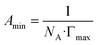Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
SurfPro – a curated database and predictive model of experimental properties of surfactants†
Stefan L.
Hödl
 ,
Luc
Hermans
,
Pim F. J.
Dankloff
,
Aigars
Piruska
,
Wilhelm T. S.
Huck
* and
William E.
Robinson
,
Luc
Hermans
,
Pim F. J.
Dankloff
,
Aigars
Piruska
,
Wilhelm T. S.
Huck
* and
William E.
Robinson
 *
*
Physical Organic Chemistry, Radboud University, Heyendaalseweg 135, 6525AJ Nijmegen, The Netherlands. E-mail: w.huck@science.ru.nl; william.robinson@ru.nl
First published on 19th March 2025
Abstract
Despite great industrial interest, modeling the physical properties of surfactants in water based on their molecular structure remains a challenge. A significant part of this challenge is in obtaining sufficient amounts of high-quality data. Experimentally determined properties such the critical micelle concentration (CMC) and surface tension at CMC (γCMC) have been reported for many surfactants. However, surfactant data are scattered across many literature sources, and reported in a manner which is often unsuitable as input for predictive models. In this work, we address this limitation by compiling the SurfPro database of surfactant properties. SurfPro consists of 1624 surfactant entries curated from 223 literature sources, containing 1395 CMC values, 972 γCMC values and more than 657 values for Γmax, C20, πCMC and Amin. However, only 647 structures have all reported properties, and for most surfactants multiple properties are missing. We trained a previously reported graph neural network architecture for single- and multi-property prediction on these incomplete data of all surfactant types in the database to accurately predict pCMC (−log10(CMC)), γCMC, Γmax and pC20. We achieved state-of-the-art performance of these four properties using an ensemble of AttentiveFP models trained on ten different folds of the training data in the multi-property setting. Finally, we leveraged the predictions and uncertainties of the ensemble model to impute all missing properties for all 977 surfactants with an incomplete set of properties. We make our curated SurfPro database, proposed test split and training datasets, the imputed database, as well as our code publicly available.
1 Introduction
Surfactants are amphiphilic molecules that consist of a hydrophilic head and a hydrophobic tail.1 They have a wide range of applications, including in pharmaceutical formulations, personal care, detergents and coatings.2 Due to their ability to modulate surface tension at the air–water or water–oil interface and form micelles, their functions lie in controlling phenomena such as wetting, emulsification, solubilization and lubrication. The variation in surface tension γ at increasing concentrations of surfactant can be fitted to a Langmuir isotherm3 using the Szyszkowski equation.4 Key characteristic parameters such as the critical micelle concentration (CMC), air–water surface tension at CMC (γCMC), surface excess concentration (Γmax) and the surfactant efficiency (C20) can be determined from this isotherm5 (Fig. 1). The surface excess concentration characterises the surface concentration of surfactant molecules at the saturated surface, which measures the effectiveness of adsorption of surfactant molecules to the interface.1 The CMC is the concentration at which the interfacial concentration of surfactant molecules is saturated and “excess” surfactant molecules self-assemble into micelles. Beyond this concentration, the surface tension is no longer sensitive to increasing surfactant concentration.1 The C20 value measures the surfactant concentration required to reduce the surface tension of a liquid by 20 mN m−1.These parameters and other important characteristics are strongly dependent on molecular structure. As the experimental determination of surfactant properties is laborious, developing models which are capable of predicting them given only molecular structure is of high interest. For example, computational methods based on molecular dynamics (MD) simulations6 or descriptor-based quantitative structure–property relationship (QSPR) models7 have been developed to this end. Whilst these approaches predict the CMC rather accurately, MD simulations are computationally costly and QSPR models tend to perform best within a single class of surfactants (cationic, anionic, etc.).
Recently, machine learning methods have been demonstrated to be effective in predicting surfactant properties given molecular structure information as input. Qin et al.8 gathered a dataset of 202 experimental CMC measurements and made their dataset of SMILES strings and CMC values publicly available. The authors trained a graph convolutional neural network to predict the log(CMC) in CMC units of μM and reported an R2 of 0.92/root mean squared error (RMSE) of 0.30 on their 10% test set (22 surfactants) using two graph convolutional layers (217 K parameters). The authors further demonstrated that a single model is able to accurately predict the log(CMC) for anionic, cationic, non-ionic and zwitterionic surfactants.
