Open Access Article
Open Access ArticleActivity recognition in scientific experimentation using multimodal visual encoding†
Gianmarco
Gabrieli
 ,
Irina
Espejo Morales
,
Dimitrios
Christofidellis
,
Mara
Graziani
,
Andrea
Giovannini
,
Federico
Zipoli
,
Amol
Thakkar
,
Irina
Espejo Morales
,
Dimitrios
Christofidellis
,
Mara
Graziani
,
Andrea
Giovannini
,
Federico
Zipoli
,
Amol
Thakkar
 ,
Antonio
Foncubierta
,
Matteo
Manica
,
Antonio
Foncubierta
,
Matteo
Manica
 and
Patrick W.
Ruch
and
Patrick W.
Ruch
 *
*
IBM Research Europe, Säumerstrasse 4, 8803 Rüschlikon, Switzerland. E-mail: ruc@zurich.ibm.com
First published on 19th December 2024
Abstract
Capturing actions during scientific experimentation is a cornerstone of reproducibility and collaborative research. While large multimodal models hold promise for automatic action (or activity) recognition, their ability to provide real-time captioning of scientific actions remains to be explored. Leveraging multimodal egocentric videos and model finetuning for chemical experimentation, we study the action recognition performance of Vision Transformer (ViT) encoders coupled either to a multi-label classification head or a pretrained language model, as well as that of two state-of-the-art vision-language models, Video-LLaVA and X-CLIP. Highest fidelity was achieved for models coupled with trained classification heads or a fine-tuned language model decoder, for which individual actions were recognized with F1 scores between 0.29–0.57 and action sequences were transcribed at normalized Levenshtein ratios of 0.59–0.75, while inference efficiency was highest for models based on ViT encoders coupled to classifiers, yielding a 3-fold relative inference speed-up on GPU over language-assisted models. While models comprising generative language components were penalized in terms of inference time, we demonstrate that augmenting egocentric videos with gaze information increases the F1 score (0.52 → 0.61) and Levenshtein ratio (0.63 → 0.72, p = 0.047) for the language-assisted ViT encoder. Based on our evaluation of preferred model configurations, we propose the use of multimodal models for near real-time action recognition in scientific experimentation as viable approach for automatic documentation of laboratory work.
1 Introduction
Validating hypotheses based on factual results of experiments is a cornerstone of the scientific method. The fact that those results can be consistently reproduced and understanding under which conditions they hold is what allows scientific advancement. However, more than 70% of researchers have reportedly failed attempts to reproduce other scientists' experiments, and over 50% fail to reproduce even their own experiments.1 In an analysis of 53 peer-reviewed publications in pre-clinical drug development, the scientific findings could only be reproduced in six (11%) of the cases.2 Reproducibility suffers in particular from flawed or missing experimental data and metadata.3,4 It is, therefore, desirable to document scientific experimentation in as much detail as possible, while automating and standardizing the process to avoid encumbering the researcher. Building on the idea of using machine learning to perform activity recognition, we propose a novel approach that improves the traceability of scientific workflows by describing the complete set of activities performed by a laboratory operator. In particular, foundation models (FMs) that are pre-trained on broad datasets under self-supervision have demonstrated their ability to be fine-tuned for specific downstream tasks incorporating multiple data modalities such as language and vision.5 The application of automated activity recognition in lab environments fosters novel exploration on how FMs may revolutionize the scientific method, and potentially accelerate the discovery process. Extending the concept of activity recognition for lifelogging,6 we propose that egocentric video recording can be used to capture step-by-step experiments to reduce and possibly eliminate the need for scientists to manually generate additional documentation. Notably, this approach places FMs at the beginning of the scientific data capture process, thereby complementing the meanwhile widespread usage of FMs for language-centric interpretation of data7–10 and generative tasks in scientific data modeling.11–14In this work, we investigate the capacity of vision-language FMs to capture sequences of actions related to scientific experimentation based on egocentric videos. In particular, we compare the performance of state-of-the-art vision-language FMs trained on large-scale image and video datasets, X-CLIP15 and Video-LLaVA,16 against related model architectures that were trained on smaller, more domain-specific datasets consisting exclusively of egocentric videos. We evaluate the performance of the models in zero-shot settings as well as when coupled to multi-class classifiers trained under supervision on the targeted action classes, studying the recognition of individual actions and action sequences as well as the model inference times.
For selected models, we further examine the effect of incorporating additional data modalities for action recognition. Egocentric video recording systems often provide multiple modalities of information, e.g. gaze, depth, and motion. Since some modalities may generally not be aligned or available in matching quantities, there are challenges constructing FMs that can analyze such data. Integrating multiple modalities, however, is inherently challenging. The dimensionality of the data is increased if the primary video modality is expanded with additional channels to incorporate gaze and depth maps. Similarly, the video length would increase if the frames from the different modalities were concatenated one after the other. In both cases, there is the risk of large input sizes that are unfeasible to fit in memory. To address some of these challenges, we propose to use visual encoding of additional modalities to incorporate those signals into egocentric videos. This way, we seamlessly integrate modalities and leverage a pre-trained FM without designing ad hoc models that accept multiple data modalities in their native formats. We study the efficacy of this approach on an egocentric laboratory action dataset augmented with gaze coordinates, depth and object masks.
