DOI:
10.1039/D4MA00958D
(Paper)
Mater. Adv., 2025,
6, 1364-1378
Assessing dual drug 9-hydroxymethyl noscapine and telmisartan-loaded stearic acid nanoparticles against (H1299) non-small cell lung cancer and their mechanistic interaction with bovine serum albumin†
Received
22nd September 2024
, Accepted 6th January 2025
First published on 8th January 2025
Abstract
Solid lipid nanoparticles are appealing to the scientific community owing to their expedient and versatile nature as systems for drug delivery and therefore are being used to treat a variety of illnesses. With parallel line of thought, herein, we have reported the synthesis and characterisation of dual drug stearic acid-loaded solid lipid nanoparticles and screened their efficacy in non-small cell lung cancer. The desired nanoparticles, namely 9-CH2OH Nos-Tel-SLNs, were prepared using the solvent diffusion method. TEM and AFM images revealed that the nanoparticles have spherical form with a mean size of 36.6 nm. The nanostructures' zeta potential and hydrodynamic size were found to be −36.23 mV and ∼406.8 nm, respectively. From RP-HPLC, the noscapine and telmisartan loaded in the nanoparticles were found to be 1.86% and 1.97% respectively. Additionally, we have probed into the interaction of BSA with the synthesized nanocomposite using UV-visible, fluorescence and CD spectroscopic techniques along with computational techniques, namely molecular docking, molecular dynamic simulations and MM-PBSA/GBSA calculations. From the fluorescence quenching of BSA upon interaction with the SLNs, we deduced that a stable ground-state complex between 9-CH2OH Nos-Tel-SLN and BSA was formed. Similarly, in silico evaluation indicated formation of a stable dual drug complex with BSA with telmisartan being more compatible for binding to the protein. To assess further, we also evaluated the anticancer property of 9-CH2OH noscapine, telmisartan and 9-CH2OH Nos-Tel-SLN against H1299 lung cancer cell line using MTT assay and the calculated IC50 of 9-CH2OH Nos-Tel-SLN was 186 μg mL−1. Overall, based on the promising results in this research, such SLNs could be a promising drug delivery tool and can be crucial in the conversion of potential anticancer drugs to marketed anticancer drugs in the near future.
1. Introduction
Of all cancers, lung cancer is particularly deadly having a high mortality rate worldwide accounting for 1.8 million deaths in 2022.1 One of the underlying reasons for huge death rates due to lung cancer is ineffective drug delivery to the tumor site. In this regard, solid lipid nanoparticles (SLNs) are a class of colloidal drug carriers which hold immense potential for controlled drug delivery systems.2,3 They offer many attractive benefits such as good efficacy, low toxicity, targeted drug delivery, high bioavailability upon oral administration, etc.4,5 The FDA-approved nano-drug named ‘Doxil’ is doxorubicin-loaded pegylated liposome used in breast and ovarian cancer treatments.6 As documented in the literature, biodegradable lipids like stearic acid, palmitic acid and oleic acid found in nanostructured lipid particles (NLPs) are being extensively used in the delivery of drugs to tumor cells.3 Additionally, fatty acids like stearic acid are approved for human consumption by the US FDA.7 Also, NLPs can be bulked up in industries due to their high stability.8 Nanoparticle-based drug delivery is advantageous due to the improved circulation times, controlled release, and improved efficacy of nanostructures as drug delivery vehicles.9–11 Recently, combination drug delivery, co-delivery of medications in conjunction with nanotechnology, has steadily become a preferred approach in research frontiers of modern drug delivery.12,13 Within this concept, at least two drugs having different mechanisms of action and different physiochemical properties can be loaded into a combination delivery system that can synergistically inhibit the proliferation of tumor cells compared to the case with free or individual drugs.14,15 Importantly, the combination of two or more drugs into the nanoparticles are amalgamated to achieve maximum therapeutic effect and pharmacokinetic properties of therapeutics.16,17 The acquisition of multi-drug resistance against a variety of cancers often requires the combination of two or more drugs that can effectively promote cell death by demonstrating a synergistic effect.18,19 Guo and co-workers have reported paclitaxel and 5-demethylnobiletin co-loaded cetuximab NLPs with synergistic effects against lung cancer.20 Recently, nanocarriers for concurrent delivery of tariquidar and docetaxel have been documented by Kim et al. for drug resistant breast cancer cells.21 Solid lipid nanomaterials are usually composed of solid lipids, active drug molecules and surfactants. Thus, we synthesised 9-hyrdoxymethyl noscapine and telmisartan-loaded stearic acid solid lipid nanoparticles (9-CH2OH Nos-Tel SLNs), where the lipid moiety in the nanocomposite is used to significantly enhance drug loading into hydrophobic components of carriers which can improve drug loading and stability and the two drugs were chosen due to their therapeutic potential widely documented in the literature.
Noscapine is a naturally occurring compound that was initially extracted from the opium poppy, Papaver somniferum, and is recognized as a marketed cough suppressant.22,23 Noscapine is a phthalideisoquinoline alkaloid that is non-addictive and non-narcotic in nature. In recent years, its potential as an anticancer agent has gained attention as a result of its inhibitory effects on various tumors. Previous studies showed that noscapine inhibited the growth of cancer cells in vitro against the breast cancer cell lines MCF-7 and MDAMB-231, with IC50 values of 29 μM and 69 μM, respectively, after 66 hours of treatment.24 Similarly, for lung cancer cell line A549, it showed an IC50 value of 73 μM.25 Noscapine's anticancer properties are attributed to its ability to bind with tubulin.26 On the other hand, telmisartan is an angiotensin II receptor inhibitor used as a common clinical antihypertensive agent.27,28 Several reports have postulated that angiogenesis is crucial for tumor progression and metastasis.29 Further, studies have reported that angiotensin receptor blockers can restrain the growth of various cancer cells. Moreover, studies suggested that treatment with telmisartan leads to apoptosis and it has anti-proliferative activity in prostate and renal cancer cells.30,31 Also, anticancer effects of telmisartan on various human melanoma cell lines have been investigated after 72 hours of treatment, with IC50 values for HTB140, FemX, 518a2, and A375 cell lines being 136.7, 116.6, 51.2, and 54.1 μM respectively.32
The synthesized 9-CH2OH Nos-Tel SLNs were extensively characterised via various techniques like FE-SEM, TEM, DLS, FT-IR, AFM, XPS etc. Also, noscapine and telmisartan drug content/drug loading in the synthesized nanoparticles were estimated using RP-HPLC.
In addition, we have explored the interaction of 9-CH2OH Nos-Tel SLNs with bovine serum albumin (BSA) using both computational and spectroscopic approaches as the drug–protein interactions are vital and can provide insights into the drug ADMET profile and overall bioavailability. We were inclined towards albumins because they are the most abundant proteins of the circulating system which are involved in binding and transportation of essential nutrients as well as drugs (both exogenous and endogenous) to cells.33 BSA is an abundant and important plasma protein found in mammals, and is 76% homologous to human serum albumin (HSA).34 BSA is chosen as the model protein to visualize the possible interaction(s) of nanocomposite and albumins due to its structural homology and its cost-effectiveness. BSA has three homologous domains (I–III), further subdivided into two sub-domains, named A and B.35 The two tryptophan residues in BSA are responsible for its characteristic wavelength in UV light and fluorescence. Trp-134 resides in sub-domain IB on the surface of the protein whereas another Trp-212 residue is positioned in the hydrophobic cavity of sub-domain IIA.36
Lastly, we have assessed the dose-dependent cellular toxicity of 9-CH2OH Nos-Tel SLNs along with 9-CH2OH Nos and telmisartan in non-small lung cancer cell line H1299 via MTT assay. These SLNs could be an interesting target for nanomedicine(s) and warrant further in vitro and in vivo assays to gain clinical success in the future.