Moriarty et al.9 used the same dataset to train graph neural networks (GNN) combined with Gaussian processes, and improved upon the predictive results obtained by Qin et al.,8 reporting a RMSE of 0.23 using a GNN – GaussianProcesses model. The authors additionally explored another validation set of 43 surfactants10 and various GNN architecture variants. Brozos et al.11 extended Qin's dataset to 429 CMC values and further collected 164 Γmax measurements from literature to train a GNN (307 K parameters). They compared single- and multi-task learning as well as model ensembles, and achieved 0.21 mean absolute error (MAE)/0.28 RMSE for log(CMC) on a test set of 66 surfactants and 0.53 MAE/0.76 RMSE for Γmax (24 surfactants) using a single-task model ensemble. Using the multi-task model ensemble trained on pCMC and Γmax, they report 0.4 MAE/0.56 RMSE on Γmax but worse performance on log(CMC) with 0.23 MAE/0.31 RMSE. In another publication the same authors further extended their database with measurements at different temperatures from literature for 492 unique surfactants and achieved accurate predictions for their test settings of 0.24 RMSE including temperature dependence.12
Recently, Chen et al.13 built upon the database of Qin et al.8 to a total of 779 CMC values and developed a descriptor-based QSPR model. The authors split the surfactants into two classes (ionic and non-ionic) and trained separate linear and tree-based machine learning models for both classes. They reported a MAE of 0.24/RMSE of 0.28 on a test set of 79 surfactants.
For γCMC, a database of 691 air–water surface tension measurements aggregated from literature was recently published by Ricardo et al.14. The authors trained a random forest model with five-fold cross validation and achieved errors of 3.38 MAE or 0.55 R2 on average over five hold-out validation sets, which corresponds to the test set errors reported in other works.
Seddon et al.15 used a surface tension dataset of 154 hydrocarbon surfactants to fit the Szyszkowski equation4 and extract the CMC, Γmax and Langmuir constant KL. Using these properties, they trained QSPR models using molecular descriptors and gradient-boosted decision trees. Their approach to extract training data was restricted to the availability of γ – log(C) data, and not all surfactants in their dataset included measurements up to the CMC.
Despite these earlier studies, generally applicable surfactant property prediction models are not available. A limiting factor is the availability of a large database of experimentally determined property measurements of the CMC, γCMC, Γmax and other key surfactant parameters of relevance to the design of novel systems, such as the efficiency of the surfactant in reducing the surface tension (C20), the minimal area occupied by surfactants (Amin) or the surface pressure at CMC (πCMC). Furthermore, database entries must also be suitable for modeling. For instance, providing molecular structure information in the form of machine-readable SMILES strings, as opposed to trade names or trivial names, is essential in facilitating structure-to-property models.
In this work, we address the scarcity of publicly available, machine-readable data suitable for modeling by curating a large database of surfactant properties with machine-readable structures, containing several surfactant classes and properties. This database contains CMC, γCMC, Γmax, C20, πCMC and Amin values derived from experimental measurements. These data and corresponding SMILES structures were digitized and curated from 223 literature sources. For 977 out of 1624 structures not all properties have been reported, and both the fraction and distribution of reported properties differ significantly between surfactant types.
We demonstrate GNNs trained for multi-property prediction can effectively learn from this incomplete data and outperform single-property models especially for properties with the fewest data points. We leverage a stratified split to obtain a representative test set for evaluation and find an ensemble from all models trained on each cross-validation fold outperforms individual models on average. Finally, we impute all missing property values using the ensemble to complete the database. We make our curated SurfPro database, proposed test split, training data sets, imputed database and code publicly available.16,17
2 Methods
2.1 Data acquisition
The results from these papers were compiled into a comprehensive database by manually digitizing and verifying the entries. Tables of property measurements reported in primary literature sources were extracted first into a comma-separated value format, alongside the chemical identifiers used to refer to the surfactants in the paper. The reported units were also extracted and converted to standardized units (Table 1). Measurements performed in the presence of any oil (e.g. paraffin) were not included the database.
A significant challenge during this phase was to generate SMILES strings26 from the source formats. Many primary sources only report trivial compound names, structure images, abbreviations or “code names” defined in the manuscript text. Trivial names were mapped to SMILES strings using PubChem27 searches where possible, and other structural references were transcribed manually. In some cases it was possible to efficiently translate from structured identifiers which were constructed according to a well-defined scheme of structural units. For example, in one publication28 gemini surfactants were encoded as sequences of tokens encoding the head, tail, and spacer groups. In this case, it was possible to write scripts to combine SMILES fragments based on the provided identifiers into surfactant SMILES strings programmatically (results were manually verified). SMILES strings were computationally verified and canonicalized using RDKit29 (version 2024.03.5). Source references are reported for each property individually, and primary sources were used where possible.
