Open Access Article
Open Access ArticleKnowledge discovery from porous organic cage literature using a large language model†
Yaoyi
Su‡
a,
Siyuan
Yang‡
ab,
Yuanhan
Liu
a,
Aiting
Kai
ab,
Linjiang
Chen
*cd and
Ming
Liu
 *ab
*ab
aDepartment of Chemistry, Zhejiang University, Hangzhou, Zhejiang 310058, China. E-mail: mingliu@zju.edu.cn
bZJU-Hangzhou Global Scientific and Technological Innovation Center, Zhejiang University, Hangzhou, Zhejiang 311200, China
cKey Laboratory of Precision and Intelligent Chemistry, University of Science and Technology of China, Hefei, Anhui 230026, China. E-mail: linjiangchen@ustc.edu.cn
dSchool of Chemistry, School of Computer Science, University of Birmingham, Birmingham B15 2TT, UK
First published on 19th December 2024
Abstract
Porous organic cages (POCs) are an emerging subclass of porous materials, drawing increasing attention due to their structural tunability, modularity and processibility, with the research in this area rapidly expanding. Nevertheless, it is a time-consuming and labour-intensive process to obtain sufficient information from the extensive literature on organic molecular cages. This article presents a GPT-4-based literature reading method that incorporates multi-label text classification and a follow-up information extraction, in which the potential of GPT-4 can be fully exploited to rapidly extract valid information from the literature. In the process of multi-label text classification, the prompt-engineered GPT-4 demonstrated the ability to label text with proper recall rates according to the type of information contained in the text, including authors, affiliations, synthetic procedures, surface area, and the Cambridge Crystallographic Data Centre (CCDC) number of corresponding cages. Additionally, GPT-4 demonstrated proficiency in information extraction, effectively transforming labeled text into concise tabulated data. Furthermore, we built a chatbot based on this database, allowing for quick and comprehensive searching across the entire database and responding to cage-related questions.
Introduction
Porous organic cages (POCs) are an emerging subclass of porous materials, distinguished by their unique structural tunability and ease of processing. Like other porous materials, POCs have adjustable pore structures, which make them suitable for a wide range of applications, including gas adsorption and separation,1–4 molecular detection,5,6 and use as catalyst carriers.7–9 The pioneering work on organic cage molecules was first reported by Lehn et al. in 1969, where they introduced a three-dimensional cryptand for cation binding.10 It is not until 2009 that Tozawa et al. discovered a series of rigid imine cages exhibiting permanent porosity in the solid state.1 Since then, significant interest has emerged in the design, synthesis, and application of POCs, which vary in their building units, shapes, and sizes.11,12 The synthesis of POCs is inherently complex, requiring a range of organic reactions and intricate experimental procedures.13 To replicate these syntheses, researchers must refer to the detailed synthetic steps outlined in the literature. Beyond the synthesis itself, information on the specific surface area, crystal structure, and topology of POCs is essential due to its relevance to their applications. However, extracting this information from the extensive body of literature is both time-consuming and labor-intensive.Large language models (LLMs) like Generative Pre-trained Transformer (GPT) can generate responses based on patterns and statistical principles learned during their pre-training phase.14 These models can interact dynamically, adapting to the context of a conversation to simulate human-like dialogue and communication. With hundreds of millions of parameters, GPT has shown exceptional performance and dominance in various fields, including natural language processing (NLP),15,16 medical imaging analysis,17,18 and chemical and biological research,19,20 garnering widespread recognition and acclaim for its capabilities.