2. Experimental section
2.1. Materials
Telmisartan (Tel) was procured from TCI, noscapine hydrochloride from Sigma-Aldrich and 9-CH2OH noscapine was synthesised in the laboratory. Stearic acid (SA) was purchased from Thomas Baker, India and Tween 20 was purchased from Himedia. All the other chemicals procured were of the highest analytical grade. BSA and all other chemicals/reagents were purchased from Sigma-Aldrich, TCI, and Alfa Aesar.
2.2. Synthesis of 9-CH2OH noscapine
For more than three decades, our team has been diligently engaged in the design and synthesis of noscapinoids, or analogues of noscapine. Noscapine is a 3-substituted phthalide derived pharmacophore that consists of methoxy-containing isoquinoline and benzofuranone rings linked by a C–C chiral bond.37 Over time, the 9′-substituted noscapine analogues have proven to be more effective than other substitutions of the parent compound but selective chemical modification at C-9 is quite complicated. In this study, ‘9-hydroxymethyl noscapine’ was synthesised using the Blanc reaction wherein noscapine hydrochloride (Scheme 1, Compound 1) was treated primarily with paraformaldehyde and HCl. The reaction mixture (RM) was stirred at ambient temperature for 1.5 hours. After confirming the reaction completion using TLC, the RM was quenched using Na2CO3. The organic fraction was then extracted using CHCl3, filtered through dried Na2SO4 and the filtrate was concentrated to give crude. The crude was later purified using column chromatography (ethyl acetate/hexane: 30/70) to give the desired product (Scheme 1, Compound 2) in 95% yield. The 1H and 13C NMR spectra are given in the ESI.†
 |
| | Scheme 1 Synthesis of 9-hydroxymethyl noscapine (9-CH2OH Nos). | |
2.3. Synthesis of 9-CH2OH Nos-Tel solid lipid nanoparticles
The 9-hydroxymethyl noscapine telmisartan-loaded nanostructured lipid particles (9-CH2OH Nos-Tel-SLNs) were prepared using a previously reported solvent diffusion method.38,39 Briefly, stearic acid (200 mg), Tween® 20, 9-CH2OH noscapine (30 mg), and telmisartan (30 mg) were dissolved in ethanol and acetone in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. After this, the organic phase was poured into 100 mL of distilled pre-heated water, kept at a temperature of 80 °C. The RM was allowed to stir for 1 hour at the same temperature. Solvent was evaporated under vacuum and then lyophilized to obtain dry product.
1 ratio. After this, the organic phase was poured into 100 mL of distilled pre-heated water, kept at a temperature of 80 °C. The RM was allowed to stir for 1 hour at the same temperature. Solvent was evaporated under vacuum and then lyophilized to obtain dry product.
2.4. Characterization of nanoparticles
2.4.1. Microscopy.
A Thermo Scientific TALOSTEM was utilised to carry out transmission electron microscopy (TEM) to determine the morphology and size of 9-CH2OH Nos-Tel-SLNs. For the experiment, Cu grids coated with carbon were used. The sample was dispersed in water and then deposited on the grids using the drop-casting method. The software used to calculate the average size of the nanoparticles was ImageJ. For FE-SEM analysis, a Zeiss Gemini SEM 500 thermal field emission type microscope was used. The surface morphology of 9-CH2OH Nos-Tel-SLNs was visualized using an atomic force microscopy (AFM) instrument, Park Systems, NX10 using AR5T-NCHR probe. The scanning was done in non-tapping mode. The nanoparticles were dispersed in DMSO and then deposited on a glass electrode which was air-dried for 3 hours at 60 °C before analysis.
2.4.2. Zeta potential and particle size.
Samples of nanoparticles were dispersed in sterile water at a concentration of 1 mg mL−1 and then sonicated for 5–10 minutes. Then, 1 mL of this solution was transferred to a quartz cuvette before analysis. Dynamic light scattering, with a Nanoplus particle size analyzer (Particulate Systems), was used to measure the particle size and a Zetasizer Nano-ZS from Malvern Instruments, UK was employed to measure the surface charge of 9-CH2OH Nos-Tel-SLNs.
2.4.3. Fourier-transform infrared (FT-IR) spectroscopy.
A Shimadzu II FT-IR spectrometer was utilized to record the FT-IR spectra of 9-CH2OH Nos, telmisartan, stearic acid as well as 9-CH2OH Nos-Tel-SLNs.
2.4.4. X-ray photoelectron spectroscopy (XPS).
The surface analysis to measure elemental composition was conducted using a Thermo Fisher Scientific instrument. The total acquisition time for XPS survey was 25.2 s and the source used was Al K alpha. The standard lens mode was used with a spot size of 400 μm.
2.5. Drug loading using RP-HPLC
The drug loading studies were carried out with a RP-HPLC instrument comprising a PDA detector (SPD-M20A, Shimadzu, Japan PDA) and Rheodyne injector with a 20-μL fixed loop. The solvents used were HPLC-grade acetonitrile and water which were sonicated and passed through a 0.45-μm nylon filter before being used. The stock solutions of 9-CH2OH noscapine and telmisartan were prepared to be 1000 μg mL−1 from which solutions of 50, 100, 150, 200 and 250 μg mL−1 each were prepared for obtaining the calibration curve. The samples of both drugs and of 9-CH2OH Nos-Tel-SLNs were run using Phenomenex® Kinetica, C18 (250 × 4.6 mm, 5 μm particle size) to be the stationary phase and optimized acetonitrile:water (65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35%, v/v) to be the mobile phase. The rate of flow was set to 1 mL minute−1, total run time was 8 minutes per sample and effluents were monitored at 254 nm. The following eqn (1) was used to determine drug loading:
35%, v/v) to be the mobile phase. The rate of flow was set to 1 mL minute−1, total run time was 8 minutes per sample and effluents were monitored at 254 nm. The following eqn (1) was used to determine drug loading:| | 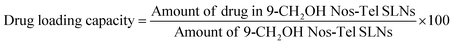 | (1) |
2.6. Binding studies with BSA and 9-CH2OH Nos-Tel-SLNs
2.6.1. UV-visible spectroscopy.
The instrument used for this study was a UV-visible spectrophotometer (Cary 300) from Varian. A BSA solution of 10 μM concentration was prepared in PBS (pH ∼ 7.4), and a stock solution of 9-CH2OH Nos-Tel-SLNs of 1 mg mL−1 in sterile water was prepared for injection. The absorption spectrum of pure BSA was recorded and then subsequently changes in the BSA absorption spectrum were studied while adding increasing amounts of 9-CH2OH Nos-Tel-SLNs (12.5–125 μg mL−1). The experiment was performed using 1× PBS (10 mM) as a reference solution and the spectra were collected over the wavelength range of 200 nm to 400 nm.
2.6.2. Steady-state fluorescence spectroscopy.
A Hitachi F-7000 fluorescence spectrophotometer was used for these investigations. The measured spectra ranged in wavelength from 200 to 500 nm while exciting the samples at 280 nm. During the experiment, the scan rate was fixed at 2400 nm min−1 with emission slit width and excitation slit width at 5 nm. We recorded a spectrum of pure BSA (10 μM) solution at 298 K and then later titrated it with gradually increasing volumes of stock solutions of 9-CH2OH Nos-Tel-SLNs (12.5–87.5 μg mL−1). During fluorescence experiments, we needed to account for the self-quenching effect in typical fluorophores (like BSA) only when the concentration is greater than 10 μM and the path length is 1 cm. Herein, we are using an optimized 10 μM concentration of BSA and do not need to focus on self-quenching of the fluorophore.40 For appropriate evaluation of the data, we accounted for the inner filter effect and corrected for it by using eqn (2):| | | Fcorrected = Fobserved·e(Aex+Aem)/2 | (2) |
In equation (2), Fcorrected denotes the fluorescence upon correction after considering the inner filter effect; Fobserved is the fluorescence that was observed for the protein (BSA); Aex is the absorption of the ligand (9-CH2OH Nos-Tel SLNs) at the excitation wavelength and Aem is the absorption of the ligand at the emission wavelength.