All surfactant properties reported in the database (Table 1) can be determined from experimental measurements of the relationship between surfactant concentration and air–water surface tension (Fig. 1). The shape of this plot can be reconstructed using values for CMC, γCMC and Γmax, and all other reported properties can be derived from them. Properties which were calculated from other properties are annotated with a “calculated” note in the corresponding reference entry to differentiate them from entries reported in the literature.
When more than one CMC value was reported in a given source, tensiometry-based measurements (e.g. Wilhelmy plate18–20 or Du Noüy ring20–22), as opposed to, for example conductivity measurements,23 were favored where possible. For duplicate entries of a given property, primary sources were prioritized over aggregated literature sources, and measurements were selected from publications with multiple reported properties over single properties for consistency between related properties. Per-property references and duplicate structures were leveraged to flag questionable entries for further manual verification. For example, entries with the same structure but significantly different experimentally reported properties and structures for which calculated properties did not match reported properties were manually verified.
2.2 Modeling
Python code used to implement, train and evaluate the models described in this section, in addition to the dataset and test split, is available on GitHub16 and Zenodo.17The AttentiveFP architecture consists of multiple “message passing”35 layers which refine these initial input feature vectors by propagating information based on the adjacency list. In each layer, every node's feature vector is updated by aggregating “messages” received from all adjacent nodes. AttentiveFP uses a learned message function based on the graph attention mechanism,36 which is parametrized by neural networks and takes as input the feature vector of both the node, its neighbor and (optionally) their edge vector. The messages from all neighbors are aggregated into the updated node vector by a “Gated Recurrent Unit” (GRU).37 Global “refinement layers” are applied after these local message passing layers, which construct a representation of the entire molecule. These attention-based layers instead connect each atom to a “virtual super node” capturing global context. The output of the encoder is a single latent vector describing the entire molecule, which is the input to the regression head. A schematic of this GNN is depicted in Fig. S1.†
These preliminary investigations on model hyperparameters indicated that the “hidden dimension” of the AttentiveFP GNN had the biggest influence on model performance. Doubling of the hidden dimension leads to a sub-quadratic increase in trainable parameters due to the linear to quadratic scaling of its constituent modules. In contrast, using more than 2 “hidden layers” and 2 “global refinement layers” significantly increases the model parameters without robust gains in performance. A “dropout” probability of p = 0.1 was found to give reliable results. Either omitting dropout or using significantly larger dropout probabilities deteriorated model performance.
Based on these investigations, we found a hidden dimension of 64 and output dimension of 128, with 2 hidden layers, 2 global refinement layers and dropout p = 0.1 consistently yielded accurate results, and used these hyperparameters for all further experiments unless stated otherwise. This configuration (AttentiveFP64d) has 116 K model parameters including the regression head, which is significantly smaller than previously proposed GNNs for surfactants (217 K8 and 307 K11) and molecules (586 K38). A smaller variant with a hidden dimension of 32 and output dimension of 64 (AttentiveFP32d, with a total of 36 K parameters) was also explored, as well as a larger architecture with 96 hidden dimensions and output dimension 192 (AttentiveFP96d, 245 K parameters).
 | (1) |
The “ensemble” MAE calculation method estimates the expected error when a prediction is made by taking the average prediction across folds (eqn (2), where K is the number of folds and ŷk is the prediction of a model trained on the kth fold of the data). Ensemble methods also allow model prediction uncertainty to be estimated. The standard deviation of the ensemble prediction is defined in eqn (3). Ensembles of machine learning models and (deep) neural networks have been theoretically40,41 and empirically42,43 shown to improve model accuracy and out-of-distribution robustness. The MAE for the ensemble prediction is given in eqn (4), with similar definitions as eqn (1).