Prompt engineering has become a crucial technique in LLMs for optimizing and fine-tuning them to perform specific tasks and achieve desired outcomes. This technique involves creating high-quality prompts that guide LLMs to generate accurate results.21,22 The process involves selecting the appropriate type of prompt, adjusting their size and structure, and sequencing them effectively according to the task requirements. Zheng et al. used prompt engineering to guide GPT-3.5 in extracting synthetic texts from the literature related to Metal–Organic Frameworks (MOFs) with a precision accuracy exceeding 90%.23 Afterwards, the same group also used a prompt-learning strategy to facilitate MOF material synthesis experiments through a symbiotic human-AI collaboration.24 They later applied a similar approach to guide the discovery and optimization of synthesis conditions for MOFs and Covalent Organic Frameworks (COFs).25 In 2024, Lu et al. successfully predicted the yield of ammonia catalytic reduction with up to 86% accuracy by incorporating pre-existing experimental data in the prompt project.26
In this study, we employed prompt engineering to guide GPT-4 in performing multi-label text classification, a task more complex than binary classification and a significant challenge for large language models. Literature paragraphs were labeled based on the information they contained, such as authors, cage names, synthetic procedures, surface area, and the CCDC number of the corresponding cages. These labeled paragraphs were then used as the input for GPT-4 to extract and tabulate information into the cage knowledge database. Each row in the database contains details such as the cage name, corresponding synthetic procedures, monomers and their synthesis procedures, cage stoichiometry, surface area, and CCDC number. The accuracy of GPT-4's multi-label classification and information extraction was assessed by comparing its results with manually curated data, which served as the ground truth. Ultimately, the cage knowledge database was used to develop a chatbot capable of reliably answering a variety of cage-related questions.
Methodology
While the GPT model has shown promising performance for various linguistic tasks,15–20 directly using it to read entire bodies of literature and extract specific information about POCs presents significant challenges. To address this, we implemented a two-step process for literature analysis using GPT-4 (Fig. 1), consisting of multi-label text classification followed by information extraction. | ||
| Fig. 1 The workflow of GPT-based information extraction from the literature. The workflow of GPT-based information extraction from the literature. | ||
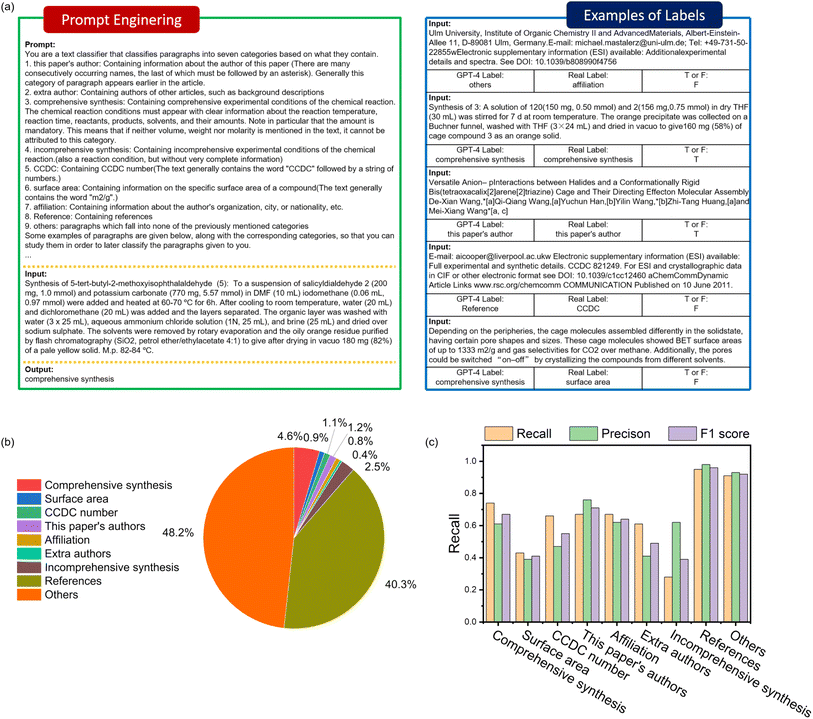
In the first step, the articles were divided into text segments. Each text segment was assigned a categorical label using a GPT-4 model trained with prompt engineering techniques (ESI, Section S2†). Since topology is described in a well-defined and fixed format, Python code was employed to identify specific sequences, as this method is more cost-effective compared to using GPT-4. In the second step, the selected text containing relevant information was further organized into tabulated data by both human experts and GPT-4. The verified answers were then compiled into a database, which was subsequently used for constructing chatbots.
Preparation of literature
We searched for literature in the Web of Science database using the keyword ‘porous organic cage’. Literature that focuses only on applications of reported POCs rather than synthesis of new POCs was excluded, resulting in 153 articles. These papers were authored by 34 different research groups and published across seven publishers to ensure diversity in writing styles and formats. The POCs covered are primarily imine-type cages, with a smaller portion consisting of alkyne-type cages, aryl ether-type cages, and others.GPT-based multi-label text classification
In this step, selected POC literature was segmented into paragraphs and labeled using a prompt-engineered GPT-4 model. We used PyPDF2 code to convert the pdf file into a split text, and then the symbols at the end of the text was used to determine whether the text is the end of a sentence.27 If it is not the end, it is connected to the next text. This operation combines several text segments in a logical manner, thereby reducing the number of segments to be processed. A set of prompts was developed to train GPT-4 to generate labels for each paragraph, focusing on key aspects required by chemists, such as authors, affiliations, synthetic procedures, surface area, and CCDC numbers. To reduce ambiguity during labeling and filter out texts with insufficient information, additional categories were included, such as incomplete synthesis, additional authors, references, and others. The explanation for each label is provided in Table 1. For each labeling task, we devised 2–3 prompts per label, as detailed in ESI, Section S2.† We manually labelled each paragraph to serve as ground truth. The accuracy of the GPT-4 labelling was then assessed by comparing it to the ground truth, and evaluated using precision, recall, and F1 score.| Category | Description | Required |
|---|---|---|
| Comprehensive synthesis | Contained comprehensive experimental conditions of the chemical reaction. The chemical reaction conditions must appear with clear information about the reaction temperature, reaction time, reactants, products, solvents, and their amounts | ✓ |
| CCDC | Contained CCDC number | ✓ |
| Surface area | Contained information on the specific surface area of a compound | ✓ |
| This paper's authors | Contained information about the authors of this paper | ✓ |
| Affiliation | Contained information about the authors' organizations, cities, nationalities etc. | ✓ |
| Extra authors | Contained authors of other articles, such as background descriptions | |
| Incomprehensive synthesis | Contained incomprehensive experimental conditions of the chemical reaction | |
| References | Contained references | |
| Others | Paragraphs that exceed all of the previously mentioned categories |
Precision and recall were calculated as follows:
where TP, FP, and FN represent True Positive, False Positive, and False Negative, respectively.
The F1 score is a reconciled average of precision and recall:
Information extraction & tabulation
All the paragraphs labeled with authors, affiliations, synthesis procedures, surface area, topology and CCDC numbers during the manual labelling process were used as textual input for GPT-4 to extract and tabulate relevant information, regardless of whether GPT-4 correctly classified the paragraphs. This process resulted in a POC database that improved the quality of the data, enabling more efficient interpretation and analysis. Each entry in the database systematically summarizes the relevant information, ensuring that key details—such as the cage name, topology, surface area, CCDC number, cage synthesis, monomer names, and monomer synthesis—are presented in a coherent and standardized format. We also manually extracted the same type of information as the cage knowledge database for the evaluation of GPT-4. The GPT-generated table was then compared with our manually created cage knowledge database using the Bidirectional Encoder Representation from Transformers (BERT) score. The BERT score is calculated as follows: the generated text and the reference text are encoded in the BERT model to obtain their respective vector representations. Subsequently, the similarity of two input texts is calculated by computing the cosine similarity of the vector representation for each of the two words.28Database utilization and analysis
Having utilized text mining techniques to construct a cage knowledge database, our aim was to leverage this resource to its fullest potential. The cage knowledge database was then fed to a prompt-engineered GPT-4 assistant, enabling it to answer questions based on the database. Additionally, a user interface was built using Tkinter, a python's open source graphical User Interface (GUI) platform.29 In order to fully explore the value of the database, we conducted statistical analysis of the synthesis strategy, topology, crystal structure from CCDC, and surface area in the database.Results and discussion
Text classification
In the multi-label text classification, as shown in Fig. 2a, all text segments were processed by GPT-4 with prompt engineering. Labels were then generated and evaluated by comparing them with manually labeled text, which served as the standard, to assess the accuracy of the GPT-4 model.Fig. 2b shows the distribution of different text categories, revealing that most of the text in the original documents falls under the categories of references and other sections. The key information we needed—such as authors, affiliations, specific surface areas, CCDC numbers, and experimental procedures—constitutes less than 10% of the total text content. This indicates that the GPT-4 classification process significantly reduces the volume of text to be processed in the tabulation step, lowering the corresponding costs.
