 Open Access Article
Open Access ArticleAn rGO-doped laser induced graphene electrochemical biosensor for highly sensitive exosome detection
Xiaoshuang
Chen
a,
Xiaohui
Yan
a,
Jiaoyan
Qiu
a,
Xue
Zhang
a,
Yunhong
Zhang
a,
Hongpeng
Zhou
a,
Yujuan
Zhao
a,
Lin
Han
 ab and
Yu
Zhang
ab and
Yu
Zhang
 *ab
*ab
aInstitute of Marine Science and Technology, Shandong University, Qingdao (266000), China. E-mail: yuzhang@sdu.edu.cn
bSchool of Integrated Circuits, Shandong University, Jinan (250100), China
First published on 2nd September 2024
Abstract
In this study, we developed a novel electrochemical sensing chip integrated with reduced graphene oxide (rGO) with laser-induced graphene (LIG) for the detection of exosomes associated with breast cancer biomarkers. Employing laser-induced technology, a three-dimensional porous graphene material is fabricated on the surface of a flexible polyimide film, which is subsequently combined with rGO through π–π stacking. This integration facilitates the doping of two-dimensional and three-dimensional material (2D/3D) structures, significantly enhancing the conductivity of the electrode material. Additionally, this approach markedly improves the surface hydrophobicity and biomolecule affinity of LIG, optimizing the immobilization of specific antibodies for exosomes. Importantly, this experiment marks the first occasion of merging two-dimensional rGO with three-dimensional LIG, resulting in the construction of a high-performance biosensing chip that enables specific capture and highly sensitive detection of exosomes. Under optimized conditions, the quantitative detection range for exosomes is established at 5 × 102 to 5 × 105 particles per μL, with a limit of detection (LOD) of 166 particles per μL. The biosensor is successfully used to analyze exosomes in breast cancer cell lines and patient serum samples, proving its practical application. This electrochemical biosensing chip offers significant practical application value in the early screening and diagnosis of diseases.
Introduction
Breast cancer stands as the most prevalent malignant tumor among women, representing a grave threat to their health and well-being. According to the latest forecasts on the global cancer burden released by the International Agency for Research on Cancer (IARC) in 2020,1 breast cancer has emerged as the leading type of malignant tumor worldwide, constituting 11.7% of all new cases of cancer. The precise mechanisms underlying the pathogenesis of breast cancer remain elusive, with the associated high-risk factors being challenging to manage. Consequently, developing methods for the early detection of breast cancer holds critical importance. Exosomes are nanometer-sized extracellular vesicles containing a large number of biomolecules, including lipids, carbohydrates, proteins, DNA fragments, and microRNA,2–5 as exosomes in various body fluids display specific molecular profiling, and therefore tumor-derived exosomes can be considered as promising biomarkers in cancer.6,7 As one of the membrane proteins on exosomes, CD63 is known as the specific marker protein of exosomes,8 and it mediates the malignancy of breast cancer through glycosylation regulation.9 Therefore, detection based on CD63 membrane protein has become an important marker for breast cancer identification.Previous research reports on exosome detection have usually focused on optimizing several traditional detection methods, such as western blotting,10 nanoparticle tracking and analysis (NTA),11 flow cytometry,12 and enzyme-linked immunosorbent assay (ELISA).13 Nonetheless, the low abundance of tumor-derived exosomes in biological fluids and the masking of their signals by heterogeneous exosomes pose significant challenges.14 Additionally, traditional detection methods suffer from drawbacks such as the need for large sample volumes, low sensitivity, and the requirement for costly instruments, which hinder the flexible detection of real samples. In recent years, innovative approaches for detecting breast cancer exosomes have been developed, including fluorescence immunoassay,15 surface enhanced Raman spectroscopy (SERS),16 and photoelectrochemical (PEC)17 and electrochemical assays.18–20 Compared with spectroscopic methods,21 electrochemical detection has lower background interference, higher detection sensitivity and lower cost. Compared with field effect transistor (FET) sensors,22 electrochemical chips are simple to fabricate, and the equipment can be miniaturized for high portability. Therefore, they are prominently used in biomolecule detection.23,24 Among electrochemical detection methods, electrochemical impedance spectroscopy (EIS) is a detection method that reflects biochemical processes and biorecognition events by measuring the capacitance of the electrode surface and the charge transfer resistance of the interface, which is non-destructive and has high sensitivity.25 [Fe(CN)6]3−/4− has been used as a redox pair for the construction of EIS biosensors in many articles in the last two years due to its redox reversibility, stable nature, high electrode compatibility, and easy preparation.26–29 This method holds considerable promise in identifying breast cancer exosomes, where the choice of materials plays a pivotal role. Utilizing materials with superior electrochemical properties can significantly enhance various facets of sensing performance.
Laser-induced graphene (LIG),30 produced via the laser-induced carbonization of polyimide (PI) films, is a foam-like carbon-based material. It is regarded as an ideal electrode material due to its straightforward processing method (no mask/reagents required), large active surface area, high cost-effectiveness, excellent biocompatibility, superior conductivity, and electrocatalytic activity. Consequently, LIG has been extensively applied in various fields, including supercapacitors,31–33 triboelectric nanogenerators,34,35 and a wide range of physical sensors.36–38 Nonetheless, compared with early metal/covalent organic frameworks and noble metal composites, LIG suffers from problems such as low conductivity and insufficient electroactive area.39 To address these issues, the addition or doping of foreign functional materials into LIG to purposefully construct suitable composite materials has emerged as an ideal solution.