 | (2) |
 | (3) |
 | (4) |
3 Results and discussion
3.1 SurfPro database
Table 2 lists the overall count of experimental property measurements per property, divided by surfactant type. The largest fraction of surfactants in the database are cationic, including 904 unique structures with one (cationic) or two (gemini cationic) counter ions. The second-largest category is non-ionic, which includes 100 sugar-based surfactants, followed by anionic surfactants which contains 242 entries, and only 3 gemini anionic surfactant entries. Zwitterionic surfactants are the lowest represented class in the database, with only 54 entries overall, which includes 17 gemini zwitterionic compounds.| Surfactant type | SMILES | pCMC | γ CMC | Γ max | pC20 | A min | πCMC | All |
|---|---|---|---|---|---|---|---|---|
| Cationic | 316 | 237 | 274 | 176 | 176 | 182 | 195 | 176 |
| Gemini cationic | 588 | 497 | 416 | 302 | 308 | 302 | 326 | 302 |
| Anionic | 239 | 239 | 39 | 45 | 31 | 45 | 39 | 30 |
| Gemini anionic | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Non-ionic | 325 | 281 | 145 | 72 | 71 | 72 | 101 | 71 |
| Sugar-based non-ionic | 100 | 100 | 74 | 71 | 71 | 71 | 74 | 68 |
| Zwitterionic | 36 | 21 | 24 | 6 | 0 | 6 | 9 | 0 |
| Gemini zwitterionic | 17 | 17 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 1624 | 1395 | 972 | 672 | 657 | 678 | 744 | 647 |
The majority of experimental measurements (1327) were recorded between 24.85 °C to 25 °C. The properties of 55 surfactants were measured below this range (20 °C to 23 °C), and 13 surfactants were measured above it (27 °C to 40 °C). No temperature was reported for γCMC measurements of 228 structures obtained from Ricardo et al.,14 but the authors only included measurements at temperatures of 20 °C to 30 °C. Surfactant properties are dependent on temperature,12 but it has also been noted that γCMC does not vary significantly in the temperature range 20 °C to 30 °C.14 However, we included all of the data that we collected in the test and train sets. We included data recorded outside the modal temperature range of the dataset with the aim of maximising the structural diversity, and we were unable to find data for these compounds at 25 °C. We thus expect that models trained on these data are more accurate predictors for properties at around 25 °C.
The fraction of reported properties varies significantly among surfactant types. The property pCMC has the highest number of entries in the database, with a pCMC value given for the majority of entries in each class of surfactants. The number of γCMC values in the data is in general the second highest for each surfactant class, and similar in proportion to the number of Γmax, pC20, Amin and πCMC values in the database. Cationic surfactants have the highest overall proportion of micellization-related properties reported in this database. Of the 647 compounds with a complete set of reported properties, cationic and gemini cationic represent 27% and 47%, respectively. In contrast, of the 632 compounds with only CMC values reported, cationic and gemini cationic represent 7% and 28% respectively, while 30% are anionic and 28% are non-ionic.
3.2 Visualization of the distribution of reported properties by surfactant type
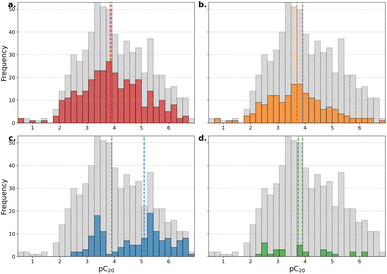
The distributions of experimental properties vary significantly between subsets of surfactant types, both compared to each other and compared to the full dataset (Fig. 2–5). pCMC measurements of gemini cationic structures (Fig. 2a, red) show a mean and standard deviation close to Gaussian and similar to the entire dataset (grey), while the distribution's mean shifts significantly higher for non-ionic (Fig. 2b, blue) and lower for cationic (Fig. 2c, orange) and anionic structures (Fig. 2d, green) with heavy tails. For γCMC the distributions of gemini cationic, cationic and non-ionic subsets (Fig. 3a–c) shift significantly relative to the entire dataset, while very few measurements are reported for anionic structures (Fig. 3d).A skewed distribution is observed for Γmax both overall and for each surfactant type (Fig. 4a–d). Heavy outliers are present in cationic structures, and a large increase of the mean Γmax is visible for non-ionic structures. pC20 measurements are normally distributed overall and for cationic and gemini cationic structures. A bi-modal distribution is observed for non-ionic (Fig. 5c) surfactants compounds, while few pC20 measurements have been reported for anionic surfactants (Fig. 5d).
3.3 Modeling results
We developed a selection of QSPR models trained to predict pCMC, γCMC, Γmax, and pC20. Conceptually, each model consisted of an encoder for molecular structure, and a regressor. The molecular structure encoder takes in molecular structure information in the form of a graph, and converts it into a numerical vector representation. This representation is then regressed onto molecular properties using the regressor part of the model. As “baselines”, we chose descriptors from RDKit fingerprint and ECFP-based representations, which algorithmically encode the molecular graph into vectors. ECFP and RDKit fingerprint (RDKFP) representations were regressed onto molecular properties using either random forest regression or ridge regression. GNNs have previously been shown to be effective in predicting surfactant properties.8,9,12 Here, we selected the AttentiveFP architecture as a GNN-based encoder for molecular structure, which has previously demonstrated state-of-the-art performance on a selection of other QSPR tasks.31,34Distributions of properties differ between different surfactant types (Fig. 2–4). Previous reports on QSPR modeling of surfactants such as Chen et al.13 developed separate models for different surfactant types, while other works developed individual models capable of predicting properties for all surfactant types.8,11 Here, we developed models which can accept any surfactant type by training on data containing surfactants of each class. The test set of 140 surfactants was selected specifically to enable evaluating models on the same set of surfactant structures for all tasks and properties. Due to the varying number of reported values for each property, ∼10% of property measurements were allocated by sampling 70 surfactants with stratification by surfactant type from two subsets of the dataset separately, respectively from structures with all reported properties, and structures with only the CMC. The distribution of surfactant type reported in the database was preserved through stratified sampling based on the surfactant type (see Methods). We included all data, regardless of the temperature they were recorded at, in training, validation and test data.