The recall and precision results are illustrated in Fig. 2c. The “comprehensive synthesis” category had the highest recall rate at 0.74, which can be attributed to distinctive markers in the text, such as frequently mentioned compound names and amounts. However, there was a significant drop in precision for this category, down to 0.61. Observing the actual-versus-predicted category matrix (ESI, Fig. S1†), the primary error came from misidentifying segments that should have been labeled as “incomprehensive synthesis” as “comprehensive synthesis”. This highlights that even with prompt engineering designed to differentiate between comprehensive and incomprehensive synthesis, some errors persist. Texts under the “this paper's authors”, “Affiliation”, and “CCDC number” categories had similar recall rates of 0.67, 0.67, and 0.66, respectively. However, the precision for “this paper's authors” was notably higher than for “Affiliation” and “CCDC number”, showing a marked difference between precision and recall. Specific surface area information had both low recall and precision, likely because, while it has the identifier “m2 g−1”, it is often confused with similar terms like “m2 s−1” and “m/z”. The recall and precision of the surface area and CCDC numbers, which should be readily identifiable due to their distinct identifiers, were found to be unsatisfactory. This outcome can be attributed to redundant texts significantly interfering with the encoding and decoding process of GPT-4. Evidence for this conclusion is present in the actual-versus-predicted category matrix (ESI, Fig S1†), which shows that the recall of CCDC is 66.44%, with 25.17% of the information misclassified as “others.” Similarly, the recall for surface area is 47.71%, while information classified as “others” accounts for 43.13%, a value comparable to the recall.
Information extraction & tabulation
A typical example of tabulated information compiled from an article by both manual work and GPT-4 is shown in Fig. 3a for comparison. As observed, there is considerable similarity between the GPT-4 output and the manual results, with many matching parts and only minor differences. The name, topology and CCDC number of the molecular cages were accurately extracted with 100% text similarity. For the other sections, text similarity was less than 100%. These differences can be easily identified by comparing the two texts visually. To facilitate this comparison, Fig. 3a highlights the differing text portions in red.For the monomers, differences arose in the naming of the same compound (BPDDP), while other differences were primarily due to redundant text being extracted by GPT-4. Specifically, GPT-4 included titles along with the synthetic routes for cages and monomers, while manual work did not. Regarding surface area, GPT-4's response included more information than the manual response, providing additional comparisons of surface area between BPPOC and another compound, BTPOC. This suggests that GPT-4 has the potential to offer additional information, enhancing researchers' understanding of cage-related knowledge.
The results of the BERT score calculation, shown in Fig. 3b, indicate that the average similarity score across all information in the articles was 0.9155. In particular, with a score of 0.9357, the CCDC numbers showed the highest similarity. The similarity scores for specific surface area, synthetic routes of molecular cages, names and synthetic routes of monomers were also relatively high, each reaching a value of around 0.90. The lowest similarity score of 0.8405 was observed for the information related to the topology, mainly because a significant part of the relevant information could not be successfully extracted from the text and was therefore labelled as “None”.
Based on the complexity of their synthesis, the articles studied were systematically categorized into three different classes: class I represents articles in which only a single POC has been reported; class II represents articles reporting multiple POCs without transformation relationships between them, usually synthesized in parallel using the same reaction type but different building blocks; class III represents articles that reported multiple POCs with transforming relationships among them. Analysis of the statistical graph shows a trend that, in general, the accuracy of information extraction gradually decreases as the complexity of the articles increases. However, in the extraction of topologies, articles in the second class had a considerably higher similarity than those in the first class, contrary to the general trend.