2 Related work
Documentation of experimental actions is a central element of scientific research, being an essential prerequisite for collaboration and to obtain machine-actionable scientific data.17 Key challenges are the fragmentation and need for interoperability of tools such as Electronic Lab Notebooks18,19 used by researchers to manually annotate experimental activities, leading to wide variations in the quality and granularity of documented experimentation. Here, we explore how egocentric video and additional modalities can be leveraged for action recognition in scientific experimentation, with the potential to greatly facilitate data capture and documentation in laboratory environments. Sasaki et al. recently reported a study of object detection and action recognition during chemical experimentation from fixed-perspective viewpoints, relying on YOLOv8 as a one-stage model for object detection and 3D ResNet for action recognition.20 In the present work, we are primarily interested in understanding the point-of-view (POV) of the experimentalist in order to evaluate the feasibility of transcribing end-to-end workflows.With the emergence of wearable recording devices, the field of action recognition in lifelogging and egocentric video received substantial attention from researchers.6,21–23 Multiple datasets have been released to support research and benchmarking in action recognition,24–26 with Ego4D26 being one of the largest. The ability to leverage such datasets for pre-training and to subsequently fine-tune models for the domain-specific semantics of scientific experimentation remains to be explored. Moreover, from a multimodal perspective, there have been efforts to combine visual appearance (RGB representations) and skeleton, infrared, depth, Inertial Measurement Unit (IMU) data, motion or other modalities.27–29 One of the modalities researchers have explored for egocentric action recognition is gaze. Gaze has been used to inform machine learning methods about the Region Of Interest (ROI) so that relevant features are extracted.30 Gaze estimation in a multitask learning setting has been found to improve the egocentric AR performance.31 However, rather counter-intuitively, using the gaze directly in an ad hoc gaze/vision model appears to have little to no effect.32
Most of the literature on multimodal video understanding encompasses the use of language and audio as additional modalities.33–37 These three are very different in representation format, with the language being symbolic, vocabulary encoded, and the audio-visual signal finding a continuous encoding along one and two dimensions, respectively. To deal with the different representation formats, most of the existing works focused on mid- and late fusion, where each modality is treated independently by unimodal systems and then combined in the middle of the model for mid-fusion and just before prediction for late fusion. Simple average,33 weighted averages,34 bilinear products35 and rank-minimization36 have been proposed as late-fusion combination strategies. Recent approaches have explored the use of large language models (LLMs) to obtain a vocabulary-based encoding of all modalities, for instance, by generating audio transcriptions and video captions and combining all text inputs as a single input to the LLM.38 Finally, purely multimodal architectures have been proposed to deal with the raw signals in the original encoding formats. Among these, the video–audio–text transformer in ref. 37 leverages contrastive losses to obtain a purely multimodal representation through modality-specific patches and positional embeddings. Similarly, unified frameworks such as VideoCLIP39 were proposed to fuse video and text captions in a unique latent representation.
As an alternative to the aforementioned approaches, this work analyzes the impact of early fusion of different visual modalities at the input level. Instead of working with audio and language signals, we focus on inputs that share the same representation in the 2D visual space and along a temporal axis. For instance, we propose a simple yet effective strategy to encode multimodal visual signals in a single visual input that does not overload the input size.
3 Experimental
3.1 Recording devices
We recorded egocentric POV videos in laboratory environments using head-mounted recording devices (Pupil Labs, Germany‡) to capture RGB video and gaze coordinates. A sampling rate of 4 Hz was chosen for both modalities in order to track the hand motions and scene manipulations in the encountered experiments with sufficient temporal granularity while supporting real-time data streaming and processing. The recording devices were provided with a wired connection to a smartphone to record and stream POV scenes via a dedicated mobile app. Users were asked to wear the recording devices integrated in their protective eyewear while performing predetermined activities in the laboratory environment (cf. Subsection 3.3). The video streams were discretized into 4 s clips which represents the temporal granularity of annotations for training and action recognition during inference. Video recording was triggered and ended through user commands.3.2 Activity recognition models
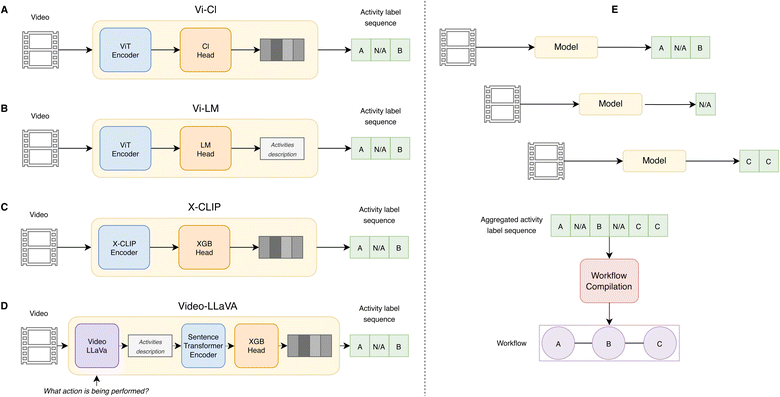
We studied four video-based model architectures (Fig. 1) with respect to their use in action recognition in scientific experimentation:3.3 Datasets
We derived a subset of videos from the Ego4D dataset26 according to keywords that relate to activities that may be encountered in laboratory environments (Section 1 in ESI†). Subsequently, we extracted 4 s segments from this subset of videos for which the accompanying annotated text contained at least one of these specific keywords. If a segment's duration fell short of 4 s, we extended the window by incorporating an additional two seconds both before and after the identified segment. The resulting set of 4 s egocentric video clips was split into 330K clips for training, 3324 clips for validation, and 3325 clips for testing.To evaluate the activity recognition models, we created two distinct datasets comprising POV videos recorded by 60 different users performing typical laboratory workflows. The individual action labels and descriptions of the activities for the two datasets are reported in Table 1 while example frames for each activity are shown in Section 2 of the ESI.† The Lab Actions dataset provides an example of diversified activities that are common in chemistry experimentation and can be combined or repeated to build workflows of various complexity levels. The activity labels are a subset of the chemical actions described previously in ref. 47. Instead, the Lab Motion dataset comprises a smaller set of activities encountered in exploratory semiconductor chip processing and experimental workflows with alternating development and rinsing steps. The peculiarity of this dataset resides in the more granular discrimination of the three development activities which only differ in terms of the hand motion pattern.