Currently, there are reports of doping LIG with noble metal nanoparticles40 and hydrophilic polymers.41 Such modifications significantly enhance the electrochemical performance of LIG, albeit necessitating intricate and laborious preparation steps, with the modification methods often encountering stability issues. The integration of 2D/3D materials into LIG for electrochemical sensing applications has been shown to substantially improve sensor stability. Presently, MoS2 and metal–organic frameworks (MOFs) are been incorporated into LIG to fabricate temperature and gas sensors,42,43 yet reports on sensors doping 2D materials with LIG for biological immunodetection remain scarce. Notable 2D materials44 include graphene oxide (GO), reduced graphene oxide (rGO), and MXenes. Among these, rGO is distinguished by its superior electron transfer capabilities, attributed to the restoration of graphene's partially sp2-hybridized carbon network. It also demonstrates commendable chemical and electrochemical stability within a certain electrochemical window, thereby enhancing the electrochemical robustness of LIG.45 Furthermore, the reduction process of rGO increases the material's porosity, thus expanding its specific surface area and enhancing the rate of biomolecule immobilization. Remarkably, rGO/LIG can be prepared in one step by π–π stacking, which avoids the complicated preparation process and facilitates the improvement of the stability and sensitivity of detection.
In this study, based on the one-step doping of rGO and LIG, a stable 2D/3D structure was formed, and a biological immune sensing chip for highly sensitive detection of exosomes was constructed, achieving highly sensitive detection of exosomes. The incorporation of rGO not only enhances the hydrophobicity of the LIG surface but also creates optimal conditions for the immobilization of biomolecules, thereby improving the conductivity and stability of the sensing chip. Utilizing [Fe(CN)6]3−/4− as a redox couple, the chip achieves highly sensitive detection of exosomes based on alterations in the mass transfer resistance of [Fe(CN)6]3−/4− on the electrode surface, consequent to the binding of antibodies and exosomes. Furthermore, this sensing chip exhibits excellent stability, accuracy, and precision, making it suitable for the analysis of real samples. It demonstrates significant potential in differentiating between healthy individuals and breast cancer patients, highlighting its applicability in clinical diagnostics.
Materials and methods
Materials
PI films (200 μm thickness) were purchased from Golden Dragon Insulation Material Co., Ltd. (Changzhou, China). Polydimethylsiloxane (PDMS) and its curing agent (Sylgard silicone elastomer kit) were obtained from Dow Corning Corporation (Midland, MI, USA). Fetal bovine serum (FBS), penicillin/streptomycin (P/S), and trypsin were sourced from Thermo Fisher Scientific Inc. (MA, USA). High-glucose Dulbecco's modified Eagle medium (DMEM) and RPMI 1640 medium were supplied by Stovovan Biopharmaceutical Co. (Logan, USA). The exosome capture antibody (Ab) was procured from Sigma-Aldrich, Inc. (St. Louis, MO, USA). Graphite powder, 30% hydrogen peroxide, polyethyleneimine (PEI), and bovine serum albumin were purchased from Macklin Biochemical Co., Ltd. (Shanghai, China). Potassium permanganate, sodium nitrate, and glutaraldehyde were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Breast cancer serum samples were provided by Qilu Hospital of Shandong University (Jinan, China), while healthy serum samples were provided by members of our research group. All other chemicals used were of analytical grade or higher and were used without further purification. Ultrapure water (>18 MΩ cm, Milli-Q) was obtained from a Millipore water purification system (Billerica, MA, USA) and used throughout the experiments.Apparatus
Exosomes were extracted using a CS150GXL ultracentrifuge (Hitachi, Japan). Characterization of exosomes via NTA was conducted using a NanoSight NS300 particle tracking analyzer (Malvern, UK). MCF-7 cells were cultured in a CO2 incubator (Panasonic, Japan). A CO2 laser engraver from Kaitai (China) was utilized for laser engraving and cutting of the PI films, as well as PDMS to construct micro–nano chips. The morphology of the nanomaterials was captured using an FEI Quanta 250 FEG field emission scanning electron microscope (FEI, USA). Transmission electron microscopy (TEM) images were obtained using a Tecnai G2 F20 transmission electron microscope (FEI, USA). Raman absorption curves and Raman homogeneity mapping images of the micro–nano chip materials were acquired using a Renishaw micro laser Raman spectrometer (Renishaw, UK). The resistance of the engraved laser-induced graphene (LIG) was tested using a Keithley 2400 source meter unit (Tektronix, USA). Cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) were carried out using a CHI 660E electrochemical workstation (CH Instruments, Shanghai, China). The experiments utilized a conventional three-electrode system, comprising a laser-induced graphene electrode (diameter of 3 mm) as the working electrode, an Ag/AgCl electrode (3 M KCl) as the reference electrode, and a platinum wire as the counter electrode.Cell culture and exosome extraction
MCF-7 cells were rapidly thawed in a 37 °C water bath, followed by centrifugation at 300g for 3 min to remove excess cryopreservation fluid. The cells were then cultured in DMEM medium supplemented with 10% FBS and 1% P/S antibiotics in a 5% CO2 incubator. When the cell density reached approximately 80%, the culture was continued for 48 h in FBS-free medium, after which the supernatant was collected. Next, differential centrifugation was employed to separate and extract exosomes. Initially, the samples were centrifuged at 2000g for 10 min to eliminate necrotic cells, followed by further centrifugation at 1000g for 30 min to clear cell debris, yielding the supernatant. Subsequently, the supernatant was subjected to centrifugation at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C for 70 min to acquire the exosome precipitate. The precipitate was then resuspended in DPBS buffer and centrifuged once more at 4 °C and 100
000g and 4 °C for 70 min to acquire the exosome precipitate. The precipitate was then resuspended in DPBS buffer and centrifuged once more at 4 °C and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min to purify the exosomes. Finally, the purified exosomes were dispersed in 200 μL DPBS buffer and stored at −80 °C for future use.