Consequent upon obtaining proper emission data, we utilized them to produce a Stern–Volmer plot by noting down the changes in fluorescence intensities and then constructed a double-logarithmic plot (modified Stern–Volmer plot).
2.6.3. Circular dichroism (CD) spectroscopy measurement.
This experiment was conducted with a JASCO J-810 CD spectrophotometer using 1 μM BSA solution at 298 K and a pH of 7.4. We recorded all the spectra from 190 nm to 250 nm at a fixed scan rate of 100 nm min−1 under continuous nitrogen flush. We used a quartz cuvette with a 1-cm path length for the experiment. The CD spectra of BSA with and without the presence of 9-CH2OH Nos-Tel-SLNs were recorded while keeping the concentration of BSA solution constant during addition of different amounts of 9-CH2OH Nos-Tel-SLNs.
2.7. Computational studies
2.7.1.
In silico docking and molecular dynamics studies.
Molecular docking was done to visualize the binding poses of the ligands (noscapine and telmisartan) to BSA and to get insights into their binding free energies. The docking was performed using the multi ligand approach41. To begin with, the 3D crystal structure of BSA (PDB ID: 4OR0) was retrieved from the Protein Data Bank (https://www.rcsb.org/) online server directly to Maestro 13.5 (Schrodinger Suite 2023-1).42,43 The preparation of protein was done using the in-build protein preparation workflow tab and used default settings but did not delete the ligands as we required the 3D coordinates of the active sites. In this structure there are 4–5 active sites present; thus as a result we have used all the sites and performed docking for the precise and most probable binding sites.44 Any missing residue was repaired as it is inbuilt in the preprocess step followed by minimization of all the residues. The structures of drugs telmisartan and noscapine were drawn and energy minimization step was done with Chem3D 16.0 using the MM2 forcefield and applying a 0.010 RMS gradient. Similarly, the ligand preparation was done using inbuilt module LigPrep using OLPS4 forcefield and used rest settings as default.45,46 Lastly, Glide version 98128 (mmshare version 61128) was used to dock the ligands one by one to all the 5 active sites present on the raw PDB file.47–50 Several other tools were also utilized for the visualization and analysis. The final docked ligands were then further used for molecular dynamics (MD) simulation studies using GROMACS-2023.1 with the AMBER99SB-ILDN protein, nucleic AMBER94 forcefield.51–56 Antechamber and Acpype.py were utilized to get the topology of ligands used.57 The simulation system was solvated using TIP3P model and then system was neutralised by replacement of the solvent molecules with Na+ or Cl− as required by the system. The energy was minimized with the MD (Leap-Frog integrator) method followed by 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps for 1000 ps NVT equilibration at 300 K using V-rescale thermostat and NPT equilibration step at 300 K using Berendsen's pressure coupling algorithm and 1 atm pressure for 500
000 steps for 1000 ps NVT equilibration at 300 K using V-rescale thermostat and NPT equilibration step at 300 K using Berendsen's pressure coupling algorithm and 1 atm pressure for 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps for 1000 ps. Lastly, MD simulation was done for a time span of 20
000 steps for 1000 ps. Lastly, MD simulation was done for a time span of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps (20 ns).
000 ps (20 ns).
2.7.2. Binding free energy calculation with MMPBSA/GBSA.
The final trajectories of the MD simulation were further employed to get the net binding free energy of the system (protein + Nos + Tel). Molecular mechanics Poisson–Boltzmann surface area/generalized-Born surface area (MM-PBSA/GBSA) tools were utilized to get the free energy values of different types of strong or weak interaction of the drugs with the active site.58–60 This is one of the most used techniques that allow one to analyse the binding free energy, understand the stability of the structure during MD and measure the role of each individual residue present on the protein by free energy decomposition studies.
In this method, the binding free energy is calculated by using the equations below:
| | | ΔGbind = 〈GCOM〉 − 〈GREC〉 − 〈GLIG〉 | (3) |
| | | 〈Gx〉 = 〈EMM〉 + 〈Gsol〉 − 〈TS〉 | (4) |
which can also be written as
where Δ
H is the binding enthalpy and −
TΔ
S is the conformational entropy upon binding of drugs with the protein. There are other steps involved but the steps shown above are the most generally used steps to get an idea regarding the output values.
2.8. Cellular experiments
Using the H1299 lung cancer cell line, the cytotoxicity of 9-CH2OH Nos, telmisartan, and 9-CH2OH Nos-Tel-SLNs was assessed. The cells were obtained from National Centre for Cell Science (NCCS), Pune, India. H1299 cells were grown in DMEM with 10% FBS along with 5% antibiotic–antimycotic supplement. A stock solution comprising 25 mg mL−1 each of 9-CH2OH Nos, telmisartan, and 9-CH2OH Nos-Tel-SLNs was prepared in DMSO solvent. A temperature of 37 °C at 5% CO2 in a CO2 incubator was set for maintaining the cell lines. After being seeded onto a 96-well plate at a density of 8 × 103 cells per well, the cells were incubated for a day. Following treatment with various drug doses (50, 75, 100, 125, and 150 μg mL−1), the cancer cells were cultivated for 48 hours in an incubator. DMSO-treated cells were used as control. After incubation, 20 μL of (5 mg mL−1) MTT reagent was added to each well, which was subsequently incubated for another three hours in the CO2 incubator. The MTT reagent was discarded after the incubation. The formazan crystals formed were dissolved using DMSO (100 μL). The absorbance was then recorded at 570 nm and reference at 630 nm. The mean ± SD of quadruplet data of both control and samples was used to represent the final result. To perform statistical comparisons, Student's t-test was utilized. With GraphPad Prism software 8.4.2, the IC50 (inhibitory concentration for 50% cell proliferation) was calculated from graphical plots of the dose–response curve at each concentration. The following eqn (3) was applied to express the cell viability percentage:| |  | (6) |
3. Results and discussion
3.1. Characterization of 9-CH2OH Nos-Tel-SLNs
3.1.1. Microscopy.
The size and morphology of the solid lipid nanoparticle formulation were studied using TEM. First, 1 mg of 9-CH2OH Nos-Tel SLNs was diluted with sterile water (1 mL) and sonicated for 2 minutes before being placing onto a Cu-r grid coated with carbon. It was then stained with 1% phosphotungstic acid. The sample was dried prior to inspection. In Fig. 1a, the TEM image revealed that the particles were roughly spherical in shape with 36.6 nm as the average size. The nanoparticle size ranged from 20 to 50 nm as seen in the associated histogram (Fig. 1b). The morphology of the nanoparticles was confirmed using FE-SEM (Fig. 1c). AFM was performed to observe the morphology of the synthesised nanocomposites in more detail. In Fig. 1d, the nanoparticles were seen to be in agglomerated form.
 |
| | Fig. 1 (a) TEM image at 100 nm; (b) histogram distribution plot depicting size range; (c) FE-SEM image at 200 nm; and (d) AFM image of synthesized 9-CH2OH Nos-Tel-SLNs. | |
3.1.2. DLS and zeta potential.
From this study, we could confirm the formation of 9-CH2OH Nos-Tel-SLNs as DLS results showed the nanostructure's hydrodynamic size to be ∼406.8 nm at 25 °C in sterile water with polydispersity index (PDI) of 0.341 (Fig. 2a). The possible aggregation of 9-CH2OH Nos-Tel-SLNs in water could be the reason for these values. The difference in the average size of the nanostructures from DLS and TEM analyses might be ascribed to the hydrodynamic diameter with swollen nanostructures in this experiment.33,34 The zeta potential came out to be −36.23 mV which might be due to charged groups in both the drugs (Fig. 2b).
 |
| | Fig. 2 (a) DLS intensity distribution and (b) zeta potential distribution of synthesized 9-CH2OH Nos-Tel-SLNs. | |
3.1.3. FT-IR analysis.
The FT-IR stretching frequencies for 9-CH2OH noscapine, telmisartan, stearic acid as well as 9-CH2OH Nos-Tel-SLNs have been recorded. The spectrum of 9-CH2OH Nos exhibited peaks at 3469 cm−1, 2927 cm−1, and 1755 cm−1 representing O–H, aromatic C–H, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch respectively. The spectrum of telmisartan exhibited weak bands at 2937 cm−1 (aromatic C–H stretch), 1681 cm−1 (C
O stretch respectively. The spectrum of telmisartan exhibited weak bands at 2937 cm−1 (aromatic C–H stretch), 1681 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching in carboxylic acid), and 1263 cm−1 (C–O stretching in carboxylic acid). The spectrum of stearic acid exhibited peaks at 2926 cm−1 (asymmetric C–H stretch) and 2864 cm−1 (symmetric C–H stretching/methylene CH2 stretching).3 The 9-CH2OH Nos-Tel-SLNs spectrum displayed peaks at 3061 cm−1 (aliphatic C–H stretch), 2926 cm−1 and 2864 cm−1 (C–H stretch in CH3/CH2 groups), 1697 cm−1 (C
O stretching in carboxylic acid), and 1263 cm−1 (C–O stretching in carboxylic acid). The spectrum of stearic acid exhibited peaks at 2926 cm−1 (asymmetric C–H stretch) and 2864 cm−1 (symmetric C–H stretching/methylene CH2 stretching).3 The 9-CH2OH Nos-Tel-SLNs spectrum displayed peaks at 3061 cm−1 (aliphatic C–H stretch), 2926 cm−1 and 2864 cm−1 (C–H stretch in CH3/CH2 groups), 1697 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch), and 1251 cm−1 (C–O stretch). The comparative IR spectral analysis indicated that Nos and Tel preserved their characteristics in the lipid matrix (Fig. 3).
O stretch), and 1251 cm−1 (C–O stretch). The comparative IR spectral analysis indicated that Nos and Tel preserved their characteristics in the lipid matrix (Fig. 3).
 |
| | Fig. 3 Comparative FTIR spectra of 9-CH2OH Nos; telmisartan; stearic acid; and 9-CH2OH Nos-Tel-SLNs. | |
3.1.4. X-ray photoelectron spectroscopy (XPS).
To determine the chemical state and composition of different elements in the nanocomposite, we have performed XPS analysis. The spectra in Fig. 4 confirm the presence of O, N, and C in the nanocomposite. The sharp peak at 286 eV corresponds to C–C; however, the C spectrum has a broad binding energy peak at 289 eV indicating O-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O components. The chemical states for C
O components. The chemical states for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O binding energy were observed around 530–536 eV for O 1s. However, for N 1s the binding energy was around 400 eV.61
O and C–O binding energy were observed around 530–536 eV for O 1s. However, for N 1s the binding energy was around 400 eV.61
 |
| | Fig. 4 XPS spectra of 9-CH2OH Nos-Tel-SLNs: (a) XPS survey spectrum; (b) N 1s scan; (c) O 1s scan; and (d) C 1s scan. | |
3.2. Estimation of 9-CH2OH noscapine and telmisartan drug loading in 9-CH2OH Nos Tel SLNs
RP-HPLC is a conventional technique which is used for estimation of loaded drugs content or encapsulation efficiency in nanoparticles. Herein, we used this technique to calculate the drug loading of 9-CH2OH noscapine as well as telmisartan in the synthesized nanoparticles. The retention times for telmisartan and 9-CH2OH noscapine were found to be 2.21 and 3.82 minutes respectively. The obtained calibration curves of 9-CH2OH noscapine (Fig. 5a) and telmisartan (Fig. 5b) were found to be linear with R2 = 0.9986 and 0.9971, respectively, in concentrations ranging from 50 to 250 μg mL−1. The obtained equation for 9-CH2OH noscapine was Y = 7987.35X – 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 362.7 and that for telmisartan was Y = 22
362.7 and that for telmisartan was Y = 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 986.16X + 317
986.16X + 317![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 787.9. Using these equations and the peak areas in the chromatogram of the nanoparticles shown in Fig. 5c, the loadings of noscapine and telmisartan were found to be 1.86% and 1.97%, respectively, in 9-CH2OH Nos-Tel SLNs.
787.9. Using these equations and the peak areas in the chromatogram of the nanoparticles shown in Fig. 5c, the loadings of noscapine and telmisartan were found to be 1.86% and 1.97%, respectively, in 9-CH2OH Nos-Tel SLNs.
 |
| | Fig. 5 Calibration curves of (a) 9-CH2OH noscapine and (b) telmisartan. (c) HPLC chromatogram of 9-CH2OH Nos-Tel SLNs depicting retention times and peak areas of the two loaded drugs. | |
3.3. Biophysical studies
3.3.1. UV-visible spectroscopy.
UV-visible absorption spectroscopy is a widely established technique to get an insight into ligand-protein interactions (9-CH2OH Nos-Tel SLNs and BSA, in our case). It also tells us about the conformational changes in the protein ensuing upon possible complex formation between nanocomposite and protein. The absorption spectrum of BSA62,63 has characteristic peaks at 198 nm and 278 nm. The peak at λmax = 198 nm represents the carbonyl group (n to π*) transitions in the backbone of BSA whereas that at λmax = 278 nm corresponds to aromatic amino acid (π to π*) transitions. The absorption spectra of BSA with and without 9-CH2OH Nos-Tel-SLNs are shown in Fig. 6d. Upon successive addition of 9-CH2OH Nos-Tel SLNs to BSA, we observed a hyperchromic shift, i.e., enhancement in the peak intensity of BSA suggesting interaction between the nanoparticles and the protein. It can be inferred that due to possible complex formation between nanoparticles and BSA, there are conformational changes in BSA which lead to increased aromatic amino acid exposure in the hydrophobic region.64
 |
| | Fig. 6 Absorption spectra of (a) 9-CH2OH noscapine; (b) telmisartan; (c) 9-CH2OH Nos-Tel SLNs; and (d) BSA against varying amounts of 9-CH2OH Nos-Tel SLNs. | |
3.3.2. Steady-state fluorescence spectroscopy.
Fluorescence or emission spectroscopy is an effective technique, being utilized to gain a deeper comprehension of ligand-protein interactions and to probe into the mechanism of complex formation between protein (BSA) and nanocomposite.65 Proteins like BSA have intrinsic fluorescence due to the presence of aromatic amino acid residues like phenylalanine, tryptophan, and tyrosine.66 Amongst these, tryptophan is the most liable to be influenced by any structural changes and thus contributes majorly towards fluorescence quenching of the protein.67,68 The fluorescence spectra of BSA with and without 9-CH2OH Nos-Tel-SLNs are shown in Fig. 7a. In the absence of the nanoparticles, BSA showed maximum emission at 344 nm when excited at 280 nm wavelength.69 In line with our absorption spectroscopy results, we noted that there was a decrease in the emission intensity of BSA (fluorescence quenching) due to stepwise addition of 9-CH2OH Nos-Tel-SLNs along with a bathochromic shift (Δλ = 5 nm). The fluorescence quenching upon addition of the nanoparticles is evidence of the interaction between BSA and 9-CH2OH Nos-Tel-SLNs.
 |
| | Fig. 7 (a) Fluorescence/emission spectra of BSA against different amounts of 9-CH2OH Nos-Tel SLNs. (b) Stern–Volmer plot. (c) Modified Stern–Volmer plot. | |
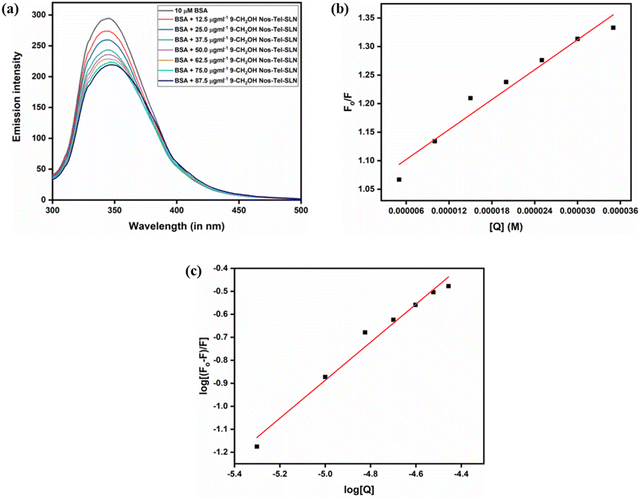
To assess the nature of the interaction, i.e., static, dynamic or mixed quenching, we utilized the Stern–Volmer eqn (7) mentioned below. Static quenching occurs when a ground-state complex is formed between a drug molecule and protein, whereas dynamic quenching refers to when collisional encounters occur between protein and drug(s). Mixed quenching is an outcome of both collisions and formation of complex between the protein and drug(s) simultaneously.70,71 The relations between Fo/F and [Q] in all three cases are given as:
| |  | (7) |
| |  | (8) |
| | 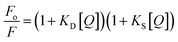 | (9) |
In the above equations,
Fo is BSA's fluorescence without the nanoparticles;
F is BSA's fluorescence with the nanoparticles; [
Q] corresponds to the drug's concentration,
Ksv is the Stern–Volmer constant,
Kq is the biomolecular quenching constant,
Ks is the association constant,
KD is the dynamic quenching constant and
τo (∼10
−8 seconds)
72 corresponds to the protein's lifetime in the absence of quencher. From the equations, it is clear that a plot of
Fo/
F against [
Q] is a straight line (linear) in the case of static quenching (
Fig. 7b,
eqn 7) and dynamic quenching (
eqn 8) whereas it is non-linear for mixed quenching (
eqn 9). In our study, the plot was found to be linear, for which
Ksv and
Kq are mentioned in
Table 1. From
Kq value of ∼0.874 × 10
12 M
−1 s
−1, we concluded that BSA follows the static mode of quenching forming a stable ground-state complex with 9-CH
2OH Nos-Tel SLNs (
Kq is of the order of 10
12,
i.e., higher than 2 × 10
10 M
−1 s
−1 or the maximum scattering collisional quenching constant).
73
Table 1 Quenching constants and binding parameters for 9-CH2OH Nos-Tel SLN binding with BSA
| Temperature (K) |
K
sv (M−1) × 104 |
K
q (M−1 s−1) × 1012 |
K
b (M−1) × 104 |
n
|
| 298 |
0.874 |
0.874 |
0.1752 |
0.82 |
Furthermore, we used the modified Stern–Volmer equation (Fig. 7c, eqn 10) to measure the binding constant and also the number of binding sites:
| |  | (10) |
In the above equation,
Kb is the binding constant and
n represents the number of binding sites. From
Table 1, we can see that 9-CH
2OH Nos-Tel SLN and BSA bind in a 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
1 stoichiometric ratio with 0.17 × 10
4 M
−1 as the value of the binding constant.
3.4. Computational studies
3.4.1. Molecular docking and MD studies.
We performed in silico assessments via molecular docking and MD simulation studies to verify our experimental observations and visualize the binding affinity of the drugs (noscapine and telmisartan) in 9-CH2OH Nos-Tel SLNs and to further investigate the stability of the formed complex. We performed the docking using Glide (an inbuilt module to the Schrodinger Suite 2023-1). As seen, there are 5 active sites present in the PDB file of BSA (PDB ID: 4OR0). We docked the drug one by one to all the active sites and compared the affinity of binding for both drugs on the bases of their respective docking scores. In Table 3, we show the details of all five active sites and the docking values to the corresponding sites. As we can see from Table 3, the best binding scores were found to be at NSP 601 for both drugs.
Table 3 Docking scores of both drugs (Nos and Tel) at different active sites in BSA
| Ligand present on the active site |
Drug |
Docking score (kcal mol−1) |
Glide score |
|
Best docking score.
Second best docking score.
|
| NSP 601 |
Nos |
−5.893a |
−6.070 |
| Tel |
−6.568a |
−8.356 |
| NSP 602 |
Nos |
−4.135 |
−4.937 |
| Tel |
−5.676b |
−5.967 |
| NSP 603 |
Nos |
−4.506b |
−4.683 |
| Tel |
−5.520 |
−5.810 |
| PEG 604 |
Nos |
−3.096 |
−3.274 |
| Tel |
−4.458 |
−6.512 |
| PEG 605 |
Nos |
−3.826 |
−4.003 |
| Tel |
−3.670 |
−3.960 |
We noted that the docking score of telmisartan was significantly better relative to that of noscapine; thus we decided to use the docking score of the best pose for Tel, i.e., −6.568 kcal mol−1 (best result), and the docking score for Nos, i.e., −4.506 kcal mol−1 (second-best result), for further simulations. The respective interactions of the system/complex containing both drugs at different sites in BSA are displayed in Fig. 9a. The individual interactions of noscapine at NSP 603 and telmisartan at NSP 601 are exhibited in Fig. 9b and c. Also, Fig. 9d displays the 2D interactions of both drugs with BSA. The obtained 3D coordinates of the drugs from docking were further utilized for molecular dynamics calculations to check the associated conformational changes to the structure of the protein when binding with the two drugs at an atomic level. Gromacs is the most frequently used tool for MD and other computational studies. Here, we did a 20-ns calculation of final complex which we obtained from molecular docking and used different inbuilt modules of Gromacs to analyse the drug and protein interactions at an atomic level. After completion of the calculations, the final trajectories were investigated to obtain the changes in conformation that occurred during simulation.
 |
| | Fig. 9 (a) Final complex of Nos and Tel with BSA. (b) 3D interaction pose of Nos with BSA at NSP 603. (c) 3D interaction pose of Tel with BSA at NSP 601. (d) 2D interaction poses of both the drugs with BSA. | |
Using the final trajectories, the root mean square fluctuation (RMSF) and root mean square deviation (RMSD) values were evaluated to get information about the conformational flexibility of the protein structure when both the drugs were bound to it. It is clear from Fig. 10a that the RMSF ranged between 0.1 and 0.4 nm indicating the stabilization of the protein structure during the simulation. From Fig. 10b, we see that the RMSD of Nos bound to BSA was initially stabilized until 9 ns but then momentarily increased to almost 3 nm and then later attained equilibrium with RMSD fluctuating between 2.5 and 3 nm for the remaining span of the simulation. On the other hand, RMSD for Tel bound to BSA was almost constant at around 0.25 nm throughout the simulation (Fig. 10b). This indicated that the binding affinity of Tel to BSA is higher and this interaction was more compatible compared to Nos-BSA binding. To confirm these observations, we further did hydrogen bond numbers analysis (H-bond analysis) of both drugs. From Fig. 10c, we note that the number of H-bonds for Tel-BSA interactions was almost thrice relative to Nos-BSA interactions during the simulation. The average number of H-bonds per time frame for Nos was found to be 1.030 out of 668![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 347 and for Tel it was 2.806 out of 688
347 and for Tel it was 2.806 out of 688![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 370. This analysis strengthened the earlier compatibility results and at the same time also validated the higher RMSD of Nos-BSA binding.
370. This analysis strengthened the earlier compatibility results and at the same time also validated the higher RMSD of Nos-BSA binding.
 |
| | Fig. 10 (a) RMSF analysis of BSA. (b) RMSD analysis of BSA when both drugs are bound to it. (c) Hydrogen bond analysis of Nos and Tel with BSA. (d) Radius of gyration (Rg) analysis. | |
Lastly, analysis of radius of gyration (Rg) was done to get information about overall dimensions of the protein. In Fig. 10d, we can clearly see that the protein stability persisted throughout the simulation along with both the drugs. In the case of Tel, the value was a little higher than that for Nos which can be due to the overall large size of the drug. The large drug interactions resulted in slight loosening of the compactness or slight increase in flexibility of the secondary structure of BSA.
3.5.
In vitro cytotoxicity assay
The cytotoxicity of 9-CH2OH Nos, telmisartan and 9-CH2OH Nos-Tel-SLNs to lung cancer cells was screened by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay. We analyzed the reduction of MTT reagent by metabolically active cells to its insoluble formazan crystal form. Dimethyl sulfoxide (DMSO) was employed to dissolve the formazan crystals that form during the process. When dissolved in an appropriate solvent, they become purple in colour. The intensity of the colour can be measured spectrophotometrically and this yields a proportionate relationship with the number of live cells. The cytotoxic effects of 9-CH2OH Nos, telmisartan and 9-CH2OH Nos-Tel-SLNs at 50, 75, 100, 125, and 150 μg mL−1 were assessed in vitro and the IC50 values were determined to be 282.5, 155.7, and 186 μg mL−1, respectively, after 48 h of treatment. The concentration-dependent cytotoxicity profiles of all compounds can be seen in Fig. 11. 9-CH2OH Nos-Tel-SLNs were successfully able to deliver drugs into the lung cells substantially inhibiting the proliferation of H1299 cancer cells as shown in Fig. 12. Based on in vitro experiments, the combination of 9-CH2OH Nos and telmisartan in 9-CH2OH Nos-Tel SLNs demonstrates a positive effect. The combination approach enhances the therapeutic potential by allowing the use of lower doses of each drug, potentially minimizing the side effects associated with high doses of a single drug. Telmisartan alone exhibits higher cytotoxicity at high doses, which could also be influenced by experimental conditions, such as differences in bioavailability or delivery in the assay. Overall, the nanoparticle formulation provides a controlled and sustained release of both drugs, which is advantageous for long-term therapeutic strategies.77,78
 |
| | Fig. 11
In vitro dose-dependent cytotoxicity profiles of (a) telmisartan, (b) 9-CH2OH Nos and (c) 9-CH2OH Nos-Tel-SLNs. (d) Cell viability comparison among telmisartan, 9-CH2OH Nos, and 9-CH2OH Nos-Tel-SLNs against H1299 cells. Data represent mean ± SD and *p ≤ 0.05 was considered statistically significant. | |
 |
| | Fig. 12 Morphological changes and decline in cell viability of H1299 cells due to treatment with different concentrations of 9-CH2OH Nos-Tel-SLNs for 48 hours. | |
4. Conclusion
In the present era of evolving nanotechnology, solid lipid nanoparticles are being explored for their uses in drug delivery and clinical medicine. Also, dual-drug systems, also known as combinatorial systems, for drug delivery offer a viable way to combat drug resistance by simultaneously delivering two or more drugs to a tumor site. According to the findings presented in this study, 9-CH2OH Nos-Tel-SLNs were successfully synthesized through the solvent diffusion method. TEM and FE-SEM results reveal the formation of nanostructures that are spherical in shape having a mean size of 36.6 nm. RP-HPLC studies indicate that loaded noscapine and telmisartan content in the nanoparticles is 1.86% and 1.97% respectively. The expanding use of nanoparticles in drug delivery and cancer therapeutics necessitates a thorough examination of their interactions with blood proteins. Therefore, interaction studies were performed using spectroscopic and computational techniques, both of which unveiled a stable complex formation between 9-CH2OH Nos-Tel-SLNs and BSA. Furthermore, the H1299 cell line was used to study the in vitro anticancer activities of 9-CH2OH Nos, telmisartan and 9-CH2OH Nos-Tel-SLNs. For 9-CH2OH Nos, telmisartan and 9-CH2OH Nos-Tel-SLNs, the corresponding values of IC50 are 282.5, 155.7, and 186 μg mL−1. Therefore, the synthesized novel 9-CH2OH Nos-Tel-SLN formulation developed as a dual-drug delivery system can offer significant advantages in the treatment of lung cancer.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its ESI.†
Conflicts of interest
The authors have no conflict of interest.
Acknowledgements
S. Singh is grateful to ICMR (45/102/2018/Nan-BMS) for providing her SRF fellowship. S. Singh and R. Chandra are grateful to Indo-Russia (DST-RFBR: INT/RUS/RFBR/389) grant. S. Sewariya is grateful to CSIR (09/045(1739)/2019-EMR-I) for senior research fellowship. T. Goel is grateful for DST-INSPIRE fellowship (IF200339). The authors are grateful to the Department of Chemistry, University of Delhi for facilities and support.
References
- F. Bray, M. Laversanne, H. Sung, J. Ferlay, R. L. Siegel, I. Soerjomataram and A. Jemal, Ca-Cancer J. Clin., 2024, 74, 229–263 CrossRef.
- O. A. Madkhali, Molecules, 2022, 27, 1543 CrossRef CAS.
- J. Akbari, M. Saeedi, F. Ahmadi, S. M. H. Hashemi, A. Babaei, S. Yaddollahi, S. S. Rostamkalaei, K. A. Addo and A. Nokhodchi, Pharm. Dev. Technol., 2022, 27, 525–544 CrossRef CAS.
- K. Mo, A. Kim, S. Choe, M. Shin and H. Yoon, Pharmaceutics, 2023, 15, 2065 CrossRef CAS PubMed.
- R. A. Paun, S. Jurchuk and M. Tabrizian, Bioeng. Transl. Med., 2024, 9, e10601 CrossRef CAS.
- B. Yechezkel, J. Controlled Release, 2012, 160, 117–134 CrossRef.
- M. A. Liebert., J. Am. Coll. Toxicol., 1987, 6, 321–401 CrossRef.
- S. Das and A. Chaudhary, AAPS PharmSciTech, 2011, 12, 62–76 CrossRef CAS PubMed.
- X. Letao, W. Xing, L. Yun, Y. Guangze, J. F. Robert and Z. Chun-Xia, Adv. NanoBiomed Res., 2022, 2, 2100109 CrossRef.
- D. Lombardo, M. A. Kiselev and M. T. Caccamo, J. Nanomater., 2019, 3702518, 1–26 CrossRef.
- S. Singh, T. Goel, A. Singh, H. Chugh, N. Chakraborty, I. Roy, M. Tiwari and R. Chandra, Artificial Cells, Nanomed., Biotechnol., 2024, 52, 46–58 CAS.
- S. S. Qia, J. H. Suna, H. H. Yuc and S. Q. Yua, Drug Delivery, 2017, 24, 1909–1926 CrossRef.
- S. Fumoto and K. Nishida, Chem. Pharm. Bull., 2020, 68, 603–612 CrossRef CAS.
- H. Zhang, Q. Xue, Z. Zhou, N. He, S. Li and C. Zhao, Front. Pharmacol., 2023, 14, 1176232 CrossRef CAS PubMed.
- C. H. Kim, S. Lee, J. Y. Choi, M. J. Lyu, H. M. Jung, Y. T. Goo, M. J. Kang and Y. W. Choi, Pharmaceuticals, 2023, 16, 349 CrossRef CAS PubMed.
- Y. Guo, W. He and S. Yang,
et al.
, Colloids Surf., B, 2017, 151, 119–127 CrossRef CAS.
- G. Garg, S. Garg, P. Patel, G. D. Gupta and B. D. Kurmi, Int. J. Polym. Mater. Polym. Biomater., 2024, 1–15 Search PubMed.
- T. B. Emran, A. Shahriar, A. R. Mahmud, T. Rahman, M. H. Abir, M. F. Siddiquee, H. Ahmed, N. Rahman, F. Nainu, E. Wahyudin, S. Mitra, K. Dhama, M. M. Habiballah, S. Haque, A. Islam and M. M. Hassan, Front. Oncol., 2022, 12, 891652 CrossRef PubMed.
- C. Pi, W. Zhao, M. Zeng, J. Yuan, H. Shen, K. Li, Z. Su, Z. Liu, J. Wen, X. Song, R. J. Lee, Y. Wei and L. Zhao., Drug Delivery, 2022, 29, 1878–1891 CrossRef CAS.
- S. Guo, Y. Zhang, Z. Wu, L. Zhang, D. He, X. Li and Z. Wang, Biomed. Pharmacother., 2019, 118, 109225 CrossRef CAS.
- C. H. Kim, S. Lee, J. Y. Choi, M. J. Lyu, H. M. Jung, Y. T. Goo, M. J. Kang and Y. W. Choi, Pharmaceuticals, 2023, 16, 349 CrossRef CAS.
- P. C. Rida, D. LiVecche, A. Ogden, J. Zhou and R. Aneja, Med. Res. Rev., 2015, 35, 1072–1096 CrossRef CAS PubMed.
- X. Chen, T. T. T. Dang and P. J. Facchini, Phytochemistry, 2015, 111, 7–13 CrossRef CAS.
- C. Kocak, F. E. Kocak, B. Ozturk, G. Tekin and H. Vatansev, Bratisl. Lek. Listy, 2020, 121, 43–50 CAS.
- A. Awasthi, N. Kumar, A. Mishra, R. Ravi, A. Dalal, S. Shankar and R. Chandra, ACS Pharmacol. Transl. Sci., 2022, 5, 1292–1304 CrossRef CAS.
- V. Tomar, S. Kukreti, S. Prakash, J. Madan and R. Chandra, Curr. Top. Med. Chem., 2017, 17, 174–188 CrossRef CAS.
- M. Sharpe, B. Jarvis and K. L. Goa, Drugs, 2001, 61, 1501–1529 CrossRef CAS.
- S. Zhang and Y. Wang, Oncol. Lett., 2018, 15, 5859–5864 Search PubMed.
- T. Matsui,
et al.
, Int. J. Mol. Sci., 2019, 20, 3197 CrossRef CAS.
- K. Tascilar, L. Azoulay, S. Dell’Aniello, D. B. Bartels and S. Suissa, Am. J. Hypertens., 2016, 29, 1358–1365 CrossRef CAS.
- E. Samukawa, S. Fujihara, K. Oura, H. Iwama, Y. Yamana, T. Tadokoro, T. Chiyo, K. Kobayashi, A. Morishita and M. Nakahara,
et al.
, Int. J. Oncol., 2017, 51, 1674–1684 CrossRef CAS.
- J. Grahovac, T. Srdić-Rajić, J. F. Santibañez, M. Pavlović, M. Čavić and S. Radulović, Cancer Biol. Med., 2019, 16, 247–263 CrossRef.
- B. Carlo and E. Domenici, Curr. Med. Chem., 2002, 9, 1463–1481 CrossRef.
- H. Chugh and R. Chandra, J. Biomol. Struct. Dyn., 2019, 37, 35–36 Search PubMed.
- S. Gandhi and I. Roy, J. Mol. Liq., 2019, 296, 111871 CrossRef CAS.
- P. Szymaszek,
et al.
, J. Mol. Liq., 2022, 347, 118262 CrossRef CAS.
- A. Awasthi, M. Singh, G. Rathee and R. Chandra, RSC Adv., 2020, 10, 12626–12652 RSC.
- R. M. Shah, F. Malherbe, D. S. Eldridge, E. A. Palombo and I. H. Harding, J. Colloid Interface Sci., 2014, 428, 286–294 CrossRef CAS.
- R. M. Shah, D. Rajasekaran, M. Ludford-Menting, D. S. Eldridge, E. A. Palombo and I. H. Harding, Colloids Surf., B, 2016, 140, 204–212 CrossRef CAS PubMed.
- W. Bae, T. Y. Yoon and C. Jeong, PLoS One, 2021, 16, e0247326 CrossRef CAS PubMed.
- S. Raghavendra, S. J. Aditya Rao, V. Kumar and C. K. Ramesh, Comput. Biol. Chem., 2015, 59, 81–86 CrossRef CAS.
- H. M. Berman, The Protein Data Bank, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS.
- A. Bujacz, K. Zielinski and B. Sekula, Proteins, 2014, 82, 2199–2208 CrossRef CAS.
- G. M. Sastry, M. Adzhigirey, T. Day, R. Annabhimoju and W. Sherman, J. Comput.-Aided Mol. Des., 2013, 27, 221–234 CrossRef PubMed.
- C. Lu, C. Wu, C. D. Ghoreishi, W. Chen, L. Wang, W. Damm, G. A. Ross, M. K. Dahlgren, E. Russell, C. D. Von Bargen, R. Abel, R. A. Friesner and E. D. Harder, J. Chem. Theory Comput., 2021, 17, 4291–4300 CrossRef CAS.
- R. C. Johnston, K. Yao, Z. Kaplan, M. Chelliah, K. Leswing, S. Seekins, S. Watts, D. Calkins, J. Chief Elk, S. V. Jerome, M. P. Repasky and J. C. Shelley, J. Chem. Theory Comput., 2023, 19, 2380–2388 CrossRef CAS.
- R. A. Friesner, J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis and P. S. Shenkin, J. Med. Chem., 2004, 4, 1739–1749 CrossRef PubMed.
- T. A. Halgren, R. B. Murphy, R. A. Friesner, H. S. Beard, L. L. Frye, W. T. Pollard and J. L. Banks, J. Med. Chem., 2004, 47, 1750–1759 CrossRef CAS.
- R. A. Friesner, R. B. Murphy, M. P. Repasky, L. L. Frye, J. R. Greenwood, T. A. Halgren, P. C. Sanschagrin and D. T. Mainz, J. Med. Chem., 2006, 49, 6177–6196 CrossRef CAS PubMed.
- Y. Yang, K. Yao, M. P. Repasky, K. Leswing, R. Abel, B. K. Shoichet and S. V. Jerome, J. Chem. Theory Comput., 2021, 17, 7106–7119 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
-
S. Páll, M. J. Abraham, C. Kutzner, B. Hess and E. Lindahl, Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS, Solving Software Challenges for Exascale: International Conference on Exascale Applications and Software, EASC 2014, Stockholm, Sweden, April 2-3, 2014, Revised Selected Papers 2, 2015, pp. 3–27.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess and E. Lindahl, Bioinformatics, 2013, 29, 845–854 CrossRef CAS.
- Y. Liu, F. Ji and R. Liu, Nanotoxicology, 2013, 7, 97–104 CrossRef CAS PubMed.
- D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef CAS.
- E. Lindahl, B. Hess and D. van der Spoel, J. Mol. Model., 2001, 7, 306–317 CrossRef CAS.
- A. W. Sousa da Silva and W. F. Vranken, BMC Res Notes, 2012, 5, 367 CrossRef.
- M. S. Valdés-Tresanco, M. E. Valdés-Tresanco, P. A. Valiente and E. Moreno, J. Chem. Theory Comput., 2021, 17, 6281–6291 CrossRef PubMed.
- S. Genheden and U. Ryde, Expert Opin. Drug Discovery, 2015, 10, 449–461 CrossRef CAS PubMed.
- E. Wang, H. Sun, J. Wang, Z. Wang, H. Liu, J. Z. H. Zhang and T. Hou, Chem. Rev., 2019, 119, 9478–9508 CrossRef CAS.
- T. Goel, N. Deshwal, S. Gusain, R. Chandra, M. Tiwari and S. Singh, Int. J. Biol. Macromol., 2024, 283, 137532 CrossRef CAS PubMed.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- Z. Chi, R. Liu, Y. Teng, X. Fang and C. Gao, J. Agric. Food Chem., 2010, 58, 10262–10269 CrossRef CAS.
- Y. Li, W. He, J. Liu, F. Sheng, Z. Hu and X. Chen, Biochim. Biophys. Acta, Gen. Subj., 2005, 1722, 15–21 CrossRef CAS.
-
J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, 3rd edn, 2006, p. 278 Search PubMed.
- V. D. Suryawanshi, L. S. Walekar, A. H. Gore, P. V. Anbhule and G. B. Kolekar, J. Pharm. Anal., 2016, 6, 56–63 CrossRef.
- A. Biswas, R. K. Swarnkar, B. Hussain, S. K. Sahoo, P. I. Pradeep, G. N. Patwari and R. Anand, J. Phys. Chem. B, 2014, 3, 10035–10042 CrossRef.
- G. Sekar, M. Haldar, D. T. Kumar, C. G. P. Doss, A. Mukherjee and N. Chandrasekaran, J. Mol. Liq., 2017, 241, 793–800 CrossRef CAS.
- S. Sewariya, H. Sehrawat, N. Mishra, M. B. Singh, P. Singh, S. Kukreti and R. Chandra, Int. J. Biol. Macromol., 2023, 247, 125791 CrossRef CAS PubMed.
- S. Gandhi and I. Roy, J. Mol. Liq., 2019, 296, 111871 CrossRef CAS.
- N. Singh, N. Kumar and G. Rathee,
et al.
, ACS Omega, 2020, 5, 2267–2279 CrossRef CAS PubMed.
- Q. Wang, C. R. Huang, M. Jiang, Y. Y. Zhu, J. Wang, J. Chen and J. H. Shi, Spectrochim. Acta, Part A, 2016, 156, 155–163 CrossRef CAS PubMed.
- F. F. Tian, F. L. Jiang, X. L. Han, C. Xiang, Y. S. Ge, J. H. Li, Y. Zhang, R. Li, X. L. Ding and Y. Liu, J. Phys. Chem. B, 2010, 114, 14842–14853 CrossRef CAS.
- K. Phopin, W. Ruankham, S. Prachayasittikul, V. Prachayasittikul and T. Tantimongcolwat, Int. J. Mol. Sci., 2020, 21, 249 CrossRef CAS PubMed.
- M. Voicescu and D. G. Angelescu, J. Nanopart. Res., 2012, 14, 1174 CrossRef.
- A. Micsonai, F. Wien, E. Bulyaki, J. Kun, E. Moussong, Y. H. Lee, Y. Goto, M. Refregiers and J. Kardos, Nucleic Acids Res., 2018, 46(W1), W315–W322 CrossRef CAS.
- B. Gupta, C. S. Yong and J. O. Kim, J. Pharm. Invest., 2017, 47, 461–473 CrossRef CAS.
- R. R. Lala, A. S. Shinde and N. Y. Nandvikar, J. Pharm. Invest., 2018, 10, 17–22 CAS.
|
| This journal is © The Royal Society of Chemistry 2025 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article e and
Ramesh
Chandra
e and
Ramesh
Chandra
 *abde
*abde
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. After this, the organic phase was poured into 100 mL of distilled pre-heated water, kept at a temperature of 80 °C. The RM was allowed to stir for 1 hour at the same temperature. Solvent was evaporated under vacuum and then lyophilized to obtain dry product.
1 ratio. After this, the organic phase was poured into 100 mL of distilled pre-heated water, kept at a temperature of 80 °C. The RM was allowed to stir for 1 hour at the same temperature. Solvent was evaporated under vacuum and then lyophilized to obtain dry product.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35%, v/v) to be the mobile phase. The rate of flow was set to 1 mL minute−1, total run time was 8 minutes per sample and effluents were monitored at 254 nm. The following eqn (1) was used to determine drug loading:
35%, v/v) to be the mobile phase. The rate of flow was set to 1 mL minute−1, total run time was 8 minutes per sample and effluents were monitored at 254 nm. The following eqn (1) was used to determine drug loading: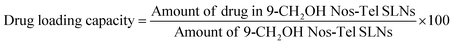
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps for 1000 ps NVT equilibration at 300 K using V-rescale thermostat and NPT equilibration step at 300 K using Berendsen's pressure coupling algorithm and 1 atm pressure for 500
000 steps for 1000 ps NVT equilibration at 300 K using V-rescale thermostat and NPT equilibration step at 300 K using Berendsen's pressure coupling algorithm and 1 atm pressure for 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps for 1000 ps. Lastly, MD simulation was done for a time span of 20
000 steps for 1000 ps. Lastly, MD simulation was done for a time span of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps (20 ns).
000 ps (20 ns).

![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch respectively. The spectrum of telmisartan exhibited weak bands at 2937 cm−1 (aromatic C–H stretch), 1681 cm−1 (C
O stretch respectively. The spectrum of telmisartan exhibited weak bands at 2937 cm−1 (aromatic C–H stretch), 1681 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching in carboxylic acid), and 1263 cm−1 (C–O stretching in carboxylic acid). The spectrum of stearic acid exhibited peaks at 2926 cm−1 (asymmetric C–H stretch) and 2864 cm−1 (symmetric C–H stretching/methylene CH2 stretching).3 The 9-CH2OH Nos-Tel-SLNs spectrum displayed peaks at 3061 cm−1 (aliphatic C–H stretch), 2926 cm−1 and 2864 cm−1 (C–H stretch in CH3/CH2 groups), 1697 cm−1 (C
O stretching in carboxylic acid), and 1263 cm−1 (C–O stretching in carboxylic acid). The spectrum of stearic acid exhibited peaks at 2926 cm−1 (asymmetric C–H stretch) and 2864 cm−1 (symmetric C–H stretching/methylene CH2 stretching).3 The 9-CH2OH Nos-Tel-SLNs spectrum displayed peaks at 3061 cm−1 (aliphatic C–H stretch), 2926 cm−1 and 2864 cm−1 (C–H stretch in CH3/CH2 groups), 1697 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch), and 1251 cm−1 (C–O stretch). The comparative IR spectral analysis indicated that Nos and Tel preserved their characteristics in the lipid matrix (Fig. 3).
O stretch), and 1251 cm−1 (C–O stretch). The comparative IR spectral analysis indicated that Nos and Tel preserved their characteristics in the lipid matrix (Fig. 3).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O components. The chemical states for C
O components. The chemical states for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O binding energy were observed around 530–536 eV for O 1s. However, for N 1s the binding energy was around 400 eV.61
O and C–O binding energy were observed around 530–536 eV for O 1s. However, for N 1s the binding energy was around 400 eV.61
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 362.7 and that for telmisartan was Y = 22
362.7 and that for telmisartan was Y = 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 986.16X + 317
986.16X + 317![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 787.9. Using these equations and the peak areas in the chromatogram of the nanoparticles shown in Fig. 5c, the loadings of noscapine and telmisartan were found to be 1.86% and 1.97%, respectively, in 9-CH2OH Nos-Tel SLNs.
787.9. Using these equations and the peak areas in the chromatogram of the nanoparticles shown in Fig. 5c, the loadings of noscapine and telmisartan were found to be 1.86% and 1.97%, respectively, in 9-CH2OH Nos-Tel SLNs.




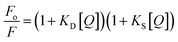

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio with 0.17 × 104 M−1 as the value of the binding constant.
1 stoichiometric ratio with 0.17 × 104 M−1 as the value of the binding constant.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 and 1
6 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10).
10).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 347 and for Tel it was 2.806 out of 688
347 and for Tel it was 2.806 out of 688![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 370. This analysis strengthened the earlier compatibility results and at the same time also validated the higher RMSD of Nos-BSA binding.
370. This analysis strengthened the earlier compatibility results and at the same time also validated the higher RMSD of Nos-BSA binding.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 391.4
391.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339.5
339.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 557.3
557.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339.5
339.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683.46
683.46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681.1
681.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.01
734.01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681.12
681.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 296.1
296.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250
250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182.9
182.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250.1
250.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 391.4
391.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339.5
339.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 557.3
557.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339.5
339.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683.46
683.46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681.1
681.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.01
734.01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 681.12
681.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 053.4
053.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 996.2
996.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 930.2
930.2