| Dataset name | Activity label | Description |
|---|---|---|
| Lab Actions | Add | Addition of liquid solutions to vials |
| AnalyticalMeasurement | Measurement with analytical instruments such as pH meters | |
| CollectLayer | Collection of organic or inorganic phases | |
| MeasureSolid | Weighing of solid powders | |
| MeasureLiquid | Measurement of liquid volume in graduated cylinders | |
| PhaseSeparation | Separation of materials in different phases | |
| Stir | Stirring of liquid solution | |
| Lab Motion | Development (circular) | Develop chip with circular motion |
| Development (figure eight) | Develop chip with figure-eight motion | |
| Development (puddle) | Develop chip with puddling motion | |
| Rinsing | Rinse chip with water jet |
The Lab Actions dataset consists of a total of 6877 video clips of 4 s each for a total recording time of ≈8 h. The number of workflows is 193, each consisting of an end-to-end sequence of actions of total average duration 143 s. The average time per action ranged between 11 s and 25 s depending on the action. The Lab Actions dataset was split into 5911/301/655 video clips for training/validation/testing. The Lab Motion dataset includes 6478 video clips of 4 s (≈7 h total time) corresponding to 139 workflows lasting on average 186 s each. Individual actions lasted between 53 s and 69 s on average. The clips were split into sets of 5799/336/343 for training/validation/testing. Each video clip was manually annotated with the corresponding activity labels in Table 1, whereby a No action label was assigned to clips in which none of the activity labels could be assigned.
3.4 Training and prompting
The Vi-Cl and Vi-LM models rely on a vision encoder based on the ViT48 backbone and joint space-time attention trained with tube masking, following the same approach as the VideoMAE40 model. We focus on the respective base variant of the model, which has 12 layers and 87M parameters in total. We pre-trained the vision encoder from scratch on the lab-oriented selection of egocentric videos from Ego4D (cf. Subsection 3.3) for 10 full epochs (∼10K steps) using a masking probability of 0.90, batch size of 32, AdamW optimizer,49 and weight decay of 0.01. The initial learning rate was 1 × 10−3, which decayed constantly for the first 3 epochs by a factor of 0.5 per epoch and then for the rest of the epochs by a factor of 0.01. In the case of Vi-Cl, the pre-trained vision encoder was coupled to a linear classifier that was trained to associate the video embeddings with the activity labels in Table 1. For Vi-LM, the encoder and decoder components were fine-tuned together using clips from the Lab Actions or Lab Motion datasets (Table 1) as inputs to generate a caption describing the action performed by the researcher, where the ground truth is defined by manual annotations (cf. Subsection 3.3). We trained for 60 epochs using an initial learning rate of 3 × 10−4, and then a constant learning rate decay by a factor of 0.04 per epoch, batch size of 64, AdamW optimizer and weight decay of 0.01.The version of X-CLIP15 we use for action recognition is based on computing a contrastive similarity score between a video and a list of labels, and is not adequate for open-ended prompting in the zero-shot task. Instead, a video and a list of labels (Table 1) is provided as input for inference. The second approach for benchmarking uses the frozen video encoder of X-CLIP to train a classifier, RF or XGBoost, with pairs of video encodings and labels. Before encoding, videos are subdivided into clips of 4 s where each clip demonstrates an action and the classifier predictions are on a per-clip basis. We tuned the hyperparameters of the classifiers using the validation split (Section 4 in the ESI†).
For action recognition with Video-LLaVA, two approaches were followed. In the first, we provided a text prompt to the pre-trained model for zero-shot predictions with the question “Which action is being performed among Add, Analytical Measurement, Collect Layer, Measure Solid, Measure Liquid, Phase Separation, Stir and none of them?”. We embedded the text outputs with the BAAI/bge-base-en-v1.5 (ref. 50) model using the SentenceTransformer framework51 and obtained a prediction of each video based on the greatest cosine similarities between the prediction embedding and all the activity label embeddings. In the second approach, we prompt the model with the question “What action is being performed?” to generate an open-ended text description of each 4 s video clip. We then embed each description by leveraging the same SentenceTransformer module. As in the case of the X-CLIP pipeline, we trained either a Random Forest or an XGBoost model on the embeddings obtained from SentenceTransformer (Fig. 1D). We tuned the hyperparameters of the model heads using the same validation splits as for the other approaches (Section 4 in the ESI†).
3.5 Performance analysis
To evaluate the action recognition performance at level of individual 4 s video clips, we compute the weighted F1 score for all modeling approaches for both laboratory video datasets in Table 1. As Vi-LM can generate labels outside the list of possible activities used to train the model, the predictions not included in the list of activities were converted to No action before computing the F1 score.To measure the workflow recognition performance at the level of action sequences, we aggregate predictions over all the videos related to the same experiment (Fig. 1E). The aggregated activity label sequence is then processed to filter out No action labels and to merge consecutive activity labels. For the Lab Motion dataset, we apply an additional filter that removes spurious predictions that are isolated in time, given the longer characteristic times of the activities in that dataset. We compute a similarity measure between workflows based on the normalized Levenshtein distance,52 where we represent each workflow by a sequence of characters mapped to the steps in the workflow. For Lab Actions, a workflow step corresponds to one of the activity labels in Table 1, while for Lab Motion there are only two workflow steps (development and rinsing). Then, we measured the similarity between the sequences derived from the model predictions and the ground truth. In particular, we report the Levenshtein ratio, which takes values in the range between 0 (absence of workflow similarity) and 1 (perfect workflow prediction) and is calculated as 1 − d, where d is the normalized Levenshtein distance. We report the average Levenshtein ratio on the test split of each dataset.
To evaluate the fitness of the system for real-time action recognition in lab settings, we measured the inference execution time for all the considered models. The timing environment contains a single NVIDIA Tesla V100-SXM2 GPU, 4 Intel Xeon Scalable Processors and a total maximum memory of 20 GB is allowed per model. Inference times are reported starting from the availability of video encodings until label prediction, with the models tested under identical conditions. For actual production deployment, we expect that greater resource allocation would further lower the inference time.
3.6 Multimodal visual encoding
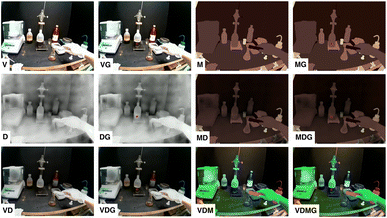
To investigate how laboratory activity recognition may benefit from additional input modalities, we extend our analysis of the Lab Actions dataset (cf. Subsection 3.3) using the Vi-Cl and Vi-LM models by incorporating additional gaze information represented by (x, y) coordinates for each RGB video frame. We introduce additional modalities by processing the RGB videos with two off-the-shelf models: a depth modality that we computed using DINOv2 (ref. 53) on each frame, and a segmentation modality, labelling all the objects per frame, that we obtained by leveraging the segmentation capabilities of SAM.54To make the various signals compatible with the vision models, we performed visual encoding as follows: the visual appearance signal (V) was kept as recorded by the device in the RGB color space, the depth signal (D) was represented in RGB gray levels, the object masks signal (M) was represented as a colored label map also in RGB. When combining V or M with D, the depth was used as a scalar factor on all RGB channels, and when combining V, M and D, these were mapped to HSV channels (M was mapped to Hue, Depth to Saturation, and the brightness of V was mapped to value). The gaze signal (G) was incorporated as constant color filled circle on each frame for any of the modality combinations. Fig. 2 contains examples of the appearance of the modalities for a video frame extracted during an Add activity taking place in a chemistry laboratory.
Different combinations of modalities were studied for training and testing, respectively, as reported in Section 4.
4 Results & discussion
4.1 Activity and workflow recognition
We report the F1 score for activity recognition and the Levenshtein ratio to assess workflow recognition in Table 2. While Vi-LM outperformed Vi-Cl for the Lab Actions dataset, the F1 score for Vi-Cl was substantially higher (+0.15) than for Vi-LM for the Lab Motion dataset. Thus, the model comprising a language component performed worse in differentiating the three development activities (cf. confusion matrices in Section 3 of the ESI†), while it could adequately resolve the two different workflow steps (development and rinsing) in sequence as evidenced by the high Levenshtein ratio. Indeed, the three development activities correspond to different motions of the same action type. For such types of activities, Vi-Cl resulted in the best F1 score (0.56) out of all model configurations, demonstrating better ability to resolve temporally correlated actions. This result emphasizes the impact of the video modality on the recognition of similar activities that are mainly distinguished by motion patterns rather than distinct scenes and objects, and underscores the benefit of domain-specific data for model fine-tuning. The zero-shot action predictions of X-CLIP and Video-LLaVA performed poorly for both datasets, since such models were not fine-tuned on scenes recorded in laboratory settings with the domain-specific classes. In fact, these models perform worse in terms of F1 score compared to a random predictor weighted by the relative occurrence of each class in the training set (Table 2). When adding classification heads to the pre-trained X-CLIP encoder or Video-LLaVA prediction embeddings, the performance metrics increase substantially. In particular, for the Lab Actions dataset, X-CLIP + XGB provided the highest performance scores, boosting the F1 score from 0.12 to 0.57 and Levenshtein ratio from 0.30 ± 0.09 to 0.75 ± 0.08 compared to the zero-shot prediction, thereby outperforming Vi-Cl and Vi-LM models. Generally, the X-CLIP model tended to predict more No action labels (Section 3 in ESI†) than the other models, resulting in less spurious activity predictions within a workflow and better Levenshtein ratios. X-CLIP was found to produce effective embeddings that can be used to train classification heads and adapt the model to laboratory applications. We attribute the enhanced performances of the X-CLIP vision encoder also to its training on the Kinetics-400 dataset,43 which comprises 400 human action classes, including human-object interactions. Instead, while Video-LLaVA coupled with Random Forest or XGBoost classifiers produced performances that approached the scores of the other models in the Lab Actions dataset, its F1 score for the Lab Motion dataset was significantly worse than the other approaches. This finding highlights the challenges in obtaining effective textual activity descriptions for very similar actions in the absence of domain-specific fine-tuning. Example video captioning using Video-LLaVA for both Lab Motion and Lab Actions is reported in Section 5 of the ESI.†| Model | Lab actions | Lab motion | ||
|---|---|---|---|---|
| F1 score | Levenshtein ratio | F1 score | Levenshtein ratio | |
| Vi-Cl | 0.49 | 0.59 ± 0.08 | 0.56 | 0.78 ± 0.08 |
| Vi-LM | 0.52 | 0.63 ± 0.09 | 0.41 | 0.82 ± 0.10 |
| X-CLIP | 0.12 | 0.30 ± 0.09 | 0.19 | 0.66 ± 0.10 |
| X-CLIP + RF | 0.46 | 0.67 ± 0.10 | 0.52 | 0.81 ± 0.07 |
| X-CLIP + XGB | 0.57 | 0.75 ± 0.08 | 0.52 | 0.86 ± 0.04 |
| Video-LLaVA | 0.03 | 0.48 ± 0.06 | 0.14 | 0.70 ± 0.07 |
| Video-LLaVA + RF | 0.44 | 0.64 ± 0.08 | 0.30 | 0.78 ± 0.08 |
| Video-LLaVA + XGB | 0.43 | 0.65 ± 0.06 | 0.29 | 0.79 ± 0.07 |
| Random weighted | 0.22 | 0.35 ± 0.03 | 0.20 | 0.37 ± 0.09 |
Regarding the feasibility of using the investigated models for real-time laboratory activity recognition, we report the inference execution times in Table 3. The fastest model was Vi-Cl followed by the pre-trained X-CLIP embedding + classifier configurations. Models comprising a generative language component were the slowest, as expected. Vi-Cl was also the most lightweight, with the least amount of parameters (87M), making such model configuration an efficient choice for a real-time applications.
| Execution time for inference | ||
|---|---|---|
| Model | # Parameters | Time/clip (s) |
| Vi-Cl | 87M | 0.46 ± 0.01 |
| Vi-LM | 147M | 1.44 ± 0.01 |
| X-CLIP + RF | 123M | 0.52 ± 0.03 |
| X-CLIP + XGB | 123M | 0.50 ± 0.04 |
| Video-LLaVA + RF | 2B | 2.05 ± 0.07 |
| Video-LLaVA + XGB | 2B | 2.46 ± 0.08 |
4.2 Multimodal visual encoding for activity and workflow recognition
Table 4 shows the F1 scores for activity recognition obtained with Vi-Cl and Vi-LM trained on multimodal visual encoding datasets. Contrary to inference on the lone RGB video modality, Vi-LM yields the best result when combining video and gaze (VG) modalities. Applied to the multimodal datasets including the original video modality (V), Vi-LM yields better F1 scores than configurations comprising only D, G, M, and their aggregations, also outperforming the lone V modality. Interestingly, Vi-LM outperforms Vi-Cl in each modality combination and yields the highest F1 scores on the multimodal datasets VG (F1 = 0.61), VDG (F1 = 0.58) and VDM (F1 = 0.57), which match or exceed the activity recognition performance of the best-performing model on the lone video modality (X-CLIP + XGB, Table 2).| Data | F1 score | Levenshtein ratio | ||
|---|---|---|---|---|
| Vi-Cl | Vi-LM | Vi-Cl | Vi-LM | |
| V | 0.49 | 0.52 | 0.59 ± 0.08 | 0.63 ± 0.09 |
| VG | 0.47 | 0.61 | 0.67 ± 0.08 | 0.73 ± 0.09 |
| VDG | 0.47 | 0.58 | 0.65 ± 0.10 | 0.65 ± 0.10 |
| VDMG | 0.44 | 0.55 | 0.58 ± 0.11 | 0.65 ± 0.08 |
| VDM | 0.48 | 0.57 | 0.65 ± 0.09 | 0.69 ± 0.08 |
| VD | 0.51 | 0.54 | 0.66 ± 0.08 | 0.69 ± 0.09 |
| D | 0.39 | 0.43 | 0.62 ± 0.11 | 0.66 ± 0.06 |
| DG | 0.42 | 0.49 | 0.65 ± 0.08 | 0.65 ± 0.09 |
| M | 0.38 | 0.50 | 0.65 ± 0.09 | 0.69 ± 0.09 |
| MG | 0.38 | 0.46 | 0.58 ± 0.11 | 0.65 ± 0.08 |
| MD | 0.37 | 0.51 | 0.62 ± 0.08 | 0.66 ± 0.10 |
| MDG | 0.39 | 0.51 | 0.62 ± 0.10 | 0.58 ± 0.08 |
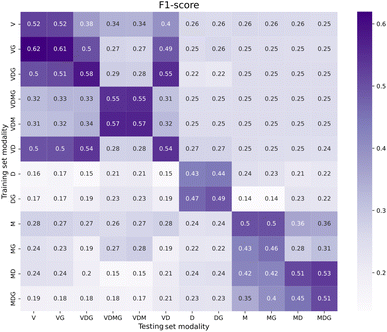
A more granular view of the test results with multimodal visual encoding is shown in Fig. 3, where we report the F1 scores for all activities in the Lab Actions dataset. The impact of modality combination on the F1 score is different depending on the activity. We observed a tendency of the Vi-Cl model to predict No action steps, resulting in higher performances for the No action label. On the contrary, Vi-LM provides more similar performances across different activities, with the AnalyticalMeasurement activity being the most recognized for all sets that include V in the data modality combination. We tentatively attributed this effect to the distinct color and location of the analytical instrument (pH meter) used in videos representing this action. Instead, the MeasureLiquid activity was the most recognized for all the other modality combinations that do not comprise the V modality. We suggest that this is due to the distinguishing shape of the objects used during this activity (e.g., the graduated cylinder and the bottle including water at the foreground of the scenes). We tested the usefulness of the data modalities in Fig. 4, where we compare the performance of the Vi-LM model trained on each data modality combination in terms of F1 score for inference on the test set of each modality combination. Interestingly, Vi-LM trained on video with gaze (VG) also performed better for inference on the test set comprising only the video modality, suggesting that gaze helps better distinguish the activities in the model latent space, in line with using gaze in multitask training.31
 | ||
| Fig. 4 F1 score of Vi-LM models for predicting activities on Lab Actions dataset videos with different data modalities. The diagonal shows the performance of the model trained and tested on the same data modalities and corresponds to Table 4. | ||
The evaluation of the similarity between workflows compiled from the predicted activity label sequences and the respective ground truths for the multimodal visual encoding datasets is reported in Table 4. Consistent with the F1 score results, the highest fidelity workflow transcription was achieved with Vi-LM on the video with gaze (VG) modalities. Despite observing variability in the results (uncertainty ranges of 95% confidence intervals), we found statistically significant differences in performances for the Vi-Cl model with VG modalities against the Vi-Cl model with only the V modality (Mann–Whitney U test, p = 0.027), as well as for the Vi-LM with VG modality against Vi-LM with only the V modality (Mann–Whitney U test, p = 0.047).