Each model was trained for three tasks: single-property, multi-property and all-property prediction. For single property prediction, models were trained to predict a single property given a molecular structure input. Multi-property AttentiveFP models were trained to simultaneously predict pCMC, γCMC and Γmax. All-property AttentiveFP models were trained to simultaneously predict pCMC, γCMC, Γmax, C20, πCMC and Amin. Ten-fold cross validation was used during model training. For each task and model, the MAE and RMSE were evaluated for each of 10 cross-validation folds, and averages were taken according to eqn (1). Additionally, the ensemble error (eqn (4)), corresponding to the errors of the average prediction from all models trained on each fold, was also calculated. We did not train single-task models to predict Amin or πCMC, since they can be calculated from pCMC, γCMC and Γmax. Table 3 lists all obtained predictive results for the ensemble (MAE and RMSE) for all tasks. Table S3† lists the average results for the same tasks, box plots of the results are provided in Fig. S2–S5.†
| pCMC | γ CMC | Γ max × 106 | pC20 | |||||
|---|---|---|---|---|---|---|---|---|
| MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | |
| AttentiveFPsingle32d | 0.275 | 0.428 | 2.685 | 3.961 | 0.488 | 0.942 | 0.461 | 0.633 |
| AttentiveFPsingle64d | 0.250 | 0.382 | 2.345 | 3.555 | 0.432 | 0.840 | 0.353 | 0.496 |
| AttentiveFPsingle96d | 0.241 | 0.365 | 2.424 | 3.561 | 0.387 | 0.784 | 0.285 | 0.405 |
| AttentiveFPmulti32d | 0.277 | 0.415 | 2.796 | 3.680 | 0.429 | 0.878 | — | — |
| AttentiveFPmulti64d | 0.239 | 0.358 | 2.621 | 3.600 | 0.358 | 0.685 | — | — |
| AttentiveFPmulti96d | 0.237 | 0.360 | 2.308 | 3.407 | 0.333 | 0.573 | — | — |
| AttentiveFPall32d | 0.279 | 0.419 | 2.711 | 3.626 | 0.479 | 0.999 | 0.349 | 0.530 |
| AttentiveFPall64d | 0.246 | 0.358 | 2.548 | 3.516 | 0.353 | 0.842 | 0.282 | 0.390 |
| AttentiveFPall96d | 0.235 | 0.346 | 2.591 | 3.531 | 0.347 | 0.707 | 0.259 | 0.363 |
| RDKFP – Ridge | 0.674 | 0.902 | 3.367 | 4.549 | 0.444 | 0.786 | 0.511 | 0.754 |
| RDKFP – RF | 0.630 | 0.840 | 2.939 | 4.211 | 0.443 | 0.773 | 0.582 | 0.743 |
| ECFP – Ridge | 0.760 | 1.010 | 3.942 | 5.124 | 0.474 | 0.876 | 0.591 | 0.753 |
| ECFP – RF | 0.737 | 0.997 | 4.142 | 5.442 | 0.453 | 0.784 | 0.588 | 0.742 |
For γCMC prediction, according to the average error calculation metric, the MAEs of all model of the same size fall within one standard deviation of each other when trained upon the three tasks (Fig. 6). For the ensemble, single-property models perform better than multi-property models for γCMC prediction, except for the AttentiveFPmulti96d model (0.33 MAE/0.57 RMSE), which outperforms both single- and all-property AttentiveFP96d models (0.35–0.39 MAE/0.71–0.78 RMSE) (Table 3). Therefore, training on multiple or all properties is in general detrimental to predicting γCMC.
For Γmax we observe more drastic improvements in performance between single-property and multi-/all-property prediction, both on average and for the ensemble (Fig. 6). For the ensemble, the single- and all-property models perform similar (0.48–0.49 MAE/0.94–1.0 RMSE) for AttentiveFP32d, whilst the multi-property model of the same size performs better (0.43 MAE/0.88 RMSE). Increasing the model size increases the performance of the multi- and all-property models relative to the single-property model. We thus consider multi-property training to be beneficial in developing models which are better predictors of Γmax.
Training on all properties also appears beneficial for pC20 prediction. This training mode introduces a moderate increase in performance for AttentiveFP32d according to the average metric, but this gain in performance closes as the model becomes larger. However, for the ensemble models, there is a greater difference in performance between AttentiveFPsingle32d (0.46 MAE/0.63 RMSE) and AttentiveFPall32d (0.35 MAE/0.53 RMSE). Again, this performance difference closes as the model size gets larger (0.29 MAE/0.41 RMSE for AttentiveFPsingle96d, 0.26 MAE/0.36 RMSE for AttentiveFPall96d). These results indicate that training on all properties can appreciably improve pC20 prediction.
The medium (AttentiveFP64d) and large (AttentiveFP96d) ensemble models score similarly in the multi-property task, and differences in predictive accuracy between the multi- and all-property task are relatively small and not consistent across properties (Fig. 6 and S2–S5†). We hypothesize this is due to the similarity of information contained in both tasks, since pC20, Amin and πCMC can be calculated given pCMC, γCMC and Γmax. Both multi- and all-property prediction tasks effectively contain the same information derived from the Langmuir isotherm, and therefore all derived models show comparable performance for the same model size.
3.4 Property prediction performance
3.5 Imputing missing properties
We selected the AttentiveFPall64d ensemble model to provide imputed properties which complete the SurfPro dataset. For pCMC prediction the model's test MAE is 0.246, which is below the average, but approximately the median test MAE, over all AttentiveFP ensemble models.Inspection of the model's parity plot for pCMC (Fig. S6a†) and the distribution of differences between the true and predicted values (Fig. S6b†) indicates that the model predicts pCMC for each surfactant type with similar accuracy. Therefore, the model's performance for pCMC prediction does not appear to be dependent on the surfactant type.
In the case of γCMC prediction, the overall test MAE is 2.55, just below the mean test MAE, and well below the median, for the AttentiveFP ensemble models. The parity plot (Fig. S7a†) and distribution of errors (Fig. S7b†) indicates that the majority of samples in each surfactant class are similarly distributed close to the mean error. However, there are some samples of the anionic, cationic and gemini surfactant classes which have significantly higher errors than average. Since these are isolated examples from the test data set, we cannot draw any inferences about any structural basis for the low prediction accuracy for these samples.
For Γmax prediction, the test MAE is 0.35, which is below the mean (and median) test MAE for all AttentiveFP ensemble models. As for pCMC and γCMC, parity plots (Fig. S8a†) and distributions of errors (Fig. S8b†) indicate similar model performance over each class of surfactants. However, there is single outlier cationic surfactant sample whose Γmax value is predicted to be significantly lower than its experimental value. Again, we cannot infer any general insight into model behaviour based on this single sample.
Finally, for pC20, the model performs with a test MAE of 0.28, which is approximately the median, but above the mean test MAE over all AttentiveFP ensemble models. The pC20 values for gemini surfactants are in general lower in error, as indicated by their relatively narrow distribution of errors about 0.0 compared to the other surfactant classes (Fig. S9a and b†). Thus, the AttentiveFPall64d model performs well in comparison to the other investigated models, performing similarly to the larger AttentiveFPall96d and AttentiveFPmulti96d ensemble models, and significantly better than all smaller AttentiveFP32d models. The model can also make predictions across variety of surfactant classes well, and in general does not appear be systematically biased to predict one surfactant class with better accuracy than any other.
Out of the 1624 structures, only 647 have all properties reported, and we fill the database with the ensemble model's predictions and uncertainties for all missing properties for those 977 structures. We used the AttentiveFPall64d ensemble model to calculate predictions for missing values in the database using the mean prediction of the ten members of the ensemble (eqn (2)). We also used these ten values to estimate the uncertainty of their mean prediction by calculating their standard deviation (eqn (3)). The entries for which the literature values are provided are assigned a standard deviation of 0.0 in the imputed database. The imputed database is available on Github16 and Zenodo.17
4 Conclusion
SurfPro is a manually curated surfactant property database of 1624 unique amphiphiles, which we have made publicly available for the community. At the time of writing, it is the largest dataset for this class of compounds, containing 1624 unique surfactant entries with corresponding measurements of the CMC, γCMC, Γmax and C20. Importantly, literature sources typically do not include experimental measurements of all properties of surfactants, often only reporting pCMC or γCMC values. Therefore, despite the size of the database, it remains incomplete as not all samples have a complete set of annotated properties. Furthermore, SurfPro contains only three anionic gemini surfactants, 21 cationic gemini with 3 or 4 counterions, and three high molecular weight compounds (≥2000 Da). Future data collection efforts could thus be targeted to address the balance of structural diversity and experimental surfactant property measurements to fill the chemical structure and property spaces of surfactants more efficiently.Using the SurfPro database, we trained an ensemble of GNN-based models for single- and multi-property prediction for pCMC, γCMC, Γmax and pC20. This model then allowed us to impute missing property values for 977 compounds in the SurfPro database, thus providing a complete database consisting of experimentally measured properties from literature sources, and a set of estimates according to the GNN ensemble model. We hope the database and modeling strategy will provide new avenues in data-driven property prediction and surfactant design.
Data availability
The SurfPro database (“surfpro_literature.csv”), reference list (“surfpro_bibliography.bib”), “imputed” database (“surfpro_imputed.csv”), test split (“surfpro_test.csv”), cross-validation folds (“surfpro_train.csv”) and Python code used to implement, train and evaluate the models described in this work are available via GitHub (https://github.com/BigChemistry-RobotLab/SurfPro16) and Zenodo (https://zenodo.org/records/14931937, DOI: https://doi.org/10.5281/zenodo.14931937).17Author contributions
Conceptualization – SH, LH, AP, WTSH, WER. Data curation – SH, LH, WER. Model development, training & evaluation – SH, WER. Supervision – WER, WTSH, AP. Visualization – SH, WER. Writing: original draft – SH, WTSH, WER. Writing: review & editing – SH, WTSH, PFJD, WER, LH, AP.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge funding from the National Growth Fund project “Big Chemistry” (1420578), funded by the Ministry of Education, Culture and Science.References
- M. Rosen and J. Kunjappu, Surfactants and Interfacial Phenomena, John Wiley & Sons, 2012 Search PubMed.
- L. L. Schramm, E. N. Stasiuk and D. G. Marangoni, Annu. Rep. Prog. Chem., Sect. C: Phys. Chem., 2003, 99, 3–48 CAS.
- I. Langmuir, J. Am. Chem. Soc., 1918, 40, 1361–1403 CAS.
- B. v. Szyszkowski, Z. Phys. Chem., 1908, 64, 385–414 Search PubMed.
- J. H. Clint, Surfactant Aggregation, Springer Science & Business Media, 1992 Search PubMed.
- A. P. Santos and A. Z. Panagiotopoulos, J. Chem. Phys., 2016, 144, 044709 Search PubMed.
- A. R. Katritzky, M. Kuanar, S. Slavov, C. D. Hall, M. Karelson, I. Kahn and D. A. Dobchev, Chem. Rev., 2010, 110, 5714–5789 CAS.
- S. Qin, T. Jin, R. C. Van Lehn and V. M. Zavala, J. Phys. Chem. B, 2021, 125, 10610–10620 CAS.
- A. Moriarty, T. Kobayashi, M. Salvalaglio, P. Angeli, A. Striolo and I. McRobbie, J. Chem. Theory Comput., 2023, 19, 7371–7386 CAS.
- P. Mukerjee and K. Mysels, Critical micelle concentrations of aqueous surfactant systems, National Bureau of Standards Technical Report NBS NSRDS 36, 1971 Search PubMed.
- C. Brozos, J. G. Rittig, S. Bhattacharya, E. Akanny, C. Kohlmann and A. Mitsos, Colloids Surf., A, 2024, 694, 134133 CAS.
- C. Brozos, J. G. Rittig, S. Bhattacharya, E. Akanny, C. Kohlmann and A. Mitsos, J. Chem. Theory Comput., 2024, 20, 5695–5707 CAS.
- J. Chen, L. Hou, J. Nan, B. Ni, W. Dai and X. Ge, Colloids Surf., A, 2024, 703, 135276 CAS.
- F. Ricardo, P. Ruiz-Puentes, L. H. Reyes, J. C. Cruz, O. Alvarez and D. Pradilla, Chem. Eng. Sci., 2023, 265, 118208 CAS.
- D. Seddon, E. A. Müller and J. T. Cabral, J. Colloid Interface Sci., 2022, 625, 328–339 CrossRef CAS PubMed.
- S. L. Hödl, L. Hermans, P. F. J. Dankloff, A. Piruska, W. T. S. Huck and W. E. Robinson, SurfPro, Github, 2025, https://github.com/BigChemistry-RobotLab/SurfPro.
- S. L. Hödl, L. Hermans, P. F. J. Dankloff, A. Piruska, W. T. S. Huck and W. E. Robinson, SurfPro, Zenodo, 2025, DOI:10.5281/zenodo.14931937.
- C.-C. Kwan and M. J. Rosen, J. Phys. Chem., 1980, 84, 547–551 CrossRef CAS.
- D. Xu, X. Ni, C. Zhang, J. Mao and C. Song, J. Mol. Liq., 2017, 240, 542–548 CrossRef CAS.
- R. A. Rahimov, G. A. Ahmadova, S. F. Hashimzade, E. Imanov, H. G. Khasiyev, N. K. Karimova and F. I. Zubkov, J. Surfactants Deterg., 2021, 24, 433–444 CrossRef CAS.
- J. Eastoe, J. S. Dalton, P. G. Rogueda, E. R. Crooks, A. R. Pitt and E. A. Simister, J. Colloid Interface Sci., 1997, 188, 423–430 CrossRef CAS.
- U. Komorek and K. A. Wilk, J. Colloid Interface Sci., 2004, 271, 206–211 CrossRef CAS PubMed.
- Kabir-ud-Din and P. A. Koya, J. Chem. Eng. Data, 2010, 55, 1921–1929 CrossRef.
- K. Shinoda, T. Yamanaka and K. Kinoshita, J. Phys. Chem., 1959, 63, 648–650 CAS.
- L.-C. Zheng and Q.-X. Tong, J. Mol. Liq., 2021, 331, 115781 CrossRef CAS.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36 CrossRef CAS.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, Nucleic Acids Res., 2023, 51, D1373–D1380 CrossRef PubMed.
- C. Guo, P. Zhou, J. Shao, X. Yang and Z. Shang, Chemosphere, 2011, 84, 1608–1616 CrossRef CAS PubMed.
- G. Landrum, RDKit (version 2024.3.5), Rdkit technical report, 2024 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn. Res, 2011, 12, 2825–2830 Search PubMed.
- Z. Xiong, D. Wang, X. Liu, F. Zhong, X. Wan, X. Li, Z. Li, X. Luo, K. Chen, H. Jiang and M. Zheng, J. Med. Chem., 2020, 63, 8749–8760 CrossRef CAS PubMed.
- M. Fey and J. E. Lenssen, Fast graph representation learning with PyTorch Geometric, 2019, https://pytorch-geometric.readthedocs.io/en/latest/.
- D. Jiang, Z. Wu, C.-Y. Hsieh, G. Chen, B. Liao, Z. Wang, C. Shen, D. Cao, J. Wu and T. Hou, J. Cheminf., 2021, 13, 1–23 Search PubMed.
- J. Born, G. Markert, N. Janakarajan, T. B. Kimber, A. Volkamer, M. R. Martínez and M. Manica, Digital Discovery, 2023, 2, 674–691 RSC.
- J. Gilmer, S. S. Schoenholz, P. F. Riley, O. Vinyals and G. E. Dahl, Proceedings of the 34th International Conference on Machine Learning, 2017, pp. 1263–1272 Search PubMed.
- P. Veličković, G. Cucurull, A. Casanova, A. Romero, P. Liò, Y. Bengio, Graph Attention Networks, arXiv, 2018, preprint, arXiv:1710.10903, DOI:10.48550/arXiv.1710.10903.
- K. Cho, B. v. Merrienboer, C. Gulcehre, D. Bahdanau, F. Bougares, H. Schwenk and Y. Bengio, Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation, arXiv, 2014, preprint, arXiv:1406.1078, DOI:10.48550/arXiv.1406.1078.
- Z. Wu, B. Ramsundar, E. Feinberg, J. Gomes, C. Geniesse, A. S. Pappu, K. Leswing and V. Pande, Chem. Sci., 2018, 9, 513–530 CAS.
- W. Falcon, PyTorch Lightning, 2019, https://github.com/Lightning-AI/lightning.
- L. Breiman, Mach. Learn., 1996, 24, 123–140 Search PubMed.
- T. G. Dietterich, International workshop on multiple classifier systems, 2000, pp. 1–15 Search PubMed.
- M. Ganaie, M. Hu, A. Malik, M. Tanveer and P. Suganthan, Eng. Appl. Artif. Intell., 2022, 115, 105151 Search PubMed.
- S. Fort, H. Hu and B. Lakshminarayanan, Deep Ensembles: A Loss Landscape Perspective, arXiv, 2020, preprint, arXiv:1912.02757, DOI:10.48550/arXiv.1912.02757.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00393d |
| This journal is © The Royal Society of Chemistry 2025 |