000g for 70 min to purify the exosomes. Finally, the purified exosomes were dispersed in 200 μL DPBS buffer and stored at −80 °C for future use.
Preparation of rGO
The preparation of rGO was based on the reduction of GO with slight modifications as reported in the literature.46 Initially, ice was added to a water bath, and a stir bar was placed in a 250 mL beaker, which was then placed within the water bath. Next, 23 mL of concentrated sulfuric acid was added to the beaker followed by 1 g of graphite powder. The mixture was stirred using a magnetic stirrer for about 10 min until homogeneous. Subsequently, 0.5 g of NaNO3 was added and stirred for another 10 min until evenly mixed. Thereafter, 3 g of KMnO4 was added and the mixture was stirred continuously for 2 h, followed by incubation at 35 °C in a water bath for 30 min. Next, 46 mL of deionized water was added and the temperature was raised to 80 °C, stirring for 15 min. An additional 140 mL of deionized water was added, and stirring was continued at 80 °C for 20 min. The beaker was then removed from the heat and concentrated hydrogen peroxide (30% by volume) was added until no more gas bubbles emerged, and the mixture was hot-filtered. Finally, the residue was rinsed with 20% HCl to remove excess potassium permanganate and centrifugally washed until the supernatant was neutral. The GO precipitate was obtained after freeze-drying.For the reduction process, sodium hydroxide was added to a 10 mL GO solution (0.5 mg mL−1) to adjust the pH from approximately 4 to 10. The mixture was then incubated in a reactor at 100 °C for 6 h, followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min, and washed with clean water. The resulting rGO nanosheets were dispersed in water, sonicated, and stored at 4 °C for further use.
000g for 10 min, and washed with clean water. The resulting rGO nanosheets were dispersed in water, sonicated, and stored at 4 °C for further use.
Amination of rGO
To aminate rGO, 400 μL of PEI was added to 5 mL of 0.1 mg mL−1 rGO and stirred at 100g for 1 h at room temperature, followed by centrifugation at 7080g for 10 min. The precipitate was washed and subsequently re-dispersed in 5 mL of ultrapure water to obtain the PEI–rGO dispersion.Construction of a bio-immunosensor chip
Initially, a complete electrode pattern was engraved using a laser engraver with optimized laser power and scanning speed settings to ensure the success of subsequent experiments. Subsequently, laser cutting technology was employed to perforate a PDMS thin layer, creating sensing microcavities with a diameter of 8 mm. The perforated PDMS layer was then precisely bonded to the engraved electrodes. Next, 5 μL of PEI–rGO dispersion was dropped onto the surface of the laser-induced graphene electrode and allowed to dry naturally at room temperature. After cleaning the electrode, 100 μL of 2.5% glutaraldehyde was added to the PEI–rGO–LIG surface and incubated for 1 h to attach antibodies. Following incubation and subsequent cleaning, 5 μL of 20 μg mL−1 Ab was applied to the electrode surface and incubated for 2 h. The working electrode was then washed with PBS buffer to remove unbound antibodies, resulting in the formation of Ab–rGO–LIG. Finally, 100 μL of 3% BSA was incubated for 1 h to occupy the unmodified sites on the electrode surface, preventing non-specific adsorption. Thus, the electrochemical immunosensing chip was successfully constructed.Exosome detection
Firstly, 10 μL of different concentrations of exosome solutions were added to the assembled BSA–Ab–rGO–LIG electrodes and incubated at 37 °C for 1 h to obtain exosomes–BSA–Ab–rGO–LIG complexes. Subsequently, the electrode was washed with PBS buffer to remove non-specifically bound exosomes from the surface. Then, 100 μL of 0.1 M KCl solution containing 5.0 mM [Fe(CN)6]3−/4− (pH 7.4) was added to the sensing microcavity, with an Ag/AgCl electrode as the reference electrode and a platinum wire electrode as the counter electrode. Alternating current impedance techniques were employed for detection. The initial potential was set at 0.21 V, with a frequency range of 100 kHz to 0.1 Hz and an amplitude of 5.0 mV. The theoretical values of impedance were fitted using the Randles equivalent circuit, and the charge-transfer resistance (Rct) was used as the analysis detection signal for quantitative exosome detection.Detection of real samples
The serum samples from breast cancer patients provided by Qilu Hospital of Shandong University and healthy members of our laboratory team were examined. Firstly, the obtained whole blood samples were left to stand at 4 °C for 3 h, after which the light yellow serum from the top layer was collected. The acquired serum samples were then centrifuged at 3000g for 15 min to purify the serum, taking the supernatant. The purified serum was stored at −80 °C for future use. The electrochemical detection process was consistent with the extracellular vesicle extraction process used in the cell lines.Results and discussion