Finally, we compare the accuracy of laboratory action recognition with the performance of state-of-the-art action classification on common video datasets. In general, the scale of domain-specific data in the Lab Actions and Lab Motion datasets is inferior compared to general activity video datasets such as Kinetics (>400 clips per class),43 Something–Something (average of 620 clips per class)55 or ActivityNet (average of 137 videos per class).56 In the present study, the diversity of data was limited to an average of 82 (Lab Motion) and 86 (Lab Actions) distinct videos per class from which all training clips were extracted. The base variant of the VideoMAE model, which shares the same backbone for vision encoding as the Vi-Cl and Vi-LM models reported here, achieves 81.5% top-1 accuracy on Kinetics-400 and 87.4% top-1 accuracy when using the ViT-Huge backbone.40 Thus, we expect that the best F1 score for action classification of 0.61 reported in the present work for Vi-LM on the VG multimodal dataset can be further improved by increasing the quantity and diversity of training data as well as increasing the number of model parameters. Both of these approaches, however, come at the expense of increased energy consumption during training, and, in the case of larger model size, also incur a penalty in terms of latency during inference. Contemplating the application of these systems to support data capture by researchers during laboratory experimentation, we propose that the trade-off between accuracy and efficiency should be evaluated as a whole given the desired levels of automation within specific constraints of the target environment.
5 Conclusions
We demonstrate how multimodal vision-language models can be adapted for near real-time action recognition during scientific experimentation in laboratory environments. Models based on pre-trained vision encoders coupled with multi-class classifiers and trained on domain-specific examples tend to perform best in terms of F1 score, tracking of scientific workflows and inference time on RGB videos. Moreover, our findings emphasize the usefulness of domain-specific training data to differentiate similar actions and capture time-resolved details of experimental procedures. Our work also proves that incorporating language models as decoders appears to offer benefits in terms of exploiting additional modalities in the input data. Notably, the best performance for individual action recognition as well as end-to-end workflow transcription was obtained by a vision-language model configuration combining video and gaze coordinates as input modalities. We propose that fine-tuning the aforementioned model configurations on relatively small, domain-specific datasets can produce useful results for action recognition in scientific experimentation to aid documentation, reproducibility and collaboration in laboratory research.Data availability
Egocentric videos from the Lab Actions and Lab Motion datasets after filtering out Personal Information and Confidential Information are available together with the corresponding annotations under https://doi.org/10.5281/zenodo.14235875. The Python code used for model benchmarking is published at https://github.com/IBM/lab-activity-recognition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Teodoro Laino for support and guidance, Carlo Baldassari, Marvin Alberts, Michael Stiefel and Oliver Schilter for their important contributions in generating the datasets, and Anel Zulji and Stefan Gamper for mechanical prototyping.References
- M. Baker, Nature, 2016, 533, 452–454 CrossRef CAS.
- C. G. Begley and L. M. Ellis, Nature, 2012, 483, 531–533 CrossRef CAS.
- R. S. Gonçalves and M. A. Musen, Sci. Data, 2019, 6, 190021 CrossRef PubMed.
- T. Miyakawa, Mol. Brain, 2020, 13, 24 CrossRef.
- R. Bommasani, D. A. Hudson, E. Adeli, R. Altman, S. Arora, S. von Arx, M. S. Bernstein, J. Bohg, A. Bosselut, E. Brunskill, E. Brynjolfsson, S. Buch, D. Card, R. Castellon, N. Chatterji, A. Chen, K. Creel, J. Q. Davis, D. Demszky, C. Donahue, M. Doumbouya, E. Durmus, S. Ermon, J. Etchemendy, K. Ethayarajh, L. Fei-Fei, C. Finn, T. Gale, L. Gillespie, K. Goel, N. Goodman, S. Grossman, N. Guha, T. Hashimoto, P. Henderson, J. Hewitt, D. E. Ho, J. Hong, K. Hsu, J. Huang, T. Icard, S. Jain, D. Jurafsky, P. Kalluri, S. Karamcheti, G. Keeling, F. Khani, O. Khattab, P. W. Koh, M. Krass, R. Krishna, R. Kuditipudi, A. Kumar, F. Ladhak, M. Lee, T. Lee, J. Leskovec, I. Levent, X. L. Li, X. Li, T. Ma, A. Malik, C. D. Manning, S. Mirchandani, E. Mitchell, Z. Munyikwa, S. Nair, A. Narayan, D. Narayanan, B. Newman, A. Nie, J. C. Niebles, H. Nilforoshan, J. Nyarko, G. Ogut, L. Orr, I. Papadimitriou, J. S. Park, C. Piech, E. Portelance, C. Potts, A. Raghunathan, R. Reich, H. Ren, F. Rong, Y. Roohani, C. Ruiz, J. Ryan, C. Ré, D. Sadigh, S. Sagawa, K. Santhanam, A. Shih, K. Srinivasan, A. Tamkin, R. Taori, A. W. Thomas, F. Tramèr, R. E. Wang, W. Wang, B. Wu, J. Wu, Y. Wu, S. M. Xie, M. Yasunaga, J. You, M. Zaharia, M. Zhang, T. Zhang, X. Zhang, Y. Zhang, L. Zheng, K. Zhou and P. Liang, arXiv, 2022, preprint, arXiv:2108.07258, DOI:10.48550/arXiv.2108.07258.
- S. Song, V. Chandrasekhar, N.-M. Cheung, S. Narayan, L. Li and J.-H. Lim, Computer Vision – ACCV 2014 Workshops, Cham, 2015, pp. 445–458 Search PubMed.
- S. Horawalavithana, E. Ayton, S. Sharma, S. Howland, M. Subramanian, S. Vasquez, R. Cosbey, M. Glenski and S. Volkova, Proceedings of BigScience Episode #5 – Workshop on Challenges & Perspectives in Creating Large Language Models, Association for Computational Linguistics, Dublin, 2022, pp. 160–172 Search PubMed.
- AI4Science, Microsoft Research and Quantum, Microsoft Azure, arXiv, 2023, preprint, arXiv:2311.07361, DOI:10.48550/arXiv.2311.07361.
- J. Yang, H. Jin, R. Tang, X. Han, Q. Feng, H. Jiang, B. Yin and X. Hu, arXiv, 2023, preprint, arXiv:2304.13712, DOI:10.48550/arXiv.2304.13712.
- G. R. Smith, C. Bello, L. Bialic-Murphy, E. Clark, C. S. Delavaux, C. F. d. Lauriere, J. v. d. Hoogen, T. Lauber, H. Ma, D. S. Maynard, M. Mirman, L. Mo, D. Rebindaine, J. E. Reek, L. K. Werden, Z. Wu, G. Yang, Q. Zhao, C. M. Zohner and T. W. Crowther, PLoS Comput. Biol., 2024, 20, e1011767 CrossRef CAS PubMed.
- H. Wang, T. Fu, Y. Du, W. Gao, K. Huang, Z. Liu, P. Chandak, S. Liu, P. Van Katwyk, A. Deac, A. Anandkumar, K. Bergen, C. P. Gomes, S. Ho, P. Kohli, J. Lasenby, J. Leskovec, T.-Y. Liu, A. Manrai, D. Marks, B. Ramsundar, L. Song, J. Sun, J. Tang, P. Veličković, M. Welling, L. Zhang, C. W. Coley, Y. Bengio and M. Zitnik, Nature, 2023, 620, 47–60 CrossRef CAS.
- Y. Liu, Z. Yang, Z. Yu, Z. Liu, D. Liu, H. Lin, M. Li, S. Ma, M. Avdeev and S. Shi, J. Materiomics, 2023, 9, 798–816 CrossRef.
- M. Manica, J. Born, J. Cadow, D. Christofidellis, A. Dave, D. Clarke, Y. G. N. Teukam, G. Giannone, S. C. Hoffman, M. Buchan, V. Chenthamarakshan, T. Donovan, H. H. Hsu, F. Zipoli, O. Schilter, A. Kishimoto, L. Hamada, I. Padhi, K. Wehden, L. McHugh, A. Khrabrov, P. Das, S. Takeda and J. R. Smith, npj Comput. Mater., 2023, 9, 1–6 CrossRef.
- S. D. Yang, Z. A. Ali and B. M. Wong, Ind. Eng. Chem. Res., 2023, 62, 15278–15289 CrossRef CAS.
- B. Ni, H. Peng, M. Chen, S. Zhang, G. Meng, J. Fu, S. Xiang and H. Ling, arXiv, 2022, preprint, arXiv:2208.02816, DOI:10.48550/arXiv.2208.02816.
- B. Lin, Y. Ye, B. Zhu, J. Cui, M. Ning, P. Jin and L. Yuan, arXiv, 2023, preprint, arXiv:2311.10122, DOI:10.48550/arXiv.2311.07361.
- K. M. Jablonka, L. Patiny and B. Smit, Nat. Chem., 2022, 14, 365–376 CrossRef CAS.
- S. Kanza, C. Willoughby, N. Gibbins, R. Whitby, J. G. Frey, J. Erjavec, K. Zupančič, M. Hren and K. Kovač, J. Cheminf., 2017, 9, 31 Search PubMed.
- S. G. Higgins, A. A. Nogiwa-Valdez and M. M. Stevens, Nat. Protoc., 2022, 17, 179–189 CrossRef CAS.
- R. Sasaki, M. Fujinami and H. Nakai, Digital Discovery, 2024, 3, 2458–2464 RSC.
- S. Song, N.-M. Cheung, V. Chandrasekhar, B. Mandal and J. Lin, arXiv, 2016, preprint, arXiv:1601.06603, DOI:10.48550/arXiv.1601.06603.
- R. Possas, S. P. Caceres and F. Ramos, 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2018, pp. 5967–5976 Search PubMed.
- K. P. Sanal Kumar and R. Bhavani, Multimed. Tool. Appl., 2020, 79, 3543–3559 CrossRef.
- M. Bock, H. Kuehne, K. Van Laerhoven and M. Moeller, arXiv, 2023, preprint, arXiv:2304.05088, DOI:10.48550/arXiv.2304.05088.
- C. Gurrin, K. Schoeffmann, H. Joho, B. Munzer, R. Albatal, F. Hopfgartner, L. Zhou and D.-T. Dang-Nguyen, Proceedings of the International Conference on MultiMedia Modeling, 2019, pp. 312–324 Search PubMed.
- K. Grauman, A. Westbury, E. Byrne, Z. Chavis, A. Furnari, R. Girdhar, J. Hamburger, H. Jiang, M. Liu, X. Liu, et al., Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2022, pp. 18995–19012 Search PubMed.
- Z. Sun, Q. Ke, H. Rahmani, M. Bennamoun, G. Wang and J. Liu, arXiv, 2020, preprint, arXiv:2012.11866, DOI:10.48550/arXiv.2012.11866.
- Z. Gao, Y. Wang, J. Chen, J. Xing, S. Patel, X. Liu and Y. Shi, Proc. ACM Interact. Mob. Wearable Ubiquitous Technol., 2023, 7, 1–26 Search PubMed.
- M. H. Lye, N. AlDahoul and H. Abdul Karim, Sensors, 2023, 23, 6804 CrossRef.
- Z. Zuo, L. Yang, Y. Peng, F. Chao and Y. Qu, IEEE Access, 2018, 6, 12894–12904 Search PubMed.
- Y. Huang, M. Cai, Z. Li, F. Lu and Y. Sato, IEEE Trans. Image Process., 2020, 29, 7795–7806 Search PubMed.
- Z. Zhang, D. Crandall, M. Proulx, S. Talathi and A. Sharma, Proceedings of the Symposium on Eye Tracking Research and Applications, 2022, 1–7 Search PubMed.
- K. Simonyan and A. Zisserman, arXiv, 2014, preprint, arXiv:1406.2199, DOI:10.48550/arXiv.1406.2199.
- P. Natarajan, S. Wu, S. Vitaladevuni, X. Zhuang, S. Tsakalidis, U. Park, R. Prasad and P. Natarajan, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2012, pp. 1298–1305 Search PubMed.
- H. Ben-Younes, R. Cadene, M. Cord and N. Thome, Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2017, pp. 2612–2620 Search PubMed.
- G. Ye, D. Liu, I.-H. Jhuo and S.-F. Chang, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2012, pp. 3021–3028 Search PubMed.
- H. Akbari, L. Yuan, R. Qian, W.-H. Chuang, S.-F. Chang, Y. Cui and B. Gong, arXiv, 2021, preprint, arXiv:2104.11178, DOI:10.48550/arXiv.2104.11178.
- L. Hanu, A. L. Verő and J. Thewlis, arXiv, 2023, preprint, arXiv:2309.10783, DOI:10.48550/arXiv.2309.10783.
- H. Xu, G. Ghosh, P.-Y. Huang, D. Okhonko, A. Aghajanyan, F. Metze, L. Zettlemoyer and C. Feichtenhofer, arXiv, 2021, preprint, arXiv:2109.14084, DOI:10.48550/arXiv.2109.14084.
- Z. Tong, Y. Song, J. Wang and L. Wang, arXiv, 2022, preprint, arXiv:2203.12602, DOI:10.48550/arXiv.2203.12602.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, J. Mach. Learn. Res., 2020, 21, 1–67 Search PubMed.
- A. Radford, J. W. Kim, C. Hallacy, A. Ramesh, G. Goh, S. Agarwal, G. Sastry, A. Askell, P. Mishkin, J. Clark, G. Krueger and I. Sutskever, arXiv, 2021, preprint, arXiv:2103.00020, DOI:10.48550/arXiv.2103.00020.
- W. Kay, J. Carreira, K. Simonyan, B. Zhang, C. Hillier, S. Vijayanarasimhan, F. Viola, T. Green, T. Back, P. Natsev, M. Suleyman and A. Zisserman, arXiv, 2017, preprint, arXiv:1705.06950, DOI:10.48550/arXiv.1705.06950.
- T. Chen and C. Guestrin, Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, pp. 785–794 Search PubMed.
- R. Luo, Z. Zhao, M. Yang, J. Dong, M. Qiu, P. Lu, T. Wang and Z. Wei, arXiv, 2023, preprint, arXiv:2306.07207, DOI:10.48550/arXiv.2306.07207.
- B. Zhu, B. Lin, M. Ning, Y. Yan, J. Cui, H. Wang, Y. Pang, W. Jiang, J. Zhang, Z. Liet al., arXiv, 2023, preprint, arXiv:2310.01852, DOI:10.48550/arXiv.2310.01852.
- A. C. Vaucher, F. Zipoli, J. Geluykens, V. H. Nair, P. Schwaller and T. Laino, Nat. Commun., 2020, 11, 3601 CrossRef PubMed.
- A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai, T. Unterthiner, M. Dehghani, M. Minderer, G. Heigold, S. Gelly, J. Uszkoreit and N. Houlsby, arXiv, 2020, preprint, arXiv:2010.11929, DOI:10.48550/arXiv.2010.11929.
- I. Loshchilov and F. Hutter, arXiv, 2017, preprint, arXiv:1711.05101, DOI:10.48550/arXiv.1711.05101.
- S. Xiao, Z. Liu, P. Zhang and N. Muennighoff, arXiv, 2023, preprint, arXiv:2309.07597, DOI:10.48550/arXiv.2309.07597.
- N. Reimers and I. Gurevych, arXiv, 2019, preprint, arXiv:1908.10084, DOI:10.48550/arXiv.1908.10084.
- L. Yujian and L. Bo, IEEE Trans. Pattern Anal. Mach. Intell., 2007, 29, 1091–1095 Search PubMed.
- M. Oquab, T. Darcet, T. Moutakanni, H. Vo, M. Szafraniec, V. Khalidov, P. Fernandez, D. Haziza, F. Massa, A. El-Nouby, et al., arXiv, 2023, preprint, arXiv:2304.07193, DOI:10.48550/arXiv.2304.07193.
- A. Kirillov, E. Mintun, N. Ravi, H. Mao, C. Rolland, L. Gustafson, T. Xiao, S. Whitehead, A. C. Berg, W.-Y. Lo, et al., arXiv, 2023, preprint, arXiv:2304.02643, DOI:10.48550/arXiv.2304.02643.
- R. Goyal, S. E. Kahou, V. Michalski, J. Materzyńska, S. Westphal, H. Kim, V. Haenel, I. Fruend, P. Yianilos, M. Mueller-Freitag, F. Hoppe, C. Thurau, I. Bax and R. Memisevic, arXiv, 2017, preprint, arXiv:1706.04261, DOI:10.48550/arXiv.1706.04261.
- F. C. Heilbron, V. Escorcia, B. Ghanem and J. C. Niebles, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015, pp. 961–970 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00287c |
| ‡ http://www.pupil-labs.com, last accessed 2024-09-04. |
| This journal is © The Royal Society of Chemistry 2025 |