The distribution of similarity was further analyzed using the molecular cage synthetic route as a representative example (Fig. 3c). The analysis shows that most similarities are above 0.8, with a significant proportion exceeding 0.9. However, a few samples had notably lower similarity. Upon reviewing these cases, we found that low similarity scores were mainly due to unsuccessful extractions, resulting in a single word “None” or very short answers. A typical example of this error was the vague description “Condensation of a pyridine system” replacing a comprehensive synthesis route. Fortunately, such instances are rare and do not significantly impact GPT-4's overall performance.
In the task of information extraction and tabulation, GPT-4 demonstrated strong capabilities in processing input text and extracting multiple categories of information simultaneously. This feature can significantly aid researchers by allowing for quicker reading and summarization of new papers. With GPT-4's assistance, researchers can save considerable time and effort in literature review, enabling them to focus more on tasks that require innovation and creativity.
Database utilization of the chatbot
Fig. 4a demonstrates that the GPT-4 model can effectively study and interpret the database to respond to user queries, as shown in a classic example. When asked about the reducing agent for the conversion of CC3 to RCC3, GPT-4 provided the correct answer “NaBH4” based on our knowledge. Additionally, GPT-4 not only provided synthetic routes with detailed information about reactants, solvents, and catalysts but also included methodologies for synthesizing reactants when detailed experimental procedures are requested. Beyond simple questions related to a single research paper, the chatbot can also address more complex queries. However, when faced with more systematic and intricate questions, the accuracy and completeness of the answers tend to decline. Fig. 4b highlights this issue: when prompted to list all the molecular cages documented by a specific scientist, the chatbot produced only 16 responses, failing to generate the complete list of 59 entries. Repeated experiments have confirmed that this outcome is not due to randomness but stems from the limitations of GPT-4. Although GPT-4 has a context limit of up to 128K, it still struggles to effectively process lengthy dialogues or texts.30These results offer substantial evidence that GPT-4 is capable of answering questions based on the information contained within the database. The types of questions it can address are diverse and not limited to a specific subset. GPT-4's responses are comprehensive enough to aid chemists in obtaining relevant information without the need to read the full text. Additionally, the system can meet practical demands in the field, such as providing detailed guidance for the synthesis of organic cages.
Database analysis
In order to summarize the information in the database, the synthetic chemistries, topologies, crystallographic structures, and surface areas of the entries were analyzed.Approximately 64% of the cages in the database were formed via imine condensation, with 11.63% being reduced from these imine precursors. This indicates that imine chemistry currently dominates the synthesis of porous organic cages (POCs). Additionally, 12.68% of the cages were synthesized via ether bonds, while other synthetic methods, such as amides and boronic esters, were also observed (Fig. 5a).
 | ||
| Fig. 5 Statistical analysis of the chemistries involved in synthesis (a), topology (b), CCDC structures (c) and surface areas (d) of cages. | ||
In terms of topologies, the analysis shows that [2 + 3]-cages account for 42.89%, which is nearly half of all entries in the database. Additionally, [4 + 6]-cages and [8 + 12]-cages are relatively prevalent, comprising 19.75% and 8.49% of the total, respectively (Fig. 5b).
Surface area provides guidance for exploring cage porosity and identifying potential applications. The density and accessible surface area (ASAs) of 253 entries were calculated using the Zeo++ software package (Fig. 5c).31 The probe radius was set as 1.82 Å, which is the kinetic radius of a nitrogen molecule. The results revealed a negative correlation between density and accessible surface area (ASA). Lower densities, around 0.5 g cm−3, correspond to ASAs exceeding 3000 m2 g−1 (red circle, Table S2†). Non-accessible surface area (NASA) values are generally lower, with significant values observed only within the density range of 1.00–1.25 g cm−3 (green circle). This is due to the inherent low surface area of high-density crystal structures.
Analysis of experimental surface area data revealed that approximately 60 POCs exhibit surface areas exceeding 500 m2 g−1, with 12 entries surpassing 1500 m2 g−1 (Fig. 5d). With the exception of a boronic ester-based cage, all high-surface-area cages were imine-based. This suggested that imine-based cages are currently one of the most promising methods for achieving high surface areas.
Conclusions
In this study, we developed a GPT-based system for extracting information from academic literature focused on organic molecular cages, resulting in a comprehensive molecular cage information database. Specifically, we evaluated the proficiency of the GPT-4 model in extracting and organizing detailed data on organic molecular cages from a large body of scientific literature. The resulting database, along with the associated interactive dialogue system, offers a valuable resource for advancing research in the design, synthesis, and application of molecular cages. Furthermore, the database created in this work provides a crucial resource for future machine learning and experimental studies aimed at discovering new POCs. However, the dialogue system shows limitations in answering more complex questions. Future efforts will focus on enhancing the search capabilities of the system. Additionally, the database can be updated dynamically, allowing newly reported organic molecular cages to be incorporated through the process outlined in this article. Looking forward, LLMs, in conjunction with other rapidly evolving AI tools and lab-automation techniques, have the potential to significantly accelerate the discovery of new molecules, such as POCs and beyond.Data availability
The data supporting this article can be found in the ESI. Raw data, the resulting database of POCs and custom codes for this work, including scripts for the directly runnable chatbot, as well as the results of text classification and information tabularization, are available at https://doi.org/10.5281/zenodo.14511583. The version of the code employed for this study is Version v1. The codes are also available at https://github.com/syy1213/LLMs-GPT-4-Cage.The codes and required python modules for text classification, information tabularization and the directly runnable chatbot can be found at https://hub.docker.com/r/syy12137059/cage_gpt/tags.
Author contributions
Yaoyi Su: investigation, formal analysis, validation and writing – original draft. Siyuan Yang: conceptualization, methodology, supervision and writing – review & editing. Yuanhan Liu and Aiting Kai: resources and validation. Linjiang Chen and Ming Liu: project administration, supervision, funding acquisition and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the National Natural Science Foundation of China (22371252) and the Zhejiang Provincial Natural Science Fund (LZ23B020005), and are thankful for the Leading Innovation Team grant from the Department of Science and Technology of Zhejiang Province (2022R01005).Notes and references
- T. Tozawa, J. T. A. Jones, S. I. Swamy, S. Jiang, D. J. Adams, S. Shakespeare, R. Clowes, D. Bradshaw, T. Hasell, S. Y. Chong, C. Tang, S. Thompson, J. Parker, A. Trewin, J. Bacsa, A. M. Z. Slawin, A. Steiner and A. I. Cooper, Nat. Mater., 2009, 8, 973–978 CrossRef PubMed.
- S. Jiang, J. Bacsa, X. F. Wu, J. T. A. Jones, R. Dawson, A. Trewin, D. J. Adams and A. I. Cooper, Chem. Commun., 2011, 47, 8919–8921 RSC.
- J. M. Lucero and M. A. Carreon, ACS Appl. Mater. Interfaces, 2020, 12, 32182–32188 CrossRef PubMed.
- K. Z. Su, W. J. Wang, S. F. Du, C. Q. Ji and D. Q. Yuan, Nat. Commun., 2021, 12, 3703 CrossRef PubMed.
- Y. X. Chen, G. C. Wu, B. B. Chen, H. Qu, T. Y. Jiao, Y. T. Li, C. Q. Ge, C. Zhang, L. X. Liang, X. Q. Zeng, X. Y. Cao, Q. Wang and H. Li, Angew. Chem., Int. Ed., 2021, 60, 18815–18820 CrossRef PubMed.
- C. Zhang, H. Y. Wang, J. Zhong, Y. Lei, R. F. Du, Y. Zhang, L. B. Shen, T. Y. Jiao, Y. L. Zhu, H. M. Zhu, H. R. Li and H. Li, Sci. Adv., 2019, 5, eaax6707 CrossRef PubMed.
- P. Bhandari and P. S. Mukherjee, ACS Catal., 2023, 13, 6126–6143 CrossRef.
- L. Qiu, R. McCaffrey, Y. H. Jin, Y. Gong, Y. M. Hu, H. L. Sun, W. Park and W. Zhang, Chem. Sci., 2018, 9, 676–680 RSC.