Construction principle of the immune biosensor chip
We integrated biosensors and microelectronics to construct a biosensing chip for exosome detection. The biosensor includes a biosensing area and an electrical connection area. The detection principle of the immunobiosensing chip is depicted in Scheme 1. Firstly, PEI–rGO was introduced into the surface of LIG. The introduction of PEI–rGO can enhance the conductivity of the electrode, and the amino group on its surface provides more sites for antibody sequestration. The observed enhancement in electrical conductivity is likely attributable to the doping interaction between LIG, a three-dimensional graphene structure, and the two-dimensional graphene rGO, forming a hybrid 2D/3D structure. This configuration not only improves the hydrophobicity of the LIG surface but also elevates its electrical conductivity, thereby enhancing the overall electrical performance of LIG and consequently increasing the sensitivity of detection. Furthermore, following the amination of rGO with PEI, antibodies are chemically bonded using glutaraldehyde as a coupling agent, which bolsters the efficiency and stability of antibody immobilization. Once immobilized, the antibodies specifically target and capture exosomes, leading to the formation of immune complexes on the electrode surface. These complexes induce changes in impedance, facilitating the highly sensitive detection of exosomes.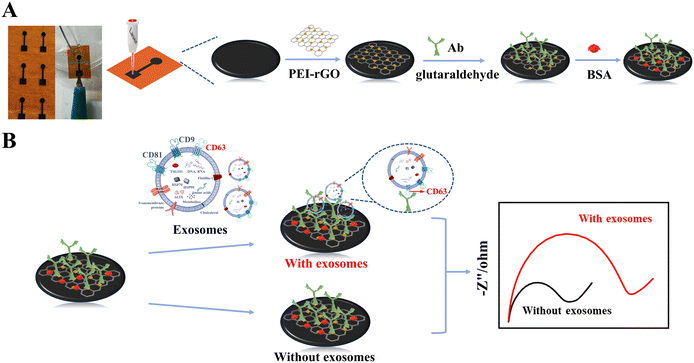 | ||
| Scheme 1 Principles of bioimmunosensing microarrays, including (A) modification of biosensing interfaces and (B) exosomes detection. | ||
Characterization of exosomes
NTA and TEM were employed to quantify and characterize the exosomes isolated from the supernatant of the human breast cancer cell line MCF-7. The results, presented in Fig. 1A, indicate a concentration of approximately 2.24 × 107 particles per μL with an average diameter of 104.3 nm, demonstrating the successful extraction of a high concentration of exosomes via ultracentrifugation. Fig. 1B displays a TEM image of the exosomes derived from MCF-7 cells, illustrating the exosomes' characteristic cup-shaped, vesicular structure with distinct boundaries. This morphology is typical of exosomes and further validates the integrity and successful isolation of the vesicles. | ||
| Fig. 1 (A) NTA results of exosomes; (B) TEM image of exosomes; (C) SEM image of rGO; (D) SEM image of LIG; (E) SEM image of rGO–LIG; (F) SEM image of exosomes–Ab–rGO–LIG. | ||
Material characterization
The assembly process of the immunobiosensor chip was elucidated using SEM. Fig. 1C highlights the characteristic two-dimensional lamellar structure of rGO, with visibly wrinkled edges. In Fig. 1D, LIG is depicted as a three-dimensional porous framework, replete with numerous micropores. Subsequent doping with rGO, as illustrated in Fig. 1E, notably intensifies the surface wrinkling of LIG. Fig. 1F shows that exosomes were captured on the surface of the modified electrode after solid-loading Ab, proving the successful construction of our immunobiosensing system.rGO–LIG, as the core material of this sensing system, was characterized for structural changes and electrical properties during the doping process using Raman spectroscopy and cyclic voltammetry. As shown in Fig. 2A, LIG (curve a) corresponds to the vicinity of 1335 cm−1, 1590 cm−1 and 2695 cm−1, and matches the characteristic bands D, G and 2D of graphene, thus qualitatively proving the existence of graphene; the existence of typical 2D spectral bands can prove that LIG is a few-layer graphene structure. For the Raman spectrum of the synthesized rGO (curve b), the characteristic spectral bands D and G can be observed, where the D peak represents the degree of disorder of graphene or the degree of its edge folds, and the G peak is attributed to the resonance of the E2g phonon, and the ratio of the intensities of the two peaks is usually taken to indicate the degree of reduction of graphene. After doping rGO with LIG, it can be observed that the ratio of the D band to the G band becomes larger, and ID/IG changes from 1.132 for LIG to 1.436, which indicates that the disorder is enhanced after doping, and there exist a large number of structural defects and edges, and these defects have a large surface energy not only to improve the electrochemical reaction activity but also to act as an effective medium for the transfer of electrons, which can endow the sensing chips with good electrical conductivity and electrocatalytic activity. The uniformity of the material plays a major role in the construction of the sensing chip. The uniformity is analyzed by randomly testing the Raman spectrum of the rGO–LIG composite material at 100 points. As shown in Fig. 2B, the relative standard deviation of its absorption peak (RSD) is <5%, indicating good material uniformity.
 | ||
| Fig. 2 (A) Raman spectra of LIG (a), rGO (b) and rGO–LIG (c); (B) representative Raman band spectrum of 100 randomly selected points of rGO–LIG. | ||
The electrochemical properties of LIG and rGO–LIG were characterized using cyclic voltammetry. In a 0.1 M KCl solution containing 5 mM [Fe(CN)6]3−/4−, the LIG electrode and rGO–LIG electrode were measured at different scanning speeds between 10 mV s−1 and 100 mV s−1. As shown in Fig. 3A and C, the corresponding redox peak current of [Fe(CN)6]3−/4− continues to increase as the scan rate increases. It was calculated that the peak current value of the LIG electrode and rGO–LIG electrode had a good linear relationship with the square root of the scan rate (Fig. 3B and D), indicating that the electron transfer process is controlled by diffusion. The effective electroactive surface area is an important factor in evaluating sensing capabilities and can be calculated by the Randles–Sevcik equation. The Randles–Sevcik equation47 is expressed as follows:
| Ip = 2.69 × 105AD1/2n3/2v1/2c | (1) |
I p represents the peak current, A represents the electroactive surface area, D represents the diffusion coefficient of [Fe(CN)6]3−/4− at 25 °C, and the value of D is 6.73 × 10−6 cm2 s−1, n represents the number of electrons transferred in the reaction, n is 1, v represents the scan rate, v is 0.05 mV s−1, and c is the [Fe(CN)6]3−/4− concentration. It can be calculated that the electroactive areas of LIG and rGO–LIG are 0.182 cm2 and 0.271 cm2, respectively. From this result, it can be seen that the effective electroactive area increases significantly, which further illustrates that doping rGO further enhances the conductivity of the biosensing interface.
Characterization of the assembly process of the immune biosensing chip
The assembly process of the immune biosensor chip was characterized using EIS and CV. As shown in Fig. 4A and B, the impedance spectrum of LIG shows a smaller semicircle with good reversibility of the redox peak (curve a). Due to the excellent conductivity of rGO, Rct decreases after LIG is doped with rGO, and the peak current increases (curve b). After the antibody is immobilized on the electrode surface, since the antibody is a macromolecular protein and has poor conductivity, Rct increases and the peak current weakens (curve c). In the same way, BSA is used to conduct the non-specific adsorption site on the electrode surface. After masking, the impedance continues to increase and the peak current decreases again (curve d). After the antibody specifically captures exosomes, an immune response complex is formed, which hinders electron transfer, and the impedance further increases, causing the peak current to further decrease (curve e). The above results show that the immune biosensing chip successfully captured and detected exosomes.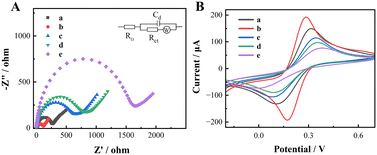 | ||
| Fig. 4 (A) EIS and (B) CV curves of LIG (a), rGO–LIG (b), Ab–rGO–LIG (c), BSA–Ab–rGO–LIG (d), and exosomes–BSA–Ab–rGO–LIG (e). Here, the exosome capture concentration is 5 × 103 particles per μL. | ||
Linearity and sensitivity
Experimental conditions were optimized and obtained as follows: antibody concentration 20 μg mL−1, Ab incubation time 120 min, exosome incubation temperature 37 °C and exosome incubation time 60 min (Fig. 5). Under the optimized experimental conditions, the analysis performance of the designed immune biosensor chip for exosomes was studied. The Rct value of the sensor chip combined with different concentrations of exosomes increases with the increase in exosome concentration (Fig. 6A). The Rct value and the logarithm of exosome concentration are in the range of 5 × 102–5 × 105 particles per μL, with a good linear relationship (Fig. 6B). The expression of the linear regression equation is y = 653.66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c −742.59, and the correlation coefficient R2 = 0.9912, where y represents the impedance value Rct (ohm), and c represents the exosome concentration (particles per μL); based on the signal-to-noise ratio S/N = 3, the detection limit is calculated to be approximately 166 particles per μL. The introduction of rGO realizes the enhanced detection sensitivity of the sensing chip. The advantages include (1) enhancing the electrochemical signal through increasing the electrochemical surface area of the LIG electrode; (2) increasing the antibody binding ability via improving the hydrophobicity of the LIG surface. In Table 1, we compared the electrochemical sensors with different modified electrodes for exosome detection, and our working detection limit was lower than that of the listed sensors. In addition to using LIG as the electrode, rGO one-step modification, the electrode preparation process and its simplicity, this sensing chip has also the advantages of high portability and cost-effectiveness.
c −742.59, and the correlation coefficient R2 = 0.9912, where y represents the impedance value Rct (ohm), and c represents the exosome concentration (particles per μL); based on the signal-to-noise ratio S/N = 3, the detection limit is calculated to be approximately 166 particles per μL. The introduction of rGO realizes the enhanced detection sensitivity of the sensing chip. The advantages include (1) enhancing the electrochemical signal through increasing the electrochemical surface area of the LIG electrode; (2) increasing the antibody binding ability via improving the hydrophobicity of the LIG surface. In Table 1, we compared the electrochemical sensors with different modified electrodes for exosome detection, and our working detection limit was lower than that of the listed sensors. In addition to using LIG as the electrode, rGO one-step modification, the electrode preparation process and its simplicity, this sensing chip has also the advantages of high portability and cost-effectiveness.
 | ||
| Fig. 5 Optimization of experimental conditions: (A) Ab concentration; (B) Ab incubation time; (C) exosome binding temperature; (D) exosome binding time. | ||
| Electrode | Modifying materials | Detection range (particles per μL) | LOD (particles per μL) | Ref. |
|---|---|---|---|---|
| GCE | PAMAM-AuNPs | 5.0 × 102–5.0 × 105 | 2.29 × 103 | 48 |
| GCE | GO-CB[7] | 4.2 × 102–4.2 × 108 | 3.68 × 102 | 49 |
| Au | AuNPs | 5.0 × 102–5.0 × 103 | 4.82 × 102 | 50 |
| Au | COFs | 1.0 × 104–1.0 × 107 | 9.66 × 103 | 51 |
| SPCE | GO-CB[7] | 1.2 × 103–1.2 × 107 | 9.46 × 102 | 52 |
| NAM | mulAu/monAu | 1.0 × 103–1.0 × 107 | 2.80 × 103 | 53 |
| LIG | rGO | 5.0 × 102–5.0 × 105 | 1.66 × 102 | This work |
Reproducibility, stability, and reliability of sensor chips
The signal reproducibility of the sensing chip was studied by analyzing exosomes at a concentration of 105 particles per μL on seven different sensing chips. Almost the same impedance signal was obtained for the seven different sensing chips, with an RSD value of only 0.98% (Fig. 6C), indicating that the constructed immune biosensor chip has good reproducibility.The stability of the constructed immune biosensor chip was studied by detecting exosomes at a concentration of 105 particles per μL on five identical sensor chips two days apart for 8 days. The results are shown in Fig. 6D, where the impedance signals decreased by 1.86%, 4.39%, 6.64%, and 8.24%, respectively, compared with the initial values, and this result indicates that the prepared sensing chip has good storage stability.
The practicability and reliability of the constructed sensor chip were proved through spike recovery experiments. Under optimal experimental conditions, the constructed sensor chip was used to analyze and detect exosomes in serum samples. The exosomes with known concentrations were added to healthy human serum to perform spike recovery experiments and compare the exosome concentration assessed by the standard curve with the known amount added. As shown in Table 2, the recovery rate of the sensing chip for detecting exosome spiked samples ranged from 96.3% to 110.8%, and the RSD ranged from 3.63% to 9.41%, indicating that the constructed sensing chip has good reliability.
| Sample no. | Added (particles per μL) | Found (particles per μL) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 1 × 103 | 1.06 × 103 | 106 | 3.68 |
| 2 | 5 × 103 | 4.73 × 103 | 94.6 | 8.34 |
| 3 | 1 × 104 | 9.63 × 104 | 96.3 | 5.59 |
| 4 | 5 × 104 | 5.54 × 104 | 110.8 | 9.41 |
Clinical competency testing
In order to verify the clinical testing capability of the present immunobiosensing biochip, we used the constructed biochip to test the clinical serum samples, including 22 breast cancer samples and 10 healthy human samples. The results are shown in Fig. 6E, which clearly shows that the expression of exosomes in the serum of breast cancer patients is significantly higher than that in the serum of healthy people, and its P < 0.0001 is derived from the statistical class analysis (Fig. 6F), which is a significant difference, thus indicating that our constructed sensing chip is able to effectively differentiate between healthy people and breast cancer patients, and it has the potential to be used for further clinical diagnosis.Conclusion
In summary, we developed an electrochemical immunobiosensing chip based on one-step doping of rGO with LIG to realize the quantitative detection of exosomes. The sensing chip has good stability, repeatability, precision, accuracy and reliability. Notably, the doping of the two materials formed a 2D/3D structure, which increases the electroactive area on the surface of the electrode and improves the conductivity of the electrode, and PEI–rGO, whose surface is rich in amino groups, provides more binding sites for the solidification of the antibody, thus providing a higher detection sensitivity. The sensing chip is also used for the detection of clinical samples, and it shows some potential applications in distinguishing between healthy people and breast cancer patients.The clinical sample ethical statement
All experiments were performed in accordance with relevant laws or guidelines and approved by the medical ethics committee of Qilu Hospital of Shandong University. Informed consent was obtained from the human participants of this study.Data availability
The data supporting this article have been fully included within the article.Author contributions
Xiaoshuang Chen: experiments, software, plotting, writing – original draft; Xiaohui Yan: investigation, experiments, formal analysis, writing – original draft; Jiaoyan Qiu: methodology, experiments, software; Xue Zhang and Yunhong Zhang: experiments; Hongpeng Zhou and Yujuan Zhao: methodology, software; Lin Han: conceptualization, supervision, writing – original draft, writing – review & editing, project administration, validation; Yu Zhang: conceptualization, investigation, resources, methodology, visualization, funding acquisition, project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Plan of China (Grant No. 2023YFB3210400), the future industries cultivate project-2022 emerging industries cultivate plan of Qingdao (22-3-4-xxgg-2-nsh), and the Collaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong. We would like to thank Xiaoju Li, Haiyan Sui, Xueyun Geng and Sen Wang from Shandong University Core Facilities for Life and Environmental Sciences for their help with the TEM and SEM.References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef.
- B. N. Hannafon and W. Q. Ding, Int. J. Mol. Sci., 2013, 14, 14240–14269 CrossRef PubMed.
- A. K. Ludwig and B. Giebel, Int. J. Biochem. Cell Biol., 2012, 44, 11–15 CrossRef CAS PubMed.
- J. M. Hu, Y. Sheng, K. J. Kwak, J. F. Shi, B. H. Yu and L. J. Lee, Nat. Commun., 2017, 8, 1683 CrossRef PubMed.
- F. Raimondo, L. Morosi, C. Chinello, F. Magni and M. Pitto, Proteomics, 2011, 11, 709–720 CrossRef CAS PubMed.
- L. Zheng, L. Zheng, B. Li, C. C. Liu and W. L. Pan, Cancer Sci., 2021, 112, 985 Search PubMed.
- Y. G. Zhou, R. M. Mohamadi, M. Poudineh, L. Kermanshah, S. Ahmed, T. S. Safaei, J. Stojcic, R. K. Nam, E. H. Sargent and S. O. Kelley, Small, 2016, 12, 727–732 CrossRef CAS.
- M. Mathieu, N. Nevo, M. Jouve, J. I. Valenzuela, M. Maurin, F. J. Verweij, R. Palmulli, D. Lankar, F. Dingli, D. Loew, E. Rubinstein, G. Boncompain, F. Perez and C. Thery, Nat. Commun., 2021, 12, 4389 CrossRef CAS PubMed.
- Z. Song, J. Mao, R. A. Barrero, P. Wang, F. Zhang and T. Wang, Molecules, 2020, 25, 5585 CrossRef CAS.
- E. J. K. Kowal, D. Ter-Ovanesyan, A. Regev and G. M. Church, Extracellular Vesicles: Methods and Protocols, 2017, vol. 1660, pp. 143–152 Search PubMed.
- C. Coughlan, K. D. Bruce, O. Burgy, T. D. Boyd, C. R. Michel, J. E. Garcia-Perez, V. Adame, P. Anton, B. M. Bettcher, H. J. Chial, M. Konigshoff, E. W. Y. Hsieh, M. Graner and H. Potter, Curr. Protoc. Cell Biol., 2020, 88, e110 CrossRef CAS.
- J. P. Nolan and J. C. Jones, Platelets, 2017, 28, 256–262 CrossRef CAS PubMed.
- E. J. Goetzl, J. B. Schwartz, E. L. Abner, G. A. Jicha and D. Kapogiannis, Ann. Neurol., 2018, 83, 544–552 CrossRef CAS PubMed.
- H. Z. Wang, J. H. Zeng, J. Huang, H. Cheng, B. Chen, X. Hu, X. X. He, Y. Zhou and K. M. Wang, Angew. Chem., Int. Ed., 2022, 61, e202116932 CrossRef CAS PubMed.
- J. L. Zhang, J. J. Shi, W. Liu, K. X. Zhang, H. J. Zhao, H. L. Zhang and Z. Z. Zhang, Sens. Actuators, B, 2018, 276, 552–559 CrossRef CAS.
- G. H. Li, N. H. Zhu, J. Zhou, K. Kang, X. X. Zhou, B. W. Ying, Q. Y. Yi and Y. Wu, J. Mater. Chem. B, 2021, 9, 2709–2716 RSC.
- Y. Zhao, J. Xiang, H. Cheng, X. Liu and F. Li, Biosens. Bioelectron., 2021, 194, 113581 CrossRef CAS.
- X. Liu, X. Gao, L. Yang, Y. Zhao and F. Li, Anal. Chem., 2021, 93, 11792–11799 CrossRef CAS.
- J. J. Zhang, L. Hao, Z. W. Zhao, D. C. Jiang and J. Chao, Sens. Actuators, B, 2022, 369, 132332 CrossRef CAS.
- L. Zhang, B. Huang, J. Jin, Y. Li and N. Gu, BMEMat, 2024, 2, e12057 CrossRef.
- D. Jin, F. Yang, Y. Zhang, L. Liu, Y. Zhou, F. Wang and G. J. Zhang, Anal. Chem., 2018, 90, 14402–14411 CrossRef CAS PubMed.
- Y. Yu, Y. T. Li, D. Jin, F. Yang, D. Wu, M. M. Xiao, H. Zhang, Z. Y. Zhang and G. J. Zhang, Anal. Chem., 2019, 91, 10679–10686 CrossRef CAS PubMed.
- L. Yang, X. Yin, B. An and F. Li, Anal. Chem., 2021, 93, 1709–1716 CrossRef CAS PubMed.
- L. Yang, H. Guo, Q. Gao, T. Hou, J. Zhang, X. Liu and F. Li, Anal. Chem., 2023, 95, 17834–17842 CrossRef CAS PubMed.
- S. H. Shamsuddin, T. D. Gibson, D. C. Tomlinson, M. J. McPherson, D. G. Jayne and P. A. Millner, Biosens. Bioelectron., 2021, 178, 113013 CrossRef CAS PubMed.
- L. Anvari, S. M. Ghoreishi, K. Khoshnevisan, M. R. Ganjali and F. Faridbod, J. Appl. Electrochem., 2023, 53, 1843–1851 CrossRef CAS.
- J. Tang, J. Qin, J. J. Li, L. P. Liu and H. S. Zeng, Biosens. Bioelectron., 2023, 222, 114961 CrossRef CAS PubMed.
- X. Xie, D. Liu, W. L. Wang, J. Xiang, M. H. Yang and G. K. Liu, Anal. Chem., 2023, 95, 2087–2093 CrossRef CAS PubMed.
- G. M. Di Mari, M. Scuderi, G. S. P. Lanza, M. G. Salluzzo, M. Salemi, F. Caraci, E. Bruno, V. Strano, S. Mirabella and A. Scandurra, Nanomaterials, 2024, 14, 170 CrossRef CAS PubMed.
- D. X. Luong, K. C. Yang, J. Yoon, S. P. Singh, T. Wang, C. J. Arnusch and J. M. Tour, ACS Nano, 2019, 13, 2579–2586 CAS.
- A. Lamberti, F. Clerici, M. Fontana and L. Scaltrito, Adv. Energy Mater., 2016, 6, 1600050 CrossRef.
- W. L. Zhang, Y. J. Lei, Q. Jiang, F. W. Ming, P. Costa and H. N. Aishareef, Small Methods, 2019, 3, 1900005 CrossRef.
- L. Zhao, S. Y. Wong, J. Y. Sim, J. Zhou, X. Li and C. Wang, BMEMat, 2023, 1, e12020 CrossRef.
- M. G. Stanford, J. T. Li, Y. Chyan, Z. Wang, W. Wang and J. M. Tour, ACS Nano, 2019, 13, 7166–7174 CrossRef CAS PubMed.
- K. H. Choi, S. Park, S. K. Hyeong, S. Bae, J. M. Hong, T. W. Kim, S. H. Lee, S. Ryu and S. K. Lee, J. Mater. Chem. A, 2020, 8, 19822–19832 RSC.
- A. F. Carvalho, A. J. S. Fernandes, C. Leitao, J. Deuermeier, A. C. Marques, R. Martins, E. Fortunato and F. M. Costa, Adv. Funct. Mater., 2018, 28, 1805271 CrossRef.
- B. Kulyk, B. F. R. Silva, A. F. Carvalho, S. Silvestre, A. J. S. Fernandes, R. Martins, E. Fortunato and F. M. Costa, ACS Appl. Mater. Interfaces, 2021, 13, 10210–10221 CrossRef CAS PubMed.
- Y. Chen, Y. Y. Zhang, F. Song, H. Y. Zhang, Q. K. Zhang, J. Xu, H. P. Wang and F. Y. Ke, Adv. Mater. Technol., 2021, 6, 2100421 CrossRef CAS.
- H. Lv, L. Z. Sun, X. Chen, D. D. Xu and B. Liu, Green Chem., 2019, 21, 2043–2051 RSC.
- X. Hui, X. Xuan, J. Kim and J. Y. Park, Electrochim. Acta, 2019, 328, 135066 CrossRef CAS.
- Z. B. Wang, Y. G. Wu, B. Zhu, Q. X. Chen, Y. Zhang, Z. J. Xu, D. H. Sun, L. W. Lin and D. Z. Wu, ACS Appl. Mater. Interfaces, 2023, 15, 4713–4723 CrossRef CAS PubMed.
- W. H. Yan, W. R. Yan, T. D. Chen, J. G. Xu, Q. Tian and D. Ho, ACS Appl. Nano Mater., 2020, 3, 2545–2553 CrossRef CAS.
- H. Lim, H. Kwon, H. Kang, J. E. Jang and H. J. Kwon, Nat. Commun., 2023, 14, 3114 CrossRef CAS PubMed.
- D. Akinwande, C. J. Brennan, J. S. Bunch, P. Egberts, J. R. Felts, H. J. Gao, R. Huang, J. S. Kim, T. Li, Y. Li, K. M. Liechti, N. S. Lu, H. S. Park, E. J. Reed, P. Wang, B. I. Yakobson, T. Zhang, Y. W. Zhang, Y. Zhou and Y. Zhu, Extreme Mech. Lett., 2017, 13, 42–77 CrossRef.
- J. N. Tiwari, V. Vij, K. C. Kemp and K. S. Kim, ACS Nano, 2016, 10, 46–80 CrossRef CAS PubMed.
- M. Wang, Y. J. Chu, L. Qiang, Y. K. Han, Y. Zhang and L. Han, Sens. Diagn., 2022, 1, 262–269 RSC.
- T. R. L. C. Paixão, Measuring Electrochemical Surface Area of Nanomaterials versus the Randles-Sevcik Equation, ChemElectroChem, 2020, 7(16), 3414–3415 CrossRef.
- H. Zhang, Z. Wang, F. Wang, Y. Zhang, H. Wang and Y. Liu, Talanta, 2021, 224, 121879 CrossRef CAS PubMed.
- Y. An, R. Li, F. Zhang and P. He, Talanta, 2021, 235, 122790 CrossRef CAS PubMed.
- Q. Liu, X. Yue, Y. Li, F. Wu, M. Meng, Y. Yin and R. Xi, Talanta, 2021, 232, 122451 CrossRef CAS PubMed.
- J. Lu, M. Wang, Y. Han, Y. Deng, Y. Zeng, C. Li, J. Yang and G. Li, Anal. Chem., 2022, 94, 5055–5061 CrossRef CAS PubMed.
- Y. An, R. Li, F. Zhang and P. He, Anal. Chem., 2020, 92, 5404–5410 CrossRef CAS PubMed.
- Y. Wang and H. Zhang, Sens. Actuators, B, 2023, 393, 134294 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |


