roxburghii seed extract-mediated AgNPs showed anti-cancerous activity against HCT-116 cell lines at a concentration of 10 mg mL−1.86,87 Another study reported Avicennia marina synthesized-AgNPs exhibited anticancer activity at 80 μg mL−1.88 Moreover, it is a well-established fact that as compared to chemically synthesized nanoparticles, green synthesized nanoparticles are usually found to be more cytotoxic.89,90 Overall, a number of studies have reported the cytotoxicity activity of green synthesized silver nanoparticles,91 wherein numerous effects have been combined to elucidate the mechanism of NP-induced cytotoxicity. Among others, one of the plausible mechanisms is the ability of nanoparticles to scavenge free radicals.92 Observed antioxidant activity of TGRAgNPs, in the present study also supports the free radical-induced cytotoxicity of the nanoparticles. Interestingly, hexadecenoic acid (palmitic acid) has been shown to exhibit potent anticancer activity.93 Palmitic acid (PA) has demonstrated efficacy in inhibiting the proliferation of tumor cells, promoting tumor cell apoptosis, and influencing the cell cycle in most of the cancers including gastric, cervical, liver, breast, colorectal and other types of tumors. It has been shown that palmitic acid influences signal transduction pathways related to membranes, leading to tumor cell proliferation inhibition, induction of apoptosis, suppression of cell invasion and migration, and increase in the efficacy of chemoradiotherapy drugs. Additionally, PA is shown to possess immune system regulating properties and induces cellular autophagy, further contributing towards its anti-cancer mechanisms.94 Interestingly, the same compound has been observed as the most abundant phytochemical in the GC-MS analysis of the TGR extract and thus might be present as a capping agent on the AgNPs (Table S1, ESI†).4.9. Anti-inflammatory activity
Protein denaturation is regarded as a primary cause of inflammation, wherein denaturation is shown to produce auto antigens in diseases like arthritis, diabetes, and cancer, which are causes of inflammation.95 Hence, inhibition of protein denaturation should prevent inflammation. In light of this, the green synthesized TGRAgNPs were evaluated for their ability to inhibit the denaturation of BSA, a model protein for such assays. Sodium diclofenac was used as a standard drug and acted as a positive control. As shown in Fig. 7b, a dose-dependent anti-inflammatory potential of TGRAgNPs was observed with the highest inhibition of 87.7 ± 1.1% at 0.4 mg mL−1 of NPs. On the other hand, TGR extract showed the maximum inhibition of 68.75 ± 0.8% at 0.4 mg mL−1. In contrast, the positive control showed a maximum level of 93% at 0.4 mg mL−1. Overall, the results infer that TGRAgNPs exhibits more potent anti-inflammatory activity as compared to the extract indicating the enhanced accessibility and activity of the nanoparticles. Similar studies with A. tribuloides root extract showed inhibition of 69% at the highest concentration of 500 μg mL−1.96A. lycoctonum mediated Ag NPs showed an anti-inflammatory effect in a dose-dependent manner with inhibition of 91.78% at the highest concentration (500 μg mL−1).97 Another study reported silver nanoparticles from Microsorum punctatum were effective in inhibiting thermally induced albumin denaturation at different concentrations with a highest inhibition of 69% at 100 μg mL−1.98 The anti-inflammatory activities of synthesized Ag nanoparticles from Justica wynaadensis, showed an inhibition of around 40% at 200 μg mL−1.99 Silver nanoparticles synthesized from leaf extract of Madhuca longifolia have been shown to exhibit protein denaturation inhibition of 53.15 ± 0.87% at a concentration of 500 μg mL−1.100 As compared to these reports, TGRAgNPs showed a significant inhibition of 87.7% at 0.4 mg mL−1 of NPs. In light of the above results, the possible coating of the biosynthesized nanoparticles by the secondary metabolites like palmitic acid, diosgenin, astaxanthin, etc., identified in GC/MS analysis, might be responsible for the anti-inflammatory effects of nanoparticles. Moreover, TGR extract, as such, has been reported to exhibit anti-inflammatory activity.101 Some of the earlier studies also suggest that the lysosomal components released by neutrophils at the site of inflammation might be suppressed by the coated secondary metabolites of the nanoparticles.1024.10. DNA damage protection assay
The green synthesized TGRAgNPs were evaluated for their potential to protect against Fenton-induced calf thymus DNA damage. As can be seen in Fig. 8A, the untreated (control) calf thymus DNA (lane 1) and TGRAgNPs treated DNA maintained their integrity. However, the Fenton reagent induced damage to the DNA is observed as the complete absence of the DNA band in the electrophoresis gel (lane 2). As seen in lanes 3–6 (Fig. 8A), TGRAgNPs were able to protect the Fenton-induced DNA damage, as evidenced by the appearance of intact DNA bands in the lanes. A concentration-dependent protection was observed as reflected by the increased intensity of the bands with increased concentration of TGRAgNPs. Catechin was used as standard (lane 7). Moreover, a concentration-dependent protective effect of the extract was observed. However, it was not as pronounced as that of the nanoparticles, as indicated by the intensity and intactness of the protein bands (lanes 3–6, Fig. 8B. Overall, the results infer that synthesized TGRAgNPs are able to protect the DNA from free radical induced damage possibly due to their hydroxyl radical scavenging potential as observed in Fig. 8. Similar results have been reported for Rhus coriaria L. mediated silver with a concentration dependent DNA damage protective effect.103In summary, TGRAgNPs demonstrated comparatively higher bioactivity for the tested activity assays when compared to the reported green-synthesized silver nanoparticles. For this, a comparative analysis of the activities exhibited by TGRAgNPs with the reported green-synthesized nanoparticles is summarized in Table S2 (ESI†).104–117 As can be inferred from Table S2 (ESI†), the observed antioxidant activity of TGRAgNPs stands out, exhibiting a maximum scavenging of 85.7%, which is notably higher than most of the other green-synthesized silver nanoparticles. For example, nanoparticles from N. leucophylla-mediated nanoparticles displayed 79% activity, while A. tribuloides and E. scaber-mediated nanoparticles showed 64% and 85%, respectively.75,76 With respect to hemocompatibility, the TGRAgNPs exhibited a hemolysis of 6.1% at 0.5 mg mL−1, which is much better than that of Cannabis sativa root extract-mediated AgNPs, with a hemolysis of 7% at 0.2 mg mL−1, and polymeric nanoparticles (>10%).79 For cytotoxicity, TGRAgNPs induced 84.4% cytotoxicity at 0.5 mg mL−1 (IC50 value of 0.17 mg mL−1), which is quite comparable to that of C. nudiflora-mediated AgNPs (IC50 = 0.1 mg mL−1) and much better than that of Putranjiva roxburghii AgNPs (IC50 = 0.5 mg mL−1).84,87 Moreover, the anti-inflammatory potential of TGRAgNPs (87.7% inhibition) exceeds the A. tribuloides root extract (69%) and Madhuca longifolia -mediated AgNPs (53.15% at 500 μg mL−1).96,100 Lastly, TGRAgNPs provided effective protection against Fenton-induced DNA damage, comparatively at lower concentration range as compared to Rhus coriaria AgNPs, showing a potent concentration-dependent protective effect.103 In summary, TGRAgNPs exhibit reasonably higher bioactivity across all the tested assays, as compared to the reported results, underlining their potential as a potent bioactive material in future therapeutic applications, particularly in the areas of antioxidant therapy, cancer and inflammation control.
5. Network pharmacology based in silico analysis
As per GC MS analysis, hexadecenoic acid (palmitic acid) is the most prominent secondary metabolite in the TGR extract (Table S1, ESI†). In order to elucidate the possible reasons for the observed activity of the AgNPs, in silico network pharmacology analysis of palmitic acid was carried out. Palmitic acid (C16H32O2) is a saturated fatty acid with a molecular weight of 256.42. It has 18 heavy atoms and no aromatic heavy atoms. The fraction of sp3 carbons is 0.94, indicating that the molecule is mostly composed of saturated carbon atoms. Palmitic acid has 14 rotatable bonds, 2 hydrogen bond acceptors, and 1 hydrogen bond donor. In terms of physicochemical properties, palmitic acid has a mean molecular refractivity (MR) of 80.80, a topological polar surface area (TPSA) of 37.30, and various logP values (iLOGP, XLOGP3, WLOGP, MLOGP, Silicos-IT Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, Consensus Log
P, Consensus Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P). These values provide insights into the molecule's lipophilicity and potential for permeation across biological membranes. The compound was analysed for its physicochemical characteristics and pharmacokinetic parameters, including ADME analysis. The ADME profile of palmitic acid indicates that it is moderately soluble, with an ESOL Log
P). These values provide insights into the molecule's lipophilicity and potential for permeation across biological membranes. The compound was analysed for its physicochemical characteristics and pharmacokinetic parameters, including ADME analysis. The ADME profile of palmitic acid indicates that it is moderately soluble, with an ESOL Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S value of −5.02. It has a relatively low solubility in both the ESOL and Ali prediction models. Palmitic acid is found to exhibit good gastrointestinal absorption and is also quite likely to cross the blood–brain barrier system.118 Moreover, it is not a P-glycoprotein substrate and does not inhibit CYP enzymes.119 Overall, based on the provided data, palmitic acid shows favourable ADMET properties in terms of solubility, absorption, and permeability, which makes it a potentially bioavailable compound with lower toxicity issues.
S value of −5.02. It has a relatively low solubility in both the ESOL and Ali prediction models. Palmitic acid is found to exhibit good gastrointestinal absorption and is also quite likely to cross the blood–brain barrier system.118 Moreover, it is not a P-glycoprotein substrate and does not inhibit CYP enzymes.119 Overall, based on the provided data, palmitic acid shows favourable ADMET properties in terms of solubility, absorption, and permeability, which makes it a potentially bioavailable compound with lower toxicity issues.
For the identification of palmitic acid-related drug targets, Web-based servers PharmMapper and Swiss Target were used for identification of possible target proteins for palmitic acid. Among the identified target proteins, the top 10 targets are shown in Fig. 9. As can be seen in the figure, nuclear receptors, fatty acid binding proteins and commonly found cytosolic enzymes are the key targets followed by G-protein coupled receptors (family AG/CG protein coupled receptor) and cytochrome P450. Overall, the results indicate the palmitic acid is able to target some key proteins of the cellular system. Moreover, to delve deep into the mechanistic insights, protein–protein interaction (PPI) network and functional gene annotation analysis was also carried out. As can be seen in the Fig. 10, PPI network analysis identified 50 proteins with a total of 439 interactions. The observed number of interactions is significantly higher than the expected number of interactions (95) with a p-value of 0.005. Results suggest that the network is densely connected with a high degree of interactivity among the proteins.
 | ||
| Fig. 10 The PPI interaction network of the common target genes of palmitic acid. Target genes are depicted as nodes while as the relationships between them are expressed as edges. | ||
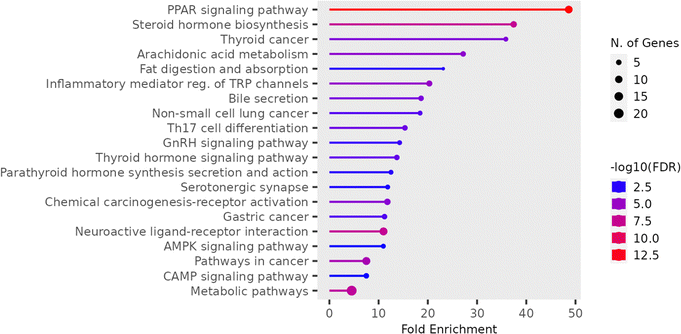
Functional analysis of palmitic acid targets was also conducted using gene ontology (GO) and pathway analysis through Shiny GO. Enrichment analysis utilized data from various annotations and ontology resources. The analysis classified GO terms and pathways, based on biological functions, into three stages: biological process (BP), molecular function (MF), and cellular component (CC) (Fig. 11). Pathway-related data was extracted from the KEGG database. As the identified pathways associated with palmitic acid encompass a diverse array of biological processes and signalling cascades. These findings suggest the multifaceted roles of palmitic acid in cellular metabolism, signalling pathways, and hormonal signalling. KEGG pathway enrichment analysis of the aforementioned common target genes is shown in Fig. 12. Post exclusion of broad pathways, the top 20 signalling pathways are listed in Table 1. Specifically, the enrichment of pathways such as the PPAR signalling pathway, steroid hormone biosynthesis, and arachidonic acid metabolism implicates palmitic acid in lipid metabolism and regulation of nuclear receptor activity. Additionally, the involvement of pathways related to cancer, including thyroid cancer, non-small cell lung cancer, and gastric cancer, underscores its potential significance in tumorigenesis and cancer progression. Furthermore, the enrichment of pathways such as inflammatory mediator regulation of TRP channels and serotonin synapse highlights its potential contributions to inflammatory responses and neurotransmitter signalling. Collectively, these findings provide valuable insights into the diverse biological functions and molecular mechanisms associated with palmitic acid, emphasizing its importance in physiological and pathological processes.
| Enrichment FDR | nGenes | Pathway Genes | Fold Enrichment | Pathways (click for details) |
|---|---|---|---|---|
| 6.4 × 10−14 | 11 | 75 | 48.6 | PPAR signalling pathway |
| 5.0 × 10−08 | 7 | 62 | 37.4 | Steroid hormone biosynthesis |
| 9.6 × 10−05 | 4 | 37 | 35.8 | Thyroid cancer |
| 3.0 × 10−05 | 5 | 61 | 27.2 | Arachidonic acid metabolism |
| 3.3 × 10−03 | 3 | 43 | 23.1 | Fat digestion and absorption |
| 1.6 × 10−05 | 6 | 98 | 20.3 | Inflammatory mediator. of TRP channels |
| 1.4 × 10−04 | 5 | 89 | 18.6 | Bile secretion |
| 9.6 × 10−04 | 4 | 72 | 18.4 | Non-small cell lung cancer |
| 3.2 × 10−04 | 5 | 108 | 15.4 | Th17 cell differentiation |
| 2.3 × 10−03 | 4 | 93 | 14.3 | GnRH signaling pathway |
| 5.1 × 10−04 | 5 | 121 | 13.7 | Thyroid hormone signaling pathway |
| 3.3 × 10−03 | 4 | 106 | 12.5 | Parathyroid hormone synthesis secretion and action |
| 3.8 × 10−03 | 4 | 112 | 11.8 | Serotonergic synapse |
| 5.1 × 10−05 | 7 | 197 | 11.8 | Chemical carcinogenesis receptor activation |
| 1.1 × 10−03 | 5 | 148 | 11.2 | Gastric cancer |
| 5.0 × 10−08 | 12 | 362 | 11 | Neuroactive ligand-receptor interaction |
| 4.5 × 10−03 | 4 | 121 | 11 | AMPK signalling pathway |
| 2.1 × 10−06 | 12 | 530 | 7.5 | Pathways in cancer |
| 4.5 × 10−03 | 5 | 221 | 7.5 | CAMP signalling pathway |
| 1.2 × 10−07 | 21 | 1538 | 4.5 | Metabolic pathways |
4. Conclusion and future perspective
The results conclude that the T. govanianum rhizome extract-mediated green approach for synthesizing TGRAgNPs is a simple, rapid, cost-effective, and eco-friendly method. Aqueous extracts of T. govanianum acted as a source of stabilizing and reducing agents for the synthesis of the silver nanoparticles. A colour change from light yellowish to dark brown was observed, implying the reduction of Ag+ to Ag0, leading to the synthesis of silver nanoparticles. UV-vis spectrum displayed a strong absorption band at 410 nm, confirming the reduction of silver nanoparticles. Further TGRAgNPs formation was confirmed by FTIR, DLS, XRD, SEM-EDX and TEM analysis. The average particle and crystallite size was found to be 38 nm and 32 nm. SEM images displayed nanoparticles in polymorphic shapes, some irregularly granulated, ellipsoidal, and highly aggregated. The nanoparticles were found to have excellent antioxidant activity with potential cytotoxicity. Moreover, the ability of TGRAgNPs to denature bovine serum albumin suggests their anti-inflammatory potential with largely being biocompatible. Furthermore, the nanoparticles were found quite effective in protecting against free radical DNA damage. In silico analysis also suggest involvement of key cellular proteins and pathways, implicated in the observed biological activities of the nanoparticles, as possible targets for the anticipated palmitic acid functionalized AgNPs. Overall, the study used an economical and promising green approach for synthesizing the biocompatible and biologically active AgNPs, using rhizome extract of Trillium govanianum. In light of these findings, future studies in this direction should primarily focus on – (i) understanding the precise molecular mechanism of green synthesis of nanoparticles and its regulation, (ii) extensive exploration of other plant-based extracts in green nanotechnology so as to enhance the repertoire of green synthesis methods, (iii) optimization of the synthesized nanoparticle for specific use in medical field leading to the development of new therapeutics with enhanced efficiency and reduced side effects, (iv) exploration of route of administration, pharmacokinetic properties, and host immune responses, (v) advanced computational analysis to predict the behaviour of the synthesized nanoparticles under various biological environments so as to enhance its targeted applications and (vi) being environment friendly, inexpensive, simple, efficient and biocompatible, the scalability of green synthesis for large scale production should be focussed upon at the earliest. Although being very promising, challenges still remain there for achieving the uniformity and consistency in the size and shape of green synthesized nanoparticles. Future studies in this direction are also highly warranted as the scalability of green synthesis process is critical for their functioning in a specific application. In fact, addressing these challenges, through optimization and standardization of the green synthesis process, is crucial for large-scale implementation of the green nanotechnology.Author contributions
SIM – Data curation, formal analysis, methodology, writing – original draft; FJ – methodology, data curation; RJ – formal analysis, conceptualization; MAR – manuscript reading, KI – data curation, data analysis, methodology; MAM – conceptualization, data curation, software, formal analysis, writing – original draft, supervision, investigation; TAD – conceptualization, data curation, formal analysis, writing – original draft, supervision, investigation, revision of the manuscript.Informed consent
Due informed consent was obtained for experimentation with human subjects.Data availability
There is no such repository or link that can be generated for the availability of data. However, the raw data generated is available on the lab instruments and will be available upon request.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This work was supported by the grant provided by Central Council for Research in Unani Medicine, Govt. of India, New Delhi to TAD with grant number: F. No. 3-67/2022-CCRUM/Tech.References
- S. Modi, R. Prajapati, G. K. Inwati, N. Deepa, V. Tirth, V. K. Yadav and B. H. Jeon, Recent trends in fascinating applications of nanotechnology in allied health sciences, Crystals, 2021, 12(1), 39 CrossRef.
- R. Tian, M. Li, H. Teng, H. Luo, D. Yan and D. M. Wei, Surface enhanced Raman scattering based on Au nanoparticles/layered double hydroxide ultrathin films, J. Mater. Chem., 2015, 3(20), 5167–5174 CAS.
- A. K. Keshari, R. Srivastava, P. Singh, V. B. Yadav and G. Nath, Antioxidant and antibacterial activity of silver nanoparticles synthesized by Cestrum nocturnum, J. Ayurveda Integr. Med., 2020, 11(1), 37–44 CrossRef PubMed.
- R. Tian, S. Zhang, M. Li, Y. Zhou, B. Lu, D. Yan and X. Duan, Localization of Au nanoclusters on layered double hydroxides nanosheets: confinement-induced emission enhancement and temperature-responsive luminescence, Adv. Funct. Mater., 2015, 25(31), 5006–5015 CrossRef CAS.
- M. A. Sofi, S. Sunitha, S. K. Pasha and D. Choi, An overview of antimicrobial and anticancer potential of silver nanoparticles, J. King Saud Univ., Sci., 2022, 34(2), 101791 CrossRef.
- T. Zhang, H. Shang, B. Zhang, D. Yan and X. Xiang, Ag/ultrathin-layered double hydroxide nanosheets induced by a self-redox strategy for highly selective CO2 reduction, ACS Appl. Mater. Interfaces, 2021, 13(14), 16536–16544 CrossRef CAS PubMed.
- S. Wei, Y. Wang, Z. Tang, H. Xu, Z. Wang, T. Yang and T. Zou, A novel green synthesis of silver nanoparticles by the residues of Chinese herbal medicine and their biological activities, RSC Adv., 2021, 11(3), 1411–1419 RSC.
- A. Dhaka, S. C. Mali and S. Sharma, Trivedi, A review on biological synthesis of silver nanoparticles and their potential applications, Results Chem., 2023, 101108 CrossRef CAS.
- P. K. Dikshit, J. Kumar, A. K. Das, S. Sadhu, S. Sharma, S. Singh, P. K. gupta and S. Kim, Green synthesis of metallic nanoparticles: Applications and limitations, Catalysts, 2021, 11(8), 902 CrossRef CAS.
- M. Tariq, K. N. Mohammad, B. Ahmed, M. A. Siddiqui and J. Lee, Biological synthesis of silver nanoparticles and prospects in plant disease management, Molecules, 2022, 27(15), 4754 CrossRef CAS PubMed.
- S. Simon, N. R. Sibuyi, A. O. Fadaka, S. Meyer, J. Josephs, M. O. Onani and A. M. Madiehe, Biomedical applications of plant extract-synthesized silver nanoparticles, Biomedicines, 2022, 10(11), 2792 CrossRef CAS PubMed.
- M. A. Kakakhel, W. Sajjad, F. Wu, N. Bibi, K. Shah, Z. Yali and W. Wang, Green synthesis of silver nanoparticles and their shortcomings, animal blood a potential source for silver nanoparticles: A review, J. Hazard. Mater. Adv., 2021, 1, 100005 CAS.
- A. K. Sidhu, N. Verma and P. Kaushal, Role of biogenic capping agents in the synthesis of metallic nanoparticles and evaluation of their therapeutic potential, Front. Nanotechnol., 2022, 3, 801620 CrossRef.
- A. Rozhin, S. Batasheva, M. Kruychkova, Y. Cherednichenko, E. Rozhina and R. Fakhrullin, Biogenic silver nanoparticles: Synthesis and application as antibacterial and antifungal agents, Micromachines, 2021, 12(12), 1480 CrossRef PubMed.
- D. Kulkarni, R. Sherkar, C. Shirsathe, R. Sonwane, N. Varpe, S. Shelke and S. Dyawanapelly, Biofabrication of nanoparticles: sources, synthesis, and biomedical applications, Front. Bioeng. Biotechnol., 2023, 11, 1159193 CrossRef PubMed.
- P. Saha, M. Mahiuddin, A. N. Islam and B. Ochiai, Biogenic synthesis and catalytic efficacy of silver nanoparticles based on peel extracts of citrus macroptera fruit, ACS Omega, 2021, 6(28), 18260–18268 CrossRef CAS PubMed.
- A. K. Alzubaidi, W. J. Al-Kaabi, A. A. Ali, S. Albukhaty, H. Al-Karagoly, G. M. Sulaiman and Y. Khane, Green synthesis and characterization of silver nanoparticles using flaxseed extract and evaluation of their antibacterial and antioxidant activities, Appl. Sci., 2023, 13(4), 2182 CrossRef CAS.
- K. D. Dejen, D. Y. Kibret, T. H. Mengesha, E. T. Bekele, A. Tedla, T. A. Bafa and F. T. Derib, Green synthesis and characterisation of silver nanoparticles from leaf and bark extract of Croton macrostachyus for antibacterial activity, Mater. Technol., 2023, 38(1), 2164647 CrossRef.
- A. K. Giri, B. Jena, B. Biswal, A. K. Pradhan, M. Arakha and S. Acharya, Green synthesis and characterization of silver nanoparticles using Eugenia roxburghii DC. extract and activity against biofilm-producing bacteria, Sci. Rep., 2022, 12(1), 8383 CrossRef CAS PubMed.
- H. B. Habeeb Rahuman, R. Dhandapani, S. Narayanan, V. Palanivel, R. Paramasivam, R. Subbarayalu and S. Muthupandian, Medicinal plants mediated the green synthesis of silver nanoparticles and their biomedical applications, IET Nanobiotechnol., 2022, 16(4), 115–144 CrossRef PubMed.
- B. Essghaier, H. Hannachi, R. Nouir, F. Mottola and L. Rocco, Green Synthesis and characterization of Novel Silver nanoparticles using Achillea maritima subsp. maritima Aqueous Extract: antioxidant and antidiabetic potential and effect on virulence mechanisms of bacterial and fungal pathogens, Nanomaterials, 2023, 13, 1964 CrossRef CAS PubMed.
- R. S. Patil, M. R. Kokate and S. S. Kolekar, Bioinspired synthesis of highly stabilized silver nanoparticles using Ocimum tenuiflorssum leaf extract and their antibacterial activity, Spectrochim. Acta, Part A, 2012, 91, 234–238 CrossRef CAS PubMed.
- R. K. Omole, R. C. George, O. I. Adeyemi, N. Torimiro, M. Saravanan, E. O. Agboluaje and M. P. Xiong, Spectral Characterization of Silver Nanoparticles Biosynthesized from Lysinibacillus fusiformis and its Antibacterial Efficacy Against Multidrug-Resistant Bacteria Isolated from Chronic Wounds, Bionanoscience, 2024, 1–11 Search PubMed.
- U. P. Manik, A. Nande, S. Raut and S. J. Dhoble, Green synthesis of silver nanoparticles using plant leaf extraction of Artocarpus heterophylus and Azadirachta indica, Results Mater., 2020, 6, 100086 CrossRef.
- A. Maniraj, M. Kannan, K. Rajarathinam, S. Vivekanandhan and S. Muthuramkumar, Green synthesis of silver nanoparticles and their effective utilization in fabricating functional surface for antibacterial activity against multi-drug-resistant Proteus mirabilis, J. Cluster Sci., 2019, 30, 1403–1414 CrossRef CAS.
- S. Priyadarshini, S. Sulava, R. Bhol and S. Jena, Green synthesis of silver nanoparticles using Azadirachta indica and Ocimum sanctum leaf extract, Curr. Sci., 2019, 117(8), 1300–1307 CrossRef CAS.
- A. Mathesh, D. S. Carmelin, A. Mohanprasanth, P. Geetha Sravanthy, R. Snega, M. Surya and M. Saravanan, Tridax procumbens–mediated one pot synthesis of silver-doped fucoidan nanoparticles and their antibacterial, antioxidant, and anti-inflammatory efficacy, Biomass Convers. Biorefin., 2024, 14(8), 9887–9896 CrossRef CAS.
- R. Manikandan, M. Beulaja, N. M. Prabhu, R. Thiagarajan, M. Anjugam, S. Palanisamy, K. Saravanan and M. Arumugam, Synthesis of silver nanoparticles using Solanum trilobatum fruits extract and its antibacterial, cytotoxic activity against human breast cancer cell line MCF 7, Spectrochim. Acta, Part A, 2015, 140, 223–228 CrossRef PubMed.
- G. Kuppurangan, B. Karuppasamy, K. Nagarajan, K. S. Rajkumar, V. Nilmini and R. Thirumurugan, Biogenic synthesis and spectroscopic characterization of silver nanoparticles using leaf extract of Indoneesiella echioides: in vitro assessment on antioxidant, antimicrobial and cytotoxicity potential, Appl. Nanosci., 2016, 6(7), 973–982 CrossRef CAS.
- P. Kannaiyan, Greener approach for synthesis of antibacterial silver nanoparticles using aqueous solution of neem gum (Azadirachta indica L.), Ind. Crops Prod., 2015, 66, 103–109 CrossRef.
- M. Ramar, B. Manikandan, P. N. Marimuthu, T. Raman, A. Mahalingam, P. Subramanian and A. Munusamy, Silver nanoparticles biosynthesized using Achillea biebersteinii flower extract: apoptosis induction in MCF-7 cells via caspase activation and regulation of Bax and Bcl-2 gene expression, Molecules, 2015, 20(2), 2693–2706 CrossRef PubMed.
- T. M. Abdelghany, A. M. Al-Rajhi, M. A. Al Abboud, M. M. Alawlaqi, A. Ganash Magdah, E. A. Helmy and A. S. Mabrouk, Recent advances in green synthesis of silver nanoparticles and their applications: about future directions, A review, Bionanoscience, 2018, 8, 5–16 CrossRef.
- R. Verma, A. Tapwal, D. Kumar and S. Puri, Antimicrobial potential and phytochemical profiling of ethnomedicinal plant Trillium govanianum Wall. ex D. Don in Western Himalaya, J. Herb. Med., 2021, 29, 100491 CrossRef.
- K. Uz-Zaman, J. Bakht, B. K. Malikovna, E. R. Elsharkawy, A. A. Khalil, S. Bawazeer and A. Rauf, Trillium govanianum Wall. Ex. Royle rhizomes extract-medicated silver nanoparticles and their antimicrobial activity, Green Process. Synth., 2020, 9, 503–514 Search PubMed.
- Z. Bedlovicova, I. Strapac, M. Balaz and A. Salayova, A brief overview on antioxidant activity determination of silver nanoparticles, Molecules, 2020, 25(14), 3191 CrossRef CAS PubMed.
- S. Andleeb, F. Tariq, A. Muneer, T. Nazir, B. Shahid, Z. Latif and D. A. Al Farraj, In vitro bactericidal, antidiabetic, cytotoxic, anticoagulant, and hemolytic effect of green-synthesized silver nanoparticles using Allium sativum clove extract incubated at various temperatures, Green Process. Synth., 2020, 9(1), 538–553 Search PubMed.
- K. Imtiyaz, A. Husain Rahmani, M. A. Alsahli, S. A. Almatroodi and M. M. A. Rizvi, Fisetin induces apoptosis in human skin cancer cells through downregulating MTH1, J. Biomol. Struct. Dyn., 2023, 4(15), 7339–7353 CrossRef PubMed.
- A. Algarni, A. Fayomi, H. A. Garalleh and A. Afand, Nanofabrication synthesis and its role in antibacterial, anti-inflammatory, and anticoagulant activities of AgNPs synthesized by Mangifera indica bark extract, Environ. Res., 2023, 231, 115983 CrossRef CAS PubMed.
- X. Wu and S. Dhanasekaran, Protective effect of leaf extract of Abutilon indicum on DNA damage and peripheral blood lymphocytes in combating the oxidative stress, Saudi Pharm. J., 2020, 28(8), 943 CrossRef CAS PubMed.
- R. Pluskota, K. Jaroch, P. Kośliński, B. Ziomkowska, A. Lewińska, S. Kruszewski and M. Koba, Selected drug-likeness properties of 2-arylidene-indan-1, 3-dione derivatives—chemical compounds with potential anti-cancer activity, Molecules, 2021, 26(17), 5256 CrossRef CAS PubMed.
- X. Wang, Y. Shen, S. Wang, S. Li, W. Zhang, X. Liu and H. Li, PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database, Nucleic Acids Res., 2017, 45, W356–W360 CrossRef CAS PubMed.
- C. Mu, Y. Sheng, Q. Wang, A. Amin, X. Li and Y. Xie, Potential compound from herbal food of Rhizoma Polygonati for treatment of COVID-19 analyzed by network pharmacology: Viral and cancer signaling mechanisms, J. Funct. Foods, 2021, 77, 104149 CrossRef CAS PubMed.
- C. Li, Z. Gao, B. Su, G. Xu and X. Lin, Data analysis methods for defining biomarkers from omics data, Anal. Bioanal. Chem., 2022, 414(1), 235–250 CrossRef CAS PubMed.
- M. R. Karim, M. N. Morshed, S. Iqbal, S. Mohammad, R. Mathiyalagan, D. C. Yang and D. U. Yang, A Network Pharmacology and Molecular-Docking-Based Approach to Identify the Probable Targets of Short-Chain Fatty-Acid-Producing Microbial Metabolites against Kidney Cancer and Inflammation, Biomolecules, 2023, 13(11), 1678 CrossRef CAS PubMed.
- L. Lv, X. Wang and H. Wu, Assessment of palmitic acid toxicity to animal hearts and other major organs based on acute toxicity, network pharmacology, and molecular docking, Comput. Biol. Med., 2017, 158, 106899 CrossRef PubMed.
- J. Peng, H. Wang, J. Lu, W. Hui, Y. Wang and X. Shang, Identifying term relations cross different gene ontology categories, BMC Bioinf., 2017, 18, 67–74 CrossRef PubMed.
- M. A. Malik, A. A. Alshehri and R. Patel, Facile one-pot green synthesis of Ag–Fe bimetallic nanoparticles and their catalytic capability for 4-nitrophenol reduction, J. Mater. Res. Technol., 2021, 12, 455–470 CrossRef CAS.
- A. Chhatre, P. Solasa, S. Sakle, R. Thaokar and A. Mehra, Color and surface plasmon effects in nanoparticle systems: Case of silver nanoparticles prepared by microemulsion route, Colloids Surf., A, 2012, 404, 83–92 CrossRef CAS.
- B. Bharath, K. Perinbam, S. Devanesan, M. S. AlSalhi and M. Saravanan, Evaluation of the anticancer potential of Hexadecanoic acid from brown algae Turbinaria ornata on HT–29 colon cancer cells, J. Mol. Struct., 2021, 1235, 130229 CrossRef CAS.
- Q. Gan, J. Wang, J. Hu, G. Lou, H. Xiong, C. Peng and Q. Huang, The role of diosgenin in diabetes and diabetic complications, The, J. Steroid Biochem. Mol. Biol., 2020, 198, 105575 CrossRef CAS PubMed.
- K. Kogure, Novel antioxidative activity of astaxanthin and its synergistic effect with vitamin E, J. Nutr. Sci. Vitaminol., 2019, 65, S109–S112 CrossRef PubMed.
- K. M. Khan, S. D. Sarker, G. A. Khan, H. Saleem, S. A. Khan and A. Mannan, Phytochemical profiling and evaluation of modified resazurin microtiter plate assay of the roots of Trillium govanianum, Nat. Prod. Res., 2020, 34(19), 2837–2841 CrossRef CAS PubMed.
- K. S. Siddiqi, A. Husen and R. A. Rao, A review on biosynthesis of silver nanoparticles and their biocidal properties, J. Nanobiotechnol., 2018, 16, 1–28 CrossRef PubMed.
- S. U. R. Qamar and J. N. Ahmad, Nanoparticles, Mechanism of biosynthesis using plant extracts, bacteria, fungi, and their applications, J. Mol. Liq., 2021, 334, 116040 CrossRef CAS.
- K. Rashid, S. Rashid, A. H. Ganie and I. A. Nawchoo, Trillium govanianum–A Promising Endemic Medicinal Herb of the Himalaya, Bioprospecting of Tropical Medicinal Plants., 2023, 381–408 Search PubMed.
- S. Mukherji, S. Bharti and G. Shukla, Mukherji, Synthesis and characterization of size-and shape-controlled silver nanoparticles, Phys. Sci. Rev., 2018, 4(1), 20170082 Search PubMed.
- M. A. Ansari, S. M. M. Asiri, M. A. Alzohairy, M. N. Alomary, A. Almatroudi and F. A. Khan, Biofabricated fatty acids-capped silver nanoparticles as potential antibacterial, antifungal, antibiofilm and anticancer agents, Pharmaceuticals, 2021, 14(2), 139 CrossRef CAS PubMed.
- J. A. Hema, R. Malaka, N. P. Muthukumarasamy, A. Sambandam, S. Subramanian and M. Sevanan, Green synthesis of silver nanoparticles using Zea mays and exploration of its biological applications, IET Nanobiotechnol., 2016, 10(5), 288–294 CrossRef PubMed.
- M. H. Ali, M. A. Azad, K. A. Khan, M. O. Rahman, U. Chakma and A. Kumer, Analysis of crystallographic structures and properties of silver nanoparticles synthesized using PKL extract and nanoscale characterization techniques, ACS Omega, 2023, 8, 28133–28142 CrossRef CAS PubMed.
- A. A. Alshehri and M. A. Malik, Phytomediated photo-induced green synthesis of silver nanoparticles using Matricaria chamomilla L. and its catalytic activity against rhodamine B, Biomolecules, 2020, 10, 1604 CrossRef PubMed.
- S. S. Albeladi, M. A. Malik and S. A. Al-thabaiti, Facile Preparation of Ag Flower-Like Nanostructures and Their Catalytic Properties in the Degradation of Organic Dyes, J. Mater. Res. Technol., 2020, 9, 10031 CrossRef.
- M. Stevanovic, I. Savanović, V. Uskoković, S. D. Škapin, I. Bračko, U. Jovanović and D. Uskoković, A new, simple, green, and one-pot four-component synthesis of bare and poly (α, γ, L-glutamic acid)-capped silver nanoparticles, Colloid Polym. Sci., 2012, 290, 221–231 CrossRef CAS PubMed.
- S. Singla, A. Jana, R. Thakur, C. Kumari and S. Goyal, Pradhan, Antioxidants and antibacterial potential of extracts of seed of Quercus leucotrichophora A. camus, Curr. Res. Green Sustainable Chem., 2023, 7, 100375 CrossRef.
- K. Saware and A. Venkataraman, Biosynthesis and characterization of stable silver nanoparticles using Ficus religiosa leaf extract: a mechanism perspective, J. Cluster Sci., 2014, 25, 1157–1171 CrossRef CAS.
- R. G. Saratale, G. Benelli, G. Kumar, D. S. Kim and E. D. Saratale, Bio-fabrication of silver nanoparticles using the leaf extract of ancient herbal medicine, dandelion (Taraxacum officinale), evaluation of their antioxidant, anticancer potential, and antimicrobial activity against phytopathogens, Environ. Sci. Pollut. Res., 2018, 25, 10392–10406 CrossRef CAS PubMed.
- S. Some, O. Bulut and K. Biswas, et al., Effect of feed supplementation with biosynthesized silver nanoparticles using leaf extract of Morus indica L. V1 on Bombyx mori L. (Lepidoptera: Bombycidae), Sci. Rep., 2019, 9(1), 14839 CrossRef PubMed.
- S. Peron, F. Hadi, F. Azarbani and H. C. Ananda Murthy, Antimicrobial, antioxidant, anti-glycation and toxicity studies on silver nanoparticles synthesized using Rosa damascena flower extract, Green Chem. Lett. Rev., 2021, 14(3), 519–533 CrossRef CAS.
- T. Desalegn, C. R. Ravikumar and H. C. A. Murthy, Eco-friendly synthesis of silver nanostructures using medicinal plant Vernonia amygdalina Del. leaf extract for multifunctional applications, Appl. Nanosci., 2021, 11, 535–551 CrossRef CAS.
- D. Kumar and V. Kumari, Metabolite profiling, antidiabetic, and antioxidant potential of different tissues of Trillium govanianum Wall. ex D. Don, S. Afr. J. Bot., 2023, 153, 102–108 CrossRef CAS.
- A. K. Sidhu, N. Verma and P. Kaushal, Role of biogenic capping agents in the synthesis of metallic nanoparticles and evaluation of their therapeutic potential, Front. Nanotechnol., 2022, 3, 801620 CrossRef.
- S. Baliyan, R. Mukherjee, A. Priyadarshini, A. Vibhuti, A. Gupta, R. P. Pandey and C. M. Chang, Determination of antioxidants by DPPH radical scavenging activity and quantitative phytochemical analysis of Ficus religiosa, Molecules, 2022, 27(4), 1326 CrossRef CAS PubMed.
- S. R. Guntur, N. S. Kumar, M. M. Hegde and V. R. Dirisala, In vitro studies of the antimicrobial and free-radical scavenging potentials of silver nanoparticles biosynthesized from the extract of Desmostachya bipinnata, Anal. Chem. Insights, 2018, 13, 1177390118782877 CrossRef PubMed.
- J. Singh and A. S. Dhaliwal, Novel green synthesis and characterization of the antioxidant activity of silver nanoparticles prepared from Nepeta leucophylla root extract, Anal. Lett., 2019, 52(2), 213–230 CrossRef CAS.
- M. S. Abdel-Aziz, M. S. Shaheen, A. A. El-Nekeety and M. A. Abdel-Wahhab, Antioxidant and antibacterial activity of silver nanoparticles biosynthesized using Chenopodium murale leaf extract, J. Saudi Chem. Soc., 2014, 18(4), 356–363 CrossRef.
- M. Sharifi-Rad, P. Pohl, F. Epifano and J. M. Álvarez-Suarez, Green synthesis of silver nanoparticles using Astragalus tribuloides delile root extract, Characterization, antioxidant, antibacterial, and anti-inflammatory activities, Nanomaterials, 2020, 10(12), 2383 CrossRef CAS PubMed.
- S. N. Kharat and V. D. Mendhulkar, Synthesis, characterization and studies on antioxidant activity of silver nanoparticles using Elephantopus scaber leaf extract, Mater. Sci. Eng., C, 2016, 62, 719–724 CrossRef CAS PubMed.
- M. Mousavi-Khattat, M. Keyhanfar and A. Razmjou, A comparative study of stability, antioxidant, DNA cleavage and antibacterial activities of green and chemically synthesized silver nanoparticles, Artif. Cells, Nanomed., Biotechnol., 2018, 46(3), 1022–1031 CrossRef PubMed.
- S. E. Arland and J. Kumar, Green and chemical syntheses of silver nanoparticles: Comparative and comprehensive study on characterization, therapeutic potential, and cytotoxicity, Eur. J. Med. Chem. Rep., 2024, 11, 100168 Search PubMed.
- S. Suman, L. Loveleen, M. Bhandari, A. Syed, A. H. Bahkali, R. Manchanda and S. Nimesh, Antibacterial, antioxidant, and haemolytic potential of silver nanoparticles biosynthesized using roots extract of Cannabis sativa plant, Artif. Cells, Nanomed., Biotechnol., 2022, 50(1), 343–351 CrossRef CAS PubMed.
- E. Parthiban, N. Manivannan, R. Ramanibai and N. Mathivanan, Green synthesis of silver-nanoparticles from Annona reticulata leaves aqueous extract and its mosquito larvicidal and anti-microbial activity on human pathogens, Biotechnol. Rep., 2019, 21, e00297 CrossRef PubMed.
- A. Mishra, N. K. Kaushik, M. Sardar and D. Sahal, Evaluation of antiplasmodial activity of green synthesized silver nanoparticles, Colloids Surf., B, 2013, 111, 713–718 CrossRef CAS PubMed.
- Y. Bian, K. Kim, T. Ngo, I. Kim, O. N. Bae, K. M. Lim and J. H. Chung, Silver nanoparticles promote procoagulant activity of red blood cells: a potential risk of thrombosis in susceptible population, Part. Fibre Toxicol., 2019, 16, 1–14 CrossRef PubMed.
- B. M. Rothen, S. Schürch, B. Haenni, N. Kapp and P. Gehr, Interaction of fine particles and nanoparticles with red blood cells visualized with advanced microscopic techniques, Environ. Sci. Technol., 2006, 40(14), 4353–4359 CrossRef PubMed.
- P. Kuppusamy, S. J. Ichwan, P. N. Al-Zikri, W. H. Suriyah, I. Soundharrajan, N. Govindan and M. M. Yusoff, In vitro anticancer activity of Au, Ag nanoparticles synthesized using Commelina nudiflora L. aqueous extract against HCT-116 colon cancer cells, Biol. Trace Elem. Res., 2016, 173, 297–305 CrossRef CAS PubMed.
- M. Madakka, N. Jayaraju and N. Rajesh, Evaluating the antimicrobial activity and antitumor screening of green synthesized silver nanoparticles compounds, using Syzygium jambolanum, towards MCF7 cell line (Breast cancer cell line), J. Photochem. Photobiol., 2021, 6, 100028 CrossRef.
- D. Acharya, S. Satapathy, P. Somu, U. K. Parida and G. Mishra, Apoptotic effect and anticancer activity of biosynthesized silver nanoparticles from marine algae Chaetomorpha linum extract against human colon cancer cell HCT-116, Biol. Trace Elem. Res., 2013, 199(5), 1812–1822 CrossRef PubMed.
- A. Balkrishna, V. K. Sharma, S. K. Das, N. Mishra, L. Bisht, A. Joshi and N. Sharma, Characterization and anti-cancerous effect of Putranjiva roxburghii seed extract mediated silver nanoparticles on human colon (HCT-116), pancreatic (PANC-1) and breast (MDA-MB 231) cancer cell lines: A comparative study, Int. J. Nanomed., 2020, 573–585 CrossRef CAS PubMed.
- S. Tian, K. Saravanan, R. A. Mothana, G. Ramachandran, G. Rajivgandhi and N. Manoharan, Anti-cancer activity of biosynthesized silver nanoparticles using Avicennia marina against A549 lung cancer cells through ROS/mitochondrial damages, Saudi J. Biol. Sci., 2020, 27(11), 3018–3024 CrossRef CAS PubMed.
- S. Kummara, M. B. Patil and T. Uriah, Synthesis, characterization, biocompatible and anticancer activity of green and chemically synthesized silver nanoparticles–a comparative study, Biomed. Pharmacother., 2016, 84, 10–21 CrossRef CAS PubMed.
- R. Amooaghaie, M. R. Saeri and M. Azizi, Synthesis, characterization and biocompatibility of silver nanoparticles synthesized from Nigella sativa leaf extract in comparison with chemical silver nanoparticles, Ecotoxicol. Environ. Saf., 2015, 120, 400–408 CrossRef CAS PubMed.
- F. B. Almukaynizi, M. H. Daghestani, M. A. Awad, A. Althomali, N. M. Merghani, W. I. Bukhari and R. S. Bhat, Cytotoxicity of green-synthesized silver nanoparticles by Adansonia digitata fruit extract against HTC116 and SW480 human colon cancer cell lines, Green Process. Synth., 2022, 11(1), 411–422 CrossRef CAS.
- F. Yousefbeyk, S. Dabirian, S. Ghanbarzadeh, D. Eghbali Koohi, P. Yazdizadeh and S. Ghasemi, Green synthesis of silver nanoparticles from Stachys byzantina K. Koch: characterization, antioxidant, antibacterial, and cytotoxic activity, Part. Sci. Technol., 2022, 40(2), 219–232 CrossRef CAS.
- S. U. Rahman, M. Ismail, M. R. Shah, M. Iriti and M. Shahid, GC/MS analysis, free radical scavenging, anticancer and β-glucuronidase inhibitory activities of Trillium govanianum rhizome, Bangladesh J. Pharmacol., 2015, 10(3), 577–583 CrossRef.
- X. Wang, C. Zhang and N. Bao, Molecular mechanism of palmitic acid and its derivatives in tumor progression, Front. Oncol., 2023, 13, 1224125 CrossRef CAS PubMed.
- G. Sangeetha and R. Vidhya, In vitro anti-inflammatory activity of different parts of Pedalium murex (L.), Inflammation, 2016, 4(3), 31–36 Search PubMed.
- M. Sharifi-Rad, P. Pohl, F. Epifano and J. M. Álvarez-Suarez, Green synthesis of silver nanoparticles using Astragalus tribuloides delile root extract: Characterization, antioxidant, antibacterial, and anti-inflammatory activities, Nanomaterials, 2020, 10(12), 2383 CrossRef CAS PubMed.
- F. S. Alatawi, M. S. Alatawi, A. M. Omran, R. M. Jame, M. Adnan, M. N. Khan and M. Rahimi, Aconitum lycoctonum L. (Ranunculaceae) mediated biogenic synthesis of silver nanoparticles as potential antioxidant, anti-inflammatory, antimicrobial and antidiabetic agents, BMC Chem., 2023, 17(1), 128 CrossRef PubMed.
- P. B. E. Kedi, C. C. Nanga, A. P. Gbambie, V. Deli, F. E. A. Meva, H. E. A. Mohamed and M. H. J. Nkors, Biosynthesis of silver nanoparticles from Microsorum punctatum (l.) copel fronds extract and an in-vitro anti-inflammation study, J. Nanotechnol. Res., 2020, 2(2), 25–41 Search PubMed.
- M. B. Lava, U. M. Muddapur, N. Basavegowda, S. S. More and V. S. More, Characterization, anticancer, antibacterial, anti-diabetic and anti-inflammatory activities of green synthesized silver nanoparticles using Justica wynaadensis leaves extract, Mater. Today: Proc., 2021, 46, 5942–5947 CAS.
- P. Salve, A. Vinchurkar, R. Raut, R. Chondekar, J. Lakkakula, A. Roy and M. F. Nur Azlina, an evaluation of antimicrobial, anticancer, anti-inflammatory and antioxidant activities of silver nanoparticles synthesized from leaf extract of Madhuca longifolia utilizing quantitative and qualitative methods, Molecules, 2022, 27(19), 6404 CrossRef CAS PubMed.
- S. Ur Rahman, A. Adhikari, M. Ismail, M. Raza Shah, M. Khurram, M. Shahid and M. Iriti, Beneficial effects of Trillium govanianum rhizomes in pain and inflammation, Molecules, 2016, 21(8), 1095 CrossRef PubMed.
- Z. U. R. Khan, N. Assad, M. Naeem-ul-Hassan, M. Sher, F. S. Alatawi, M. S. Alatawi and M. Rahimi, Aconitum lycoctonum L. (Ranunculaceae) mediated biogenic synthesis of silver nanoparticles as potential antioxidant, anti-inflammatory, antimicrobial and antidiabetic agents, BMC Chem., 2023, 17(1), 128 CrossRef CAS PubMed.
- T. Gur, Green synthesis, characterizations of silver nanoparticles using sumac (Rhus coriaria L.) plant extract and their antimicrobial and DNA damage protective effects, Front. Chem., 2022, 10, 968280 CrossRef CAS PubMed.
- J. Singh and A. S. Dhaliwal, Novel green synthesis and characterization of the antioxidant activity of silver nanoparticles prepared from Nepeta leucophylla root extract, Anal. Lett., 2019, 52(2), 213–230 CrossRef CAS.
- A. S. Shinde and V. D. Mendhulkar, Antiproliferative activity of Elephantopus scaber mediated silver nanoparticles against MCF-7, A-549, SCC-40 and COLO-205 human cancer cell lines, Asian J. Pharm. Clin. Res., 2020, 13(2), 163–167 CAS.
- F. Sharifi, F. Sharififar, S. Soltanian, M. Doostmohammadi and N. Mohamadi, Synthesis of silver nanoparticles using Salvia officinalis extract: Structural characterization, cytotoxicity, antileishmanial and antimicrobial activity, Nanomed. Res. J., 2020, 5(4), 339–346 CAS.
- J. Baharara, T. Ramezani, M. Mousavi and M. Asadi-Samani, Antioxidant and anti-inflammatory activity of green synthesized silver nanoparticles using Salvia officinalis extract, Ann. Trop Med Int Health, 2017, 10(5), 1265–1270 CrossRef.
- M. Kumara Swamy, K. M. Sudipta, K. Jayanta and S. Balasubramanya, The green synthesis, characterization, and evaluation of the biological activities of silver nanoparticles synthesized from Leptadenia reticulata leaf extract, Appl. Nanosci., 2015, 5, 73–81 CrossRef CAS.
- N. S. Kumaran, In vitro anti-inflammatory activity of silver nanoparticle synthesized Avicennia marina (Forssk.) Vierh.: A green synthetic approach, Int. J. Green Pharm., 2018, 12(3), 528–536 Search PubMed.
- O. Hotan, A. Alhaj, A. Al-quhaim, K. Alburaihi, Y. Ahmed, Q. Munasser and M. Almoiliqy, Evaluating the pharmacological activities of Aloe perryi–Silver nanoparticles induced apoptosis against colon cancer cells (HCT-116), Food Sci. Nutr., 2024, 12(8), 5890–5906 CrossRef CAS PubMed.
- D. Ayodhya, A. Ambala, G. Balraj, M. P. Kumar and P. Shyam, Green synthesis of CeO2 NPs using Manilkara zapota fruit peel extract for photocatalytic treatment of pollutants, antimicrobial, and antidiabetic activities, Results Chem., 2022, 4, 100441 CrossRef CAS.
- P. Moteriya and S. Chanda, Biosynthesis of silver nanoparticles formation from Caesalpinia pulcherrima stem metabolites and their broad spectrum biological activities, J. Genet. Eng. Biotechnol., 2018, 16(1), 105–113 CrossRef PubMed.
- H. S. Hussein, C. Ngugi, F. M. Tolo and E. N. Maina, Anticancer potential of silver nanoparticles biosynthesized using Catharanthus roseus leaves extract on cervical (HeLa229) cancer cell line, Sci. Afr., 2024, e02268 CAS.
- H. S. Al-Shmgani, W. H. Mohammed, G. M. Sulaiman and A. H. Saadoon, Biosynthesis of silver nanoparticles from Catharanthus roseus leaf extract and assessing their antioxidant, antimicrobial, and wound-healing activities, Artif. Cells, Nanomed., Biotechnol., 2017, 45(6), 1234–1240 CrossRef CAS PubMed.
- J. A. Badmus, S. A. Oyemomi, O. T. Adedosu, T. A. Yekeen, M. A. Azeez, E. A. Adebayo and J. L. Marnewick, Photo-assisted bio-fabrication of silver nanoparticles using Annona muricata leaf extract: exploring the antioxidant, anti-diabetic, antimicrobial, and cytotoxic activities, Heliyon, 2020, 6(11), 1–9 CrossRef PubMed.
- J. A. Badmus, S. A. Oyemomi, J. O. Fatoki, T. A. Yekeen, O. T. Adedosu, P. I. Adegbola and A. Lateef, Anti-haemolytic and cytogenotoxic potential of aqueous leaf extract of Annona muricata (L.) and its bio-fabricated silver nanoparticles, Caryologia, 2022, 75(1), 3–13 CrossRef.
- M. Z. Gharari, S. Sadighian, A. Yazdinezhad and A. Sharafi, Green Synthesized Silver Nanostructure Using Rhus coriaria Fruit Extract Inhibits the Growth of Malignant MCF-7 Cell Line, Braz. Arch. Biol. Technol., 2021, 64, e21210069 CrossRef.
- F. Pifferi, B. Laurent and M. Plourde, Lipid transport and metabolism at the blood-brain interface: Implications in health and disease, Front. Physiol., 2021, 12, 645646 CrossRef PubMed.
- H. T. Yao, Y. W. Chang, S. J. Lan, C. T. Chen, J. T. Hsu and T. K. Yeh, The inhibitory effect of polyunsaturated fatty acids on human CYP enzymes, Life Sci., 2006, 79(26), 2432–2440 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00959b |
| This journal is © The Royal Society of Chemistry 2025 |