- S. Jiang, H. J. Cox, E. I. Papaioannou, C. Y. Tang, H. Y. Liu, B. J. Murdoch, E. K. Gibson, I. S. Metcalfe, J. S. O. Evans and S. K. Beaumont, Nanoscale, 2019, 11, 14929–14936 RSC.
- B. Dietrich, J. M. Lehn and J. P. Sauvage, Tetrahedron Lett., 1969, 10, 2885–2888 CrossRef.
- T. Hasell and A. I. Cooper, Nat. Rev. Mater., 2016, 1, 16053 CrossRef.
- D. Y. Hu, J. J. Zhang and M. Liu, Chem. Commun., 2022, 58, 11333–11346 RSC.
- X. C. Yang, Z. Ullah, J. F. Stoddart and C. T. Yavuz, Chem. Rev., 2023, 123, 4602–4634 CrossRef CAS PubMed.
- OpenAI, https://openai.com/, accessed December 2024.
- C. C. Chiang, M. Luo, G. Dumkrieger, S. Trivedi, Y. C. Chen, C. J. Chao, T. J. Schwedt, A. Sarker and I. Banerjee, Headache, 2024, 64, 400–409 CrossRef PubMed.
- S. Nath, A. Marie, S. Ellershaw, E. Korot and P. A. Keane, Br. J. Ophthalmol., 2022, 106, 889–892 CrossRef PubMed.
- S. Ziegelmayer, A. W. Marka, N. Lenhart, N. Nehls, S. Reischl, F. Harder, A. Sauter, M. Makowski, M. Graf and J. Gawlitza, J. Med. Internet Res., 2023, 25, e50865 CrossRef PubMed.
- I. Civettini, A. Zappaterra, B. M. Granelli, G. Rindone, A. Aroldi, S. Bonfanti, F. Colombo, M. Fedele, G. Grillo, M. Parma, P. Perfetti, E. Terruzzi, C. Gambacorti-Passeríni, D. Ramazzotti and F. Cavalca, Br. J. Haematol., 2024, 204, 1523–1528 CrossRef PubMed.
- G. V. Ye and J. Comput, Aid, Mol. Des., 2024, 38, 20 Search PubMed.
- Z. L. Zheng, Z. G. He, O. Khattab, N. Rampal, M. A. Zaharia, C. Borgs, J. T. Chayes and O. M. Yaghi, Digital Discovery, 2024, 3, 491–501 RSC.
- P. Korzynski, G. Mazurek, P. Krzypkowska and A. Kurasinski, Entrep. Bus. Econ. Rev., 2023, 11, 25–37 Search PubMed.
- S. Ayad and F. AlSayoud, Lect. Note. Netw. Syst., 2024, vol. 987, pp. 412–422 Search PubMed.
- Z. L. Zheng, O. F. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef PubMed.
- Z. L. Zheng, Z. C. Rong, N. Rampal, C. Borgs, J. T. Chayes and O. M. Yaghi, Angew. Chem., Int. Ed., 2023, 62, e202311983 CrossRef PubMed.
- Z. L. Zheng, O. F. Zhang, H. Nguyen, N. Rampal, A. H. Alawadhi, Z. C. Rong, T. Head-Gordon, C. Borgs, J. T. Chayes and O. M. Yaghi, ACS Cent. Sci., 2023, 9, 2161–2170 CrossRef PubMed.
- M. Y. Lu, F. Y. Gao, X. L. Tang and L. J. Chen, Iscience, 2024, 27, 109451 CrossRef PubMed.
- PyPDF2, https://pypi.org/project/PyPDF2/, accessed December 2024 Search PubMed.
- J. Y. Lee, K. Jung and Pr. Mach, Learn. Res., 2019, 101, 1081–1093 Search PubMed.
- Tkinter, https://docs.python.org/3/library/tkinter.html, accessed December 2024 Search PubMed.
- OpenAI, https://gpt40mni.com/safety-and-limitations/, accessed December 2024 Search PubMed.
- T. F. Willems, C. H. Rycroft, M. Kazi, J. C. Meza and M. Haranczyk, Microporous Mesoporous Mater., 2012, 149, 134–141 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00337c |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |