Open Access Article
Open Access ArticleGeneral data management workflow to process tabular data in automated and high-throughput heterogeneous catalysis research†‡
Erwin
Lam§
 *a,
Tanguy
Maury§
a,
Sebastian
Preiss§
a,
Yuhui
Hou
a,
Hannes
Frey
a,
Caterina
Barillari
b and
Paco
Laveille
*a,
Tanguy
Maury§
a,
Sebastian
Preiss§
a,
Yuhui
Hou
a,
Hannes
Frey
a,
Caterina
Barillari
b and
Paco
Laveille
 *a
*a
aETH Zurich, Swiss Cat+ East, Vladimir-Prelog-Weg 1-5, 8093 Zurich, Switzerland. E-mail: elam@ethz.ch; plaveille@ethz.ch
bETH Zurich, ID SIS, Klingelbergstrasse 48, 4056 Basel, Switzerland
First published on 10th January 2025
Abstract
Data management and processing are crucial steps to implement streamlined and standardized data workflows for automated and high-throughput laboratories. Electronic laboratory notebooks (ELNs) have proven to be effective to manage data in combination with a laboratory information management system (LIMS) to connect data and inventory. However, streamlined data processing does still pose a challenge on an ELN especially with large data. Herein we present a Python library that allows streamlining and automating data management of tabular data generated within a data-driven, automated high-throughput laboratory with a focus on heterogeneous catalysis R&D. This approach speeds up data processing and avoids errors introduced by manual data processing. Through the Python library, raw data from individual instruments related to a project are downloaded from an ELN, merged in a relational database fashion, processed and re-uploaded back to the ELN. Straightforward data merging is especially important, since information stemming from multiple devices needs to be processed together. By providing a configuration file that contains all the data management information, data merging and processing of individual data sources is executed. Having established streamlined data management workflows allows standardization of data handling and contributes to the implementation and use of open research data following Findable, Accessible, Interoperable and Reusable (FAIR) principles in the field of heterogeneous catalysis.
Introduction
The emergence of automation, digitalization and high-throughput experimentation in chemistry and chemical engineering leads to the generation of large amounts of experimental data in digital format.1 Using statistical and machine learning algorithms, these large datasets can be analyzed in deeper detail to extract and correlate relevant chemical information compared to what was previously possible through manual data processing and analysis.2,3 Understanding such large datasets will further accelerate the generation of knowledge and scientific discoveries.2,4To manage data within a group or infrastructure, the usage of electronic laboratory notebooks and laboratory information management systems (ELN/LIMS) has aided data digitalization and allows unique identification and tracing of any changes in data. Furthermore, it allows sharing procedures, avoid duplicating documentation, provide a timestamp on procedure/data creation, and to mitigate the risk of data loss, especially in comparison to handwritten notes.5 Experimental procedures can be connected to an inventory system to track the instruments/consumables that were used.6 Furthermore, the use of an ELN/LIMS enables standardized data management and processing of experimental data from several instruments and users. Having standardized procedures facilitates the replication and tracing of data complying with the Findable, Accessible, Interoperable and Reusable (FAIR) principles and open research data requirements.7–11 Many options for ELN/LIMS are currently available. However, finding the ideal ELN is often a challenge as individual research facilities require specific features from an ELN. A specific example of an ELN/LIMS is openBIS, which is open-source and tailored for experimental data management that allows uploading and sharing experimental data.6,12
In the field of catalysis, various types of data related to the synthesis, characterization and testing of catalysts are created during an experimental workflow.3 Recent initiatives led to the emergence of facilities and research laboratories performing accelerated catalysis research and development through automation and high-throughput experimentation (Fig. 1a).1,13,14 Such facilities generate large amounts of data from individual instruments, and therefore having a LIMS to manage chemicals and instruments, and the connection to an ELN with streamlined and standardized data management procedures has become a requirement. Due to the heterogeneity of the instruments used, the datasets come in various forms and data structures (e.g., csv, excel based formats with varying header rows, or instrument specific data formats). Thus, the merging of data from individual instruments into a holistic dataset is a challenge and important to further process the experimental dataset. For example, to obtain turnover numbers of a given catalytic sample, data stemming from a test reactor need to be combined with data stemming from the catalyst synthesis details, and its characterization. Moreover, having properly processed and cleaned datasets allows seamless downstream utilization in combination with statistical methods and machine learning algorithms to accelerate closed loop catalyst discovery and optimization (Fig. 1b).15 This is enabled by including all experimental information, and tracking the data processing activities that were performed on the raw data.
Several key criteria for streamlined and traceable data management workflows are required. These include data being uploaded in a structured way to a centralized database that is backed up and secure. The datasets from individual instruments should contain embedded information to interconnect and merge them into a unified/combined data file. In the context of open research data (ORD),16,17 the ability to easily push whole datasets to open research data repositories10,18–20 is another important feature.21 Many of these criteria should be generally featured within an ELN/LIMS. However, implementing holistic data processing within the ELN/LIMS is often lacking which is becoming more relevant when large data are created.
Most catalysis related data (e.g., synthesis parameters, characterization fingerprints, catalyst performance) can be summarized in a tabular matter (csv format/Excel sheets). This allows implementation of data processing workflows that utilize relationships between columns of individual datasets: e.g., sample vials can be equipped with a barcode which is scanned at each individual processing step in the laboratory. The barcode column can then be used to relate individual datasets to each other to combine them into one holistic dataset.
To streamline and to standardize data processing, we present in this article a Python library for catalysis data management (PyCatDat) that allows processing of tabular data from openBIS, an open source ELN/LIMS (Fig. 1c).6,12 openBIS is developed and continuously updated since 2007 at ETH Zurich which is used by many research institutions globally.6,12 The Python library allows the download, processing and reupload of processed data to openBIS, thus bypassing the limited data processing functionality of ELNs as discussed above. The code requires a configuration file with instructions on the expected number of files and how to merge them enabling high flexibility on different data architectures without the need to directly manipulate the code or require extensive coding expertise. At the same time, the configuration file also provides traceability on how data were processed and can be adapted to different data processing methods (see data management demonstration in the Results and discussion section). For heterogeneous catalysis, the same dataset might be used in different data processing pipelines (normalize performance by catalyst weight, atom loading, composition, dispersion etc.). Usage of a configuration file for each processing pipeline can facilitate traceability and the application of FAIR principles.
Results and discussion
Data management architecture
The data management infrastructure is designed for integration with an electronic laboratory notebook/laboratory information management system (ELN/LIMS), in this case openBIS. It is used to store data generated from individual instruments in a structured way. The data management workflow is initiated by uploading experimental data consisting of files in tabulated data formats (e.g., csv or from Excel sheets) to openBIS. An application programming interface (API) allows automating data-upload (Fig. 2). Data are uploaded to openBIS either manually or automatically using the openBIS API “Dropbox function” (see ESI‡ Note 1 for further discussion) in combination with a Python script. This function allows tracing output files generated from instruments and uploads them automatically once an experiment is completed (see ESI‡ Note 2 for further discussion). This feature further highlights the importance of instrument suppliers providing options to automatically generate interpretable output files upon completion of an experiment.The type of data can consist of information related to synthesis, weighting of materials and their performance in a chemical reaction. All these data files relate to each other and the aim is to translate these relationships into a relational database structure. This allows data from individual instruments to be merged and to be processed in an automated, traceable and standardized way.
Data management codes
The Python library for catalysis data management (PyCatDat) developed in this work manages data generated from a multitude of lab equipment, e.g., automated synthesis robots, automated solid dispense tools, plate barcode readers and high-throughput fixed-bed reactors with integrated online gas chromatograph (GC) data (see Data management demonstration section).Herein we define the notation of “Project” and “Task”. A “Project” consists of the entirety of all experimental activities and is split into multiple “Tasks”. A “Task” consists of an experimental workflow, typically comprising synthesis and testing with all intermediate activities, (e.g.) thermal treatment, fixed-bed reactor preparation (see Data management demonstration section).
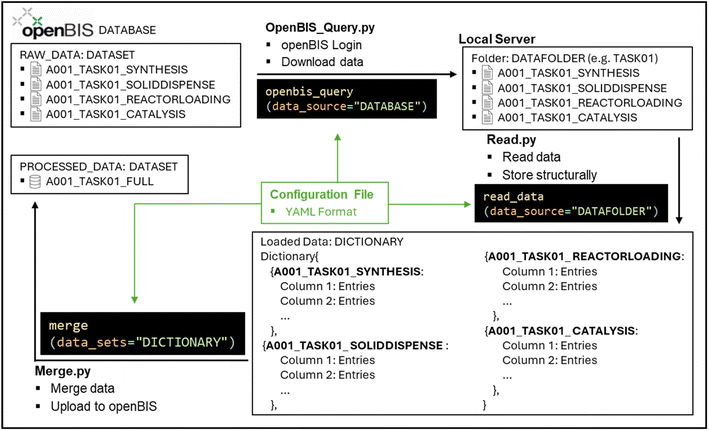
Upon collecting all raw data of an experimental “Task” from each individual equipment on openBIS, its processing can be initiated. This activity is performed with the PyCatDat library which contains functions to securely login to openBIS and download data (openBIS_Query.py), to read and merge data (Read.py and Merge.py) and to process them (e.g., Process_FixedBed.py) (Fig. 2) (see ESI‡ Note 3 for further discussion).
To execute the library, a configuration file in YAML format is required which contains human-readable serialized information. The information includes the ELN platform, the expected data structure, and the warranted data merging/processing procedure. All functions are run by providing the location of the data and configuration file. The configuration file stores information that is accessed by the Python library and therefore offers reproducibility of the data management steps in the form of a structured file. The configuration file can be modified to perform data management steps with different input variables, for example a change in the openBIS platform or different file structures that vary with projects. In addition, different types of analysis can be performed on the same raw data by modifying the inputs of the configuration file.
The openBIS_Query.py function initiates the data download step from openBIS (Fig. 3a). To run this function, the “OPENBIS” section in the configuration file requires information about the openBIS platform's URL and additional optional information such as the destination directory name, default username and size limit of the files to download (Fig. 3b). Thereafter, username and password inputs are required to ensure data are accessed by the person with the appropriate rights. The login is saved as a token to stay logged in without re-authentication for a set period. CPU parallelization to download files simultaneously and a data size limit can also be configured. Through the function, data are downloaded from the ELN to the user's local workstation. To download only the selected data, openBIS follows a defined data folder path structure where all the projects/data of a facility are located. It is possible to choose whether all the data within a “Project” or only individual “TASKS” are downloaded. Once data have been downloaded from the ELN/LIMS, further data processing is performed locally. Therefore, the library can also be expanded to access data from other sources.
 | ||
| Fig. 3 (a) OpenBIS_Query function to access an openBIS instance and download data from there. (b) Information in the configuration file related to accessing an openBIS instance and downloading data. | ||
Data downloaded from openBIS (or local data) are read and loaded into a dictionary by matching the information in the configuration file with the folder structure using the function Read.py. In the comment section of the configuration file, further relevant information can be provided. For example, information such as interruptions during experiments can be included. In the data folder, tasks are split into subfolders, where the subfolder names are provided in the configuration file's “TASKS” section (e.g., TASK01, TASK02 and TASK03). The individual raw data files are located within each subfolder. The FILE_ID in the configuration file indicates the filename/string that should be recognized. For example, the file containing information about solid weighting can be called “SOLIDDISPENSE”, which is the FILE_ID in the configuration file to recognize the filename (Fig. 4). In the case of multiple files with the same FILE_ID, these files will all be captured (see ESI‡ Note 4 for further discussion).
 | ||
| Fig. 4 Configuration file structure to provide information about expected file and filenames to be read based on the folder structure. | ||
Further details such as the separator of the tabular data with comma as default and data sheet/tab for multiple sheets in an Excel document can also be provided. The data are then stored in a dictionary which is accessed to merge raw data files in the next step. The dictionary stores all data grouped by “TASKS”, “FILE_ID” and filename. Within a project, different instructions per task can be provided to allow higher flexibility. Tasks with identical instructions can be grouped together (see ESI‡ Note 5 for further discussion).
For the subsequent merging step (Merge.py), instructions are provided in the “MERGING_ORDER” section of the configuration file (Fig. 5). The column names to perform the merging are provided in the “TASKS” section as “SOURCE_ID” and “TARGET_ID”. The “MERGING_ORDER” section contains the instructions on how and in which order the “SOURCE_ID” and “TARGET_ID” should be used for merging. For example, the file “A001_TASK01_SOLIDDISPENSE” contains a column with the barcodes of the vial (“TARGED_ID” = Destination Barcode) that matches the barcode column (“SOURCE_ID” = Catalysis Barcode) in another file (“A001_TASK01_REACTORLOADING”). The content of both files will then be combined into one single file through the selected column names.
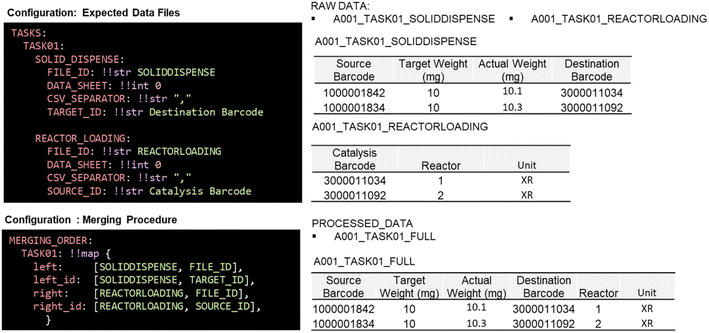 | ||
| Fig. 5 Example of the configuration file sections to provide information about merging data. Two data files with the name SOLIDDISPENSE and REACTORLOADING are merged together through their barcode. | ||
Within the “MERGING_ORDER” section, all the single raw files listed will be merged into one combined file. Merge.py will go through all the files provided in the “MERGING_ORDER” section using the provided “FILE_ID”, “TARGET_ID” and “SOURCE_ID”. The merging occurs step-by-step until every individual file is combined into one file (see ESI‡ Note 6). The merging order reflects the sequence of the experimental workflow and enables tracing the order of experiments performed.
Finally, data processing (field specific calculations) can be performed with additional instructions to the configuration file. Based on the experimental workflows, different data processing procedures can be tailored to the project therefore maximizing flexibility. This includes calculating performance metrics of fixed-bed testing such as formation rates, reactant conversion or product selectivity. An example of processing catalysis data from fixed-bed testing by accessing the configuration file and the merged datasets generated from Merge.py is illustrated in the next section (Process_FixedBed.py). This approach allows transparent data processing procedures such as data normalization, enhancing interoperability.
Data management demonstration
To illustrate the complete data management workflow, an example of performing data management tasks is presented. The data are based on a previously published study with real experimental data used (Fig. S6 and S7‡).22 In this dataset, a closed-loop high-throughput experimental workflow was conducted (Fig. 6). It consists of the synthesis of 144 heterogeneous catalysts (6 generations of 24 catalysts) followed by their testing in fixed-bed mode under CO2 hydrogenation conditions. The dataset includes data that were generated from several instruments including synthesis robots, balances, barcode readers, fixed-bed reactors and gas chromatographs. | ||
| Fig. 6 Experimental workflow to perform machine learning driven optimization for synthesizing heterogeneous catalysts for CO2 hydrogenation. | ||
The synthesis robot output files contain information about the type and amount of solid and liquid chemicals dispensed, reaction/stirring time, stirring rate and temperature. Barcode readers consist of handheld single vial barcode readers and plate barcode readers reading multiple vial barcodes simultaneously. The high-throughput fixed-bed reactors generate files that contain process and set values for pressure, temperature, testing time, gas flowrates and the gas concentrations of each reactant and product measured by online-GCs (see the ESI‡).
The experimental workflow consists of the parallel synthesis of 24 catalysts by incipient wetness impregnation using an automated synthesis robot, followed by thermal treatment under air at 550 °C, automatic dispense of the catalyst at the milligram scale and catalyst performance evaluation in fixed-bed mode for the hydrogenation of CO2 at 225–300 °C and 50 bar. The workflow is guided by artificial intelligence where synthesis parameters were provided by a software that performs Bayesian optimization to optimize the catalyst composition for its catalytic performance.
In total, 9–10 files per experimental loop/task were generated with 6 experimental loops being performed. This leads to 59 raw data files (Table 1). Using the Python library described above, all 9–10 files per task were first matched, merged and processed together. The datasets from each task were then combined onto one full dataset containing information from all 59 files. Raw data were accessed from the openBIS ELN and processed data were reuploaded there. It can be expected that performing such data processing tasks manually would be more time consuming with the risk of introducing manual error during data processing.
| Instrument | File no. | Features | Data files |
|---|---|---|---|
| a TASK06 did not contain any cost calculation of the metals. | |||
| Chemspeed Swing XL | 4 × 6 | Mixed metal solution preparation | CombVialPrep |
| Solid dispense of metal oxide | SolidDispense | ||
| Mixed metal solution addition to solid | Impregnation | ||
| Vial barcode reading | ImpregnationBarcode | ||
| Hobersal oven | 0 | — | — |
| Unchained labs junior | 1 × 6 | Vial barcode reading and catalyst weighting | CatalystDispenseBarcode |
| Zebra DS9308 barcode reader | 1 × 6 | Barcoded vial loading to which reactor | ReactorLoading |
| Avantium fixed-bed reactor XR/XD & Agilent 8890 GC | 3 × 6 | Result of catalysis performance XR unit | Catalysis_XR |
| Result of catalysis performance XD unit 1 | Catalysis_XDB | ||
| Result of catalysis performance XD unit 2 | Catalysis_XDC | ||
| Manually calculated | 1 × 5a | Market cost of metal | Cost |
Per experimental loop (Table 1), 4 individual synthesis files were extracted from a Chemspeed Swing XL. (1) The “CombVialPrep” file contains the amount of various liquid sources (metal nitrate solutions) that were mixed to prepare a mixed metal nitrate solution, (2) the “SolidDispense” file contains information about the choice and mass of solid support (metal oxide) that was dispensed, (3) the “Impregnation” file contains the amount of mixed metal nitrate solution from the “CombVialPrep” file that was added to the support described in the “SolidDispense” file, and (4) the “ImpregnationBarcode” file contains the barcode of the vials (referred to as impregnation vial) that were recorded by a barcode reader on the synthesis robot. Each file contains an entry corresponding to the vial index (index 1–24) as the identifier.
A Hobersal furnace was used to calcine the heterogeneous catalyst samples (550 °C for 4 h, 5 °C min−1) for which no data files were created. The catalysts were calcined inside their barcoded impregnation vial, keeping the same barcode during this step. Following calcination, each vial is fitted with a specific dispense head and an Unchained Labs Junior was used to dispense the targeted quantity of catalysts to a barcoded vial (referred to as “catalysis vial”). The “CatalystDispenseBarcode” file contains information about the dispensed weight and the barcode of the source (impregnation vial) and destination (catalysis vial) vial.
In the next step, the dispensed catalysts from the barcoded catalysis vial are loaded into the fixed-bed reactor. This step is recorded in a new file (“ReactorLoading”) containing the catalysis vial's barcode and the fixed-bed reactor number. Results from the catalytic testing using 3 Avantium parallel fixed-bed units 1 XR having 16 reactors and 2 XDs (XDB and XDC) having 4 reactors each are stored in the “CATALYSIS_X” (X = XR, XDB or XDC) files containing the fixed-bed reactor number, reaction conditions from process values (mass flow controllers, pressure indicators, temperature probes), and gas concentrations from online-GC analysis. Finally, the market costs of the metals used to synthesize the catalysts were manually calculated and are listed in the COST file. More details about the experimental set-up can be found in the reference article.22
To merge the individual files, five files (“CombVialPrep”, “SolidDispense”, “Impregnation”, “ImpregnationBarcode” and “COST”) are merged by matching vial indexes (Fig. 7). Per sample, two vials were used during synthesis: (i) for mixed metal nitrate solution preparation and (ii) for synthesizing the catalyst by incipient wetness impregnation (impregnation vial). Within one task, the vial indexes range from 1 to 24 where files are merged based on the vial indexes (Fig. 7). Each index within a task corresponds to a sample resulting in 24 samples per task.
 | ||
| Fig. 7 Content of the individual files and identifier to merge individual files together. The filenames and the column content required to merge the files are shown. | ||
The “CatalystDispenseBarcode” file contains two barcodes per sample (i) barcode of the impregnation vial and (ii) barcode of the catalyst vial. Therefore, the “CatalystDispenseBarcode” file connects synthesis data (impregnation vial) with catalytic testing data (catalyst vial) (Fig. 7). The “ReactorLoading” file contains the barcode for the catalyst vial, and information about which reactor was loaded with which catalyst (reactor number 1–16 and unit XR, XDB or XDC). The “Reactor Loading” file relates to the catalysis results file (“CATALYSIS_X” (X = XR, XDB or XDC)) through the fixed-bed reactor number and unit (Fig. 7).
An additional feature is implemented to allow data merging in case the generated files do not contain unique column entries. This was the case for fixed-bed testing on 3 different Avantium units. Their output files only contain information on the reactor number.1–16 Therefore, the reactor number is not sufficient to distinguish between the units in the “CATALYSIS_X” (X = XR, XDB or XDC) file and additional information about the unit is required. The feature incorporates information from the filename to identify which file corresponds to which unit. Having the exact tracing of which unit and reactor was used also allows evaluation of batch effects.23
In this example, the internal unit abbreviations were included into the filename (e.g., A001_TASK01_CATALYSIS_XR_RUN01). The data processing workflow takes the string “XR” and adds it as a new column with the column name “Unit” in the file. The other units will have as “Unit” column entry XDB or XDC. The configuration file provides the new column name and the location of the string in the filename as the column entry. It uses “_” to first separate the filename into individual strings and then selects the right string based on the configuration file (see ESI‡ Note 7 for further discussion). After including the new column entry, merging can be performed with the “ReactorLoading” file on multiple column entries for the reactor number1–16 and the unit (XR, XDB or XDC).
Based on this file structure, automated data merging is performed. An example Jupyter notebook with data already downloaded from openBIS is provided in the ESI‡ to run the data management steps without the need to access an openBIS platform (see ESI‡ Note 8 for further discussion). All the processed files are uploaded to an openBIS instance used by ETHZ SwissCAT+ with A001 as the project name and tasks named TASK0X (X = 1, 2, 3, 4, 5, 6). All the data within the project are downloaded and stored in a local folder. A001 is the main folder, and the subfolders contain data from each task (one task being an iteration/loop of 24 catalyst synthesis/testing). Data are read and stored in a structured dictionary with the structure and number of files being provided in the configuration file (see the ESI‡).
Upon merging, data processing can be executed. For project A001, information about synthesis (combination of metal elements and metal oxide support), the weights that were loaded into the reactor, and the catalytic performance originate from different files and instruments. Hence prior to merging individual files, parts of the data processing workflow would not be possible. Data are processed with the merged database, inputs from the configuration file and the data processing script (Process_FixedBed.py). The Process_FixedBed.py script along with a configuration file allows processing output files originating from automated and/or high-throughput instruments from Chemspeed, Unchained Labs and Avantium.
Data processing includes taking synthesis parameters from the Chemspeed synthesis robot to calculate nominal metal loadings of catalysts. Output data from three Avantium fixed-bed reactors are processed, and gas flow rates normalized by an internal standard, reactant conversion and individual product selectivity are calculated (see ESI‡ Note 9 for further discussion). To calculate conversion and selectivity, the expected reactants and products, including their number of carbon atoms are provided in the configuration file (see ESI‡ Note 9 for further discussion). Furthermore, depending on their configuration a set of Avantium instruments may have the same column name for different attributes. For example, the gas of the first mass flow controller (MFC) of one unit may not correspond to the same gas for another unit. Therefore, a renaming process using information from the configuration file is performed within the data processing step (see ESI‡ Note 10 for further discussion).
By incorporating tailored features based on the instruments, data are processed in a standardized way. Combining data from 3 individual instruments (synthesis data from a Chemspeed robot, catalyst dispense from Unchained Labs Junior and catalytic data from 3 individual Avantium Flowrence units), metrics such as formation rates normalized by the nominal metal loading can be calculated seamlessly. This approach is of utmost importance for a FAIR and ORD approach in the field of catalysis, allows avoiding human calculation mistakes, tracing back each performed calculation and allowing simple modification/adjustment of the calculations by external researchers if needed.24
The processed data are then uploaded back to the ELN as part of the processing activities and can subsequently be visualized to compare the data points. To effectively visualize large amounts of data, interactive figures/dashboards are generated through Tableau (see ESI‡ Note 11 for further discussion). Having data from synthesis, solid dispense and catalytic performance combined and processed allows better assessment of the data. In this case study, the experimental design for each task is based on the results from the previous tasks and were suggested through Bayesian optimization (Atinary SDLabs).25 For example, the catalyst composition or catalytic performance per task indicates the convergence of the experiments toward a target (e.g., optimization for catalytic performance). Having a broad visual inspection of the data enabled by the data management workflow allows users to navigate more effectively through the data and therefore foster its understanding and reuse especially in the context of open research data. Additionally, such merged and processed data can be used by other groups in combination with other databases to apply various AI/ML algorithms.
Conclusion
To facilitate data management and processing on electronic laboratory notebooks/laboratory information management systems, a Python library was implemented. The library takes raw data and a configuration file as input to perform a broad range of data management tasks often critical in data-driven automated and high-throughput laboratories. It includes up/download, merging and processing of tabular data related to heterogeneous catalysis research. The data management workflow is connected to an ELN, enabling data tracking and tracing subsequent data processing. This is demonstrated with a case study using real experimental data consisting of 59 individual files with 144 catalysts that were synthesized and tested for CO2 hydrogenation. Data are merged and processed together into one database where catalyst performance metrics requiring inputs from multiple files are extracted. The automated data processing workflow ultimately leads to a reduction of human error and increased efficiency. Relying on a configuration file to manage data enhances the data management transparency. In combination with a version-controlled code library allows tracing and reproduction of data processing workflows going from raw to processed data without any risk of human error. Consequently, this work contributes to paving the way to FAIR and ORD approaches in the field of heterogeneous catalysis.Data availability
All data and processing scripts for this paper are all included in the ESI‡ and can also be accessed under https://doi.org/10.5281/zenodo.14614047.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was sponsored by the ETH Domain through the Forschungsinfrastrukturen Program. This work was supported by the Open Research Data Program of the ETH Board and the Swiss Universities Open Science Program.References
- M. Abolhasani and E. Kumacheva, The rise of self-driving labs in chemical and materials sciences, Nat. Synth., 2023, 2(6), 483–492, DOI:10.1038/s44160-022-00231-0.
- R. Miyazaki, K. S. Belthle, H. Tüysüz, L. Foppa and M. Scheffler, Materials Genes of CO2 Hydrogenation on Supported Cobalt Catalysts: An Artificial Intelligence Approach Integrating Theoretical and Experimental Data, J. Am. Chem. Soc., 2024, 146(8), 5433–5444, DOI:10.1021/jacs.3c12984.
- M. Suvarna and J. Pérez-Ramírez, Embracing data science in catalysis research, Nat. Catal., 2024, 7(6), 624–635, DOI:10.1038/s41929-024-01150-3.
- M. Suvarna, T. Zou, S. H. Chong, Y. Ge, A. J. Martín and J. Pérez-Ramírez, Active learning streamlines development of high performance catalysts for higher alcohol synthesis, Nat. Commun., 2024, 15(1), 5844, DOI:10.1038/s41467-024-50215-1.
- S. G. Higgins, A. A. Nogiwa-Valdez and M. M. Stevens, Considerations for implementing electronic laboratory notebooks in an academic research environment, Nat. Protoc., 2022, 17(2), 179–189, DOI:10.1038/s41596-021-00645-8.
- C. Barillari, D. S. M. Ottoz, J. M. Fuentes-Serna, C. Ramakrishnan, B. Rinn and F. Rudolf, openBIS ELN-LIMS: an open-source database for academic laboratories, Bioinformatics, 2016, 32(4), 638–640, DOI:10.1093/bioinformatics/btv606.
- A. Trunschke, G. Bellini, M. Boniface, S. J. Carey, J. Dong and E. Erdem, et al., Towards Experimental Handbooks in Catalysis, Top. Catal., 2020, 63(19), 1683–1699, DOI:10.1007/s11244-020-01380-2.
- M. Suvarna, A. C. Vaucher, S. Mitchell, T. Laino and J. Pérez-Ramírez, Language models and protocol standardization guidelines for accelerating synthesis planning in heterogeneous catalysis, Nat. Commun., 2023, 14(1), 7964, DOI:10.1038/s41467-023-43836-5.
- B. A. Nosek, G. Alter, G. C. Banks, D. Borsboom, S. D. Bowman and S. J. Breckler, et al., Promoting an open research culture, Science, 1979, 348(6242), 1422–1425 CrossRef PubMed . Available fromhttps://www.science.org/doi/10.1126/science.aab2374.
- L. Sbailò, Á. Fekete, L. M. Ghiringhelli and M. Scheffler, The NOMAD Artificial-Intelligence Toolkit: turning materials-science data into knowledge and understanding, npj Comput. Mater., 2022, 8(1), 250 CrossRef . Available from https://www.nature.com/articles/s41524-022-00935-z.
- M. D. Wilkinson, M. Dumontier, I. Aalbersberg, G. Appleton, M. Axton and A. Baak, et al., The FAIR Guiding Principles for scientific data management and stewardship, Sci. Data, 2016, 3(1), 160018, DOI:10.1038/sdata.2016.18.
- A. Bauch, I. Adamczyk, P. Buczek, F. J. Elmer, K. Enimanev and P. Glyzewski, et al., openBIS: a flexible framework for managing and analyzing complex data in biology research, BMC Bioinf., 2011, 12(1), 468, DOI:10.1186/1471-2105-12-468.
- P. Laveille, P. Miéville, S. Chatterjee, E. Clerc, J. C. Cousty and F. de Nanteuil, et al., Swiss CAT+, a Data-driven Infrastructure for Accelerated Catalysts Discovery and Optimization, Chimia, 2023, 77(3), 154 CrossRef CAS PubMed . Available from https://www.chimia.ch/chimia/article/view/6241.
- P. Czember, F. Weber, A. Kokollari, H. Spreitzer and N. Fleck, Streamlining Catalysis Screenings on a Budget, Org. Process Res. Dev., 2024, 28(4), 1145–1150, DOI:10.1021/acs.oprd.3c00506.
- N. Artrith, K. T. Butler, F. X. Coudert, S. Han, O. Isayev and A. Jain, et al., Best practices in machine learning for chemistry, Nat. Chem., 2021, 13(6), 505–508, DOI:10.1038/s41557-021-00716-z.
- P. Araujo CBMH, Recognising Open Research Data in Research Assessment: Overview of Practices and Challenges, Zenodo, 2024 Search PubMed.
- P. H. P. Jati, Y. Lin, S. Nodehi, D. B. Cahyono and M. van Reisen, FAIR Versus Open Data: A Comparison of Objectives and Principles, Data Intell, 2022, 4(4), 867–881, DOI:10.1162/dint_a_00176.
- European Organization For Nuclear Research, OpenAIRE, CERN, 2013, available from https://www.zenodo.org/ Search PubMed.
- L. Talirz, S. Kumbhar, E. Passaro, A. V. Yakutovich, V. Granata and F. Gargiulo, et al., Materials Cloud, a platform for open computational science, Sci. Data, 2020, 7(1), 299, DOI:10.1038/s41597-020-00637-5.
- S. Herres-Pawlis, J. C. Liermann and O. Koepler, Research Data in Chemistry-Results of the first NFDI4Chem Community Survey, 2020, available from https://www.zaac.wiley-vch.de Search PubMed.
- P. S. F. Mendes, S. Siradze, L. Pirro and J. W. Thybaut, Open Data in Catalysis: From Today's Big Picture to the Future of Small Data, ChemCatChem, 2021, 13(3), 836–850 CrossRef CAS.
- A. Ramirez, E. Lam, D. P. Gutierrez, Y. Hou, H. Tribukait and L. M. Roch, et al., Accelerated exploration of heterogeneous CO2 hydrogenation catalysts by Bayesian-optimized high-throughput and automated experimentation, Chem Catal., 2024, 100888 CrossRef CAS . Available from: https://www.sciencedirect.com/science/article/pii/S266710932300489X.
- J. T. Leek, R. B. Scharpf, H. C. Bravo, D. Simcha, B. Langmead and W. E. Johnson, et al., Tackling the widespread and critical impact of batch effects in high-throughput data, Nat. Rev. Genet., 2010, 11(10), 733–739, DOI:10.1038/nrg2825.
- A. Trunschke, Prospects and challenges for autonomous catalyst discovery viewed from an experimental perspective, Catal. Sci. Technol., 2022, 12(11), 3650–3669, 10.1039/D2CY00275B.
- https://atinary.com/ .
Footnotes |
| † The most recent version of the python library can be found on the GitLab repository: https://gitlab.ethz.ch/swisscat_east_public/pycatdat. |
| ‡ Electronic supplementary information (ESI) available: Additional details (ESI Note 1–11) and figures (Fig. S1–S7). Python codes for the PyCatDat library (version 1.0). Tableau workbook for Fig. S3–S5. Python scripts for automated data upload to OpenBIS from Avantium Flowrence and Chemspeed Swing XL outputs (see ESI Note 1–2). See DOI: https://doi.org/10.1039/d4dd00350k |
| § These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |