 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A genetically encoded probe for monitoring and detection of iron in real-time†
Neha
Soleja
and
Mohd.
Mohsin
 *
*
Metabolic Engineering Laboratory, Department of Biosciences, Jamia Millia Islamia, New Delhi-110025, India. E-mail: mmohsin1@jmi.ac.in; Fax: +91 (11)2698 0229; Tel: +91 (11)26981717
First published on 20th August 2024
Abstract
Iron, the most abundant transition metal in the body, regulates cellular function but can be harmful in excess, leading to reactive oxygen species production and cellular damage. Intracellular Fe2+ exerts a significant impact on cellular function, potentially contributing to various critical diseases. To address this, detection methods need high selectivity, sensitivity, and real-time monitoring capabilities, essential for comprehending disease progression. This necessitates advancements beyond conventional detection approaches. Frataxin, a crucial mitochondrial protein, is indispensable for sustaining life, contributing not only to iron metabolism but also to the formation of iron–sulfur clusters critical for cellular function. Its deficiency is implicated in neurodegenerative diseases. We have developed a nanosensor, based on fluorescence resonance energy transfer (FRET), designed to probe iron efflux mechanisms and facilitate dynamic monitoring of iron concentration and its spatial distribution within living cells. To construct this nanosensor, we strategically positioned CyaY, a bacterial frataxin ortholog, between ECFP and Venus, forming a FRET pair. This innovative nanosensor, designated as FeOS (iron optical sensor), demonstrates exceptional selectivity for iron and maintains stability under physiological pH conditions. Additionally, we engineered three mutant variants: I17C, AD10-I17C, and D76H, with A10D-I17C displaying the highest affinity for iron and a broad detection range. The distinguishing feature of this sensor is that it is genetically encoded, facilitating real-time detection of iron levels within living cells.
Introduction
Iron, the predominant transition metal element within the human body, is abundant in the bloodstream, bone marrow, muscle tissue, and the liver, where it contributes significantly to vital biological functions such as oxygen and electron transfer, as well as enzymatic reactions.1,2 Its presence is integral to organismal metabolism.3 It is vital for various cellular processes, including oxygen transport, metabolism, and DNA repair, regulated meticulously to avoid cellular damage.4–6Emerging research suggests that abnormal iron metabolism is intricately linked to the development of numerous diseases.7 Abnormal iron metabolism appears in two primary forms: excess and insufficiency. Excessive iron levels have been associated with conditions such as malignancies, liver inflammation, and diverse neurological ailments.8 Conversely, insufficient iron often results in iron deficiency anemia, particularly prevalent among expectant mothers and young children.9 Additionally, iron deficiency has been linked to impaired cognitive growth, compromised heart health, and renal disorders.10,11 The majority of iron within organisms is tightly associated with specific enzymes, carrier proteins, or storage molecules. Labile iron exhibits notable reactivity and can catalyze the Fenton reaction, generating highly active free radicals that oxidize and damage various cellular constituents, including genetic material, cellular proteins, and fatty acids, ultimately resulting in considerable cellular damage and the possibility of cell demise.8
Previous research shows that frataxins in Homo sapiens, yeast, and Escherichia coli (E. coli) can bind Fe(II) and Fe(III) with modest affinities, suggesting a significant role in their function. CyaY, the bacterial counterpart to human frataxin, is a notable model due to its shared homology and unique characteristics.12 Despite having 27% similarity and 45% homology with human frataxin, CyaY is a 106-residue protein exclusive to the conserved domain without mitochondrial import signals. It exhibits similar iron-binding properties to yeast frataxin but with greater stability in terms of both thermal resilience and structural fold. Utilizing a bacterial model simplifies frataxin exploration due to its confined machinery within operons.5
Over the past few decades, various volumetric and gravimetric methods have been developed, each based on distinct physical and chemical principles, and have been established for quantifying iron levels. Nevertheless, the potential for errors looms, particularly when estimating trace amounts of iron.13 One of the most precise means of measuring iron concentration involves absorption atomic spectroscopy. There is a scarcity of iron-selective sensors that offer a straightforward, swift, and dependable analytical solution. Among these sensors, some rely on chalcogenide glass systems, while others are based on plasticized polymeric electrodes. The former category poses challenges due to their idiosyncratic and incompletely understood interfacial chemistry. Poly(vinyl chloride) (PVC)-based sensors are advantageous due to their capacity for compositional optimization, enhanced selectivity, and ease of miniaturization. One study explored the feasibility of utilizing Schiff bases as ionophores for iron sensors. Several Schiff bases from the literature were evaluated for their potential in developing iron-selective sensors, revealing the effectiveness of a PVC-based membrane containing a μ-bis(tridentate) ligand as an iron sensor. The operational principle of PVC-based membranes entails selective recognition by the embedded carrier through complex formation with the analyte ion. Notably, the μ-bis(tridentate) ligand is reported to form robust complexes with iron, eliciting a potentiometric response. However, these approaches involve time-intensive procedures and substantial costs, and necessitate specialized equipment that is not commonly available in standard laboratories. As a result, there exists a pressing demand for a more straightforward yet equally precise method suitable for routine iron detection and related cellular biological events.13,14
This study presents a genetically encoded FRET-based nanosensor capable of monitoring real-time changes in iron concentration within living cells. In this study, CyaY was employed as the iron-sensing domain and served as the recognition element. CyaY was linked with two distinct variants of green fluorescent protein (GFP) functioning as the reporter genes. Enhanced cyan fluorescent protein (ECFP) was chosen as the donor fluorophore, while Venus, a derivative of yellow fluorescent protein (YFP), was employed as the acceptor. These fluorophores, ECFP and Venus, were strategically placed at the N and C termini of the nanosensor, respectively, to create the sensor construct. The nanosensor was employed in both prokaryotic and eukaryotic systems, where it hinged on the rate of energy transfer between the donor and acceptor fluorophores as a diagnostic measure for assessing iron levels, both in vitro and in living cells.
Materials and methods
Nanosensor design and construction
The design and construction of the nanosensor began with the retrieval of the CyaY protein sequence and its three-dimensional structure from databases: Kyoto Encyclopedia of Genes and Genomes (KEGG) and the RCSB Protein Data Bank (PDB). The nanosensor was constructed by amplifying the CyaY gene using polymerase chain reaction (PCR), utilizing genomic DNA extracted from E. coli K12. This process utilized a forward primer (5′-CGGggtaccATGAACGACAGTGAATTTCATC-3′) and a reverse primer (5′-CGGggtaccGAAACTGACTGTTTCACCCGCC-3′), with the lowercase sequence designating the KpnI restriction site. Strategically, one stop codon was removed from the gene's 3′ end to ensure the integrity of the resulting protein and avoid truncation. The nanosensor harnessed the intrinsic fluorescence properties of fluorescent proteins (FPs), ECFP and Venus. To achieve this, PCR was employed to amplify the ECFP and Venus fluorescent proteins from the pDH18 vector (YRC, Washington, USA). Subsequently, the amplified ECFP, CyaY, and Venus genes were sequentially integrated into the bacterial expression vector pRSET-B (Invitrogen, USA) that introduces an in-frame His tag at the amino terminus of the genetic sequence. This resulted in the development of the construct known as pRSET-B_ECFP_CyaY_Venus (FeOS). The accuracy of the genetic sequences for ECFP, Venus, and CyaY was confirmed using Sanger sequencing. To facilitate the expression of the nanosensor in yeast, the chimeric sequence was transferred to the destination vector pYES-DEST52 (Invitrogen, USA). This shuttle was facilitated by Gateway technology and the LR-clonase-II enzyme, adhering to the manufacturer's provided guidelines. Saccharomyces cerevisiae (S. cerevisiae)/URA3 strain BY4742 was employed as the host organism and was cultured on YEPD agar (consisting of 1% yeast extract, 2% peptone, and 2% glucose). Subsequently, a liquid YEPD medium was utilized for growth under controlled conditions at 30 °C with aeration, and the yeast cells were transformed with the nanosensor construct. Furthermore, to enable the expression of the sensor within human embryonic kidney (HEK)-293T mammalian cells, the sensor protein sequence was excised from the pRSET-B vector and subsequently sub-cloned into pcDNA3.1(−) (Invitrogen, USA) utilizing BamHI and HindIII restriction sites.Expression and purification of the recombinant protein
FeOS was introduced into E. coli Rosetta cells via heat shock, followed by selection with 100 μg ml−1 ampicillin. The cells were cultivated in Luria Bertani (LB) medium at a temperature of 20 °C until reaching an optical density (OD600) of 0.6. To induce protein expression, isopropyl-D-1-thiogalactopyranoside (IPTG, Himedia) was added to the cells, resulting in a final concentration of 1 mM. The cells were then cultured overnight at 16 °C for a duration of 21 hours. Afterward, the expressed cells were harvested through centrifugation at 4500 rpm for 20 minutes, and the pellet obtained was re-suspended in a Tris-Cl buffer (20 mM, pH 8.0). Subsequently, the sensor protein was purified using nickel–nitrilotriacetic acid (Ni–NTA, Qiagen, Germany) resin beads via His-tag affinity chromatography. This purification process followed the initial step of cell lysis, which was achieved using sonication equipment (Labsonics, USA). The binding of the protein to the resin beads occurred at a temperature of 4 °C over the course of approximately an hour. After the beads had settled, washing was performed with 20 mM Tris-Cl and 10 mM imidazole (pH 8.0), and elution was achieved with 20 mM Tris-Cl and 250 mM imidazole (pH 8.0). Protein folding was ensured by storing overnight at 4 °C, and purity was confirmed using 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).Fluorescence emission spectra and stability assessment
Fluorescence emission spectra of the sensor protein were obtained using a microplate reader (Biotek, USA), exciting ECFP at 420 nm and capturing emission fluorescence between 460 nm and 600 nm. Stability assessment involved dilution of the sensor protein in 20 mM buffers with pH ranging from 5.0 to 8.0, including tris buffer saline (TBS), 3-(N-morpholino)propane sulfonic acid (MOPS), phosphate-buffered saline (PBS), and Tris-Cl. Stability in 20 mM PBS across pH 5.0 to 8.0, with and without iron, was also evaluated. FRET ratio change (Em540/485 nm) was measured using a microplate reader with emission filters at 485/20 nm for ECFP and 540/20 nm for Venus, in triplicate with a 96-well plate.Site-directed mutagenesis
It has previously been established that frataxins from diverse species exhibit binding to iron within corresponding protein regions, albeit with a relatively modest affinity, suggesting the importance of iron binding for their biological function. Nonetheless, these proteins display low specificity for cations and possess multiple binding sites with a propensity to chelate metals with low affinity.5 To explore potential modifications in CyaY's binding capacity and specificity for iron, affinity mutants were systematically created. These mutants, designated as I17C, double mutant A10D-I17C, and D76H of FeOS, were created by introducing point mutations within the CyaY gene using the QuikChange site-directed mutagenesis kit (Agilent, USA). Subsequently, the mutant sensor proteins were expressed and purified using the previously described method.Assessment of the selectivity of wild-type and mutant FeOS proteins
To assess the specificity and selectivity of both the wild-type (WT) sensor and its mutant counterparts, the purified sensor proteins were diluted in 20 mM PBS buffer and subjected to incubation with various metal ions at physiologically relevant concentrations. The tested metal salts encompassed iron (Fe2+ and Fe3+), copper (Cu2+), zinc (Zn2+), silver (Ag+), cadmium (Cd2+), cobalt (Co2+), and nickel (Ni2+). Changes in the FRET ratio were monitored to determine the metal specificity demonstrated by the sensor proteins.Ligand binding isotherms
To evaluate the iron-binding affinity of FeOS proteins, FRET ratio measurements were conducted in triplicates using a 96-well microtiter plate. These measurements were recorded until reaching equilibrium, signifying saturation in the ligand titration. Subsequently, GraphPad Prism software was employed to calculate the binding constants (Kd) for both the WT and sensor variants. The ligand titration curves were fitted to a binding isotherm equation as follows:15| S = (r − rapo)/(rsat − rapo) = [Fe2+]/(Kd + [Fe2+]) |
Detection of intracellular Fe2+ in bacterial cells
Fe2+ uptake within E. coli was continuously monitored over time. The sensor protein A10D-I17C was expressed in E. coli BL21-Rosetta strain while the cells were in their logarithmic growth phase, corresponding to an OD600 of 0.6. Induction was achieved with 1 mM IPTG at 16 °C under light-deprived conditions for 21 hours. After expression, cells were resuspended in 20 mM PBS buffer (pH 7.0) along with Fe2+ and dispensed into microplate wells in triplicates. FRET ratio measurements were taken every 5 minutes for 2 hours, with a control experiment performed without added Fe2+. Throughout the predefined timeframe, ratiometric changes were monitored. As a negative control, the ECFP-Venus construct was cloned in pRSET-B. It lacked the binding domain CyaY. The control experiment was carried out simultaneously using the ECFP-Venus system and the FRET ratio was monitored after addition of 1 μM Fe2+. The bacterial cell suspension (180 μl) was dispensed and incubated with metal ions such as Cu2+ and Zn2+ at a concentration of 1 μM each to study the effect of these metal ions on the sensor at the in vivo level. The FRET ratio changes were recorded at 0 minute and after 60 minutes of incubating the cell suspension with these metal ions. For these measurements, an excitation filter was set at 420/20 nm, while emission filters were adjusted to 485/20 nm and 540/20 nm for ECFP and Venus, respectively, thus enabling the precise capture of dual emission intensities in the form of FRET signals. Confocal images of the bacterial cells expressing the sensor were also acquired on a confocal microscope using LAS-AF software (Leica, Wetzlar, Germany) equipped with a TCS-SPE confocal head.Real-time monitoring of the sensor A10D-I17C within live eukaryotic cells
The sensor A10D-I17C was introduced into the S. cerevisiae BY4742 strain to study its internalization, subcellular localization, and distribution within yeast cells. Transformed cells were selected on synthetic defined (SD) agar plates and cultured in YEPD media supplemented with 3% galactose and 2% sucrose for 3–5 days. Galactose served as an inducer, while sucrose ensured nutrient availability. Yeast cells were then immobilized onto glass slides, and FRET ratio measurements (540/485 nm) were performed after adding 1 μM Fe2+. In parallel, a control experiment was conducted to record ratiometric changes in the absence of externally added Fe2+ ions. As a negative control, the ECFP-Venus construct was cloned in pYEST-DEST52. 1 μM Fe2+ was added to the ECFP-Venus system and FRET ratio changes were obtained for the defined period. Images of the yeast cells were captured using a 63× oil immersion objective on a confocal microscope over 10 minutes. Yeast cells were also incubated with Cu2+ and Zn2+, each at a concentration of 1 μM. This aimed to investigate the impact of these metal ions on the sensor in vivo. FRET ratio changes were recorded both at the start (0 minute) and after 10 minutes of incubation with these metal ions.The HEK-293T cell line was maintained in Dulbecco's modified Eagle's media (DMEM, Sigma, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco-Life Technologies, Thermo Fisher Scientific, USA) and 1% antibiotic cocktail. Cells were cultured on sterile polylysine-coated coverslips in 6-well cell culture plates in a humidified incubator at 37 °C with 5% CO2 after a serum starvation period of 3–5 hours. Transfection with sensor A10D-I17C using lipofectamine was performed, followed by 48 hours of incubation with media replacement. For imaging, cells were washed with 20 mM PBS buffer (pH 7.0), and confocal microscopy was used to capture images. FRET ratio changes were recorded after adding 1 μM Fe2+ with excitation at 420 nm, continuously for 10 minutes. Control recordings were performed without 1 μM Fe2+. A negative control was also established wherein 1 μM Fe2+ was added to the ECFP-Venus system and ratio changes were recorded for the set period. HEK cells were exposed to 1 μM Cu2+ and Zn2+, and FRET ratio changes were monitored at 0 minute and after 10 minutes of incubation to determine the effect of these metal ions on the sensor.
Results and discussion
Nanosensor construction
Iron is an essential nutrient vital for cellular energy production and serves as a fundamental component in structures such as hemoglobin, myoglobin, heme enzymes, and various enzyme cofactors. However, iron can also become detrimental when it accumulates excessively, resulting in conditions like hemochromatosis. Maintaining the proper balance of iron is crucial for the normal functioning of cells. The precise mechanism through which frataxin regulates the balance of oxidative stress is intricate and not yet fully comprehended. Hence, it is imperative to elucidate the regulatory pathways responsible for the detection and measurement of iron levels and associated cellular processes.16–18 To meet this imperative need, there is a pressing requirement for the development of a straightforward, reliable, non-invasive, cost-effective, and highly sensitive method to detect trace levels of iron in both simple and complex cellular environments. Consequently, the demand for iron detection in clinical, medical, environmental, and industrial samples has driven the development of various iron measurement techniques and methods.In this work, we have devised a genetically encoded nanosensor utilizing fluorescence sensing technology. This nanosensor allows for real-time, non-invasive detection of iron levels. Our approach involved the creation of a recombinant protein where we fused fluorescent variants, ECFP (as the donor) and Venus (as the acceptor), to the N- and C-termini of CyaY, respectively, to form a FRET pair, enabling the precise measurement of ratiometric changes in cellular iron flux rates. The nanosensor was successfully constructed within the pRSET-B vector which includes a hexahistidine (6× His) tag positioned at the N-terminus, facilitating the rapid purification of fusion proteins using nickel-chelating resin. The accuracy and integrity of the nanosensor construct were verified through sequencing, as depicted in Fig. S1.† All frataxins share common structural features, including a hydrophobic region situated within the β-sheet stretch and a negatively charged region located on helix 1. These regions play a pivotal role in their metal binding capabilities, which primarily involve clustered aspartic acid Asp and glutamic acid residues.5 CyaY, a specific frataxin variant, consists of two helices (3–22 [α1] and 87–99 [α2]) that are tightly packed against a six-stranded, anti-parallel β sheet (30–35 [β1], 38–43 [β2], 48–53 [β3], 60–63 [β4], 69–73 [β5], 78–80 [β6]).19 CyaY exhibits a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 binding stoichiometry with Fe2+. Additionally, it has been observed that the CyaY–Fe3+ complex can have a stoichiometry of 1
2 binding stoichiometry with Fe2+. Additionally, it has been observed that the CyaY–Fe3+ complex can have a stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7.17
7.17
The genetically encoded nanosensor developed in this study represents a powerful tool for advancing our understanding of neurodegenerative disorder like Friedreich's ataxia (FRDA), the role played by frataxin, and the intricate involvement of iron in this pathological condition. By enabling real-time, non-invasive detection of iron levels and flux rates, the sensor provides researchers with a valuable tool to monitor how iron is dysregulated in FRDA-affected cells. Moreover, the successful transfer of the sensor protein to specific vectors, such as pYES-DEST52 and pcDNA3.1 (−) for cellular expression, demonstrates its versatility and applicability in different cellular systems. Fig. 1 displays the linear representation of FeOS constructs within various vectors designed for expression in both prokaryotic and eukaryotic systems. The arrangement of genes within nanosensor constructs across various expression vectors designed for enhanced cellular expression is depicted in a circular diagram in Fig. S2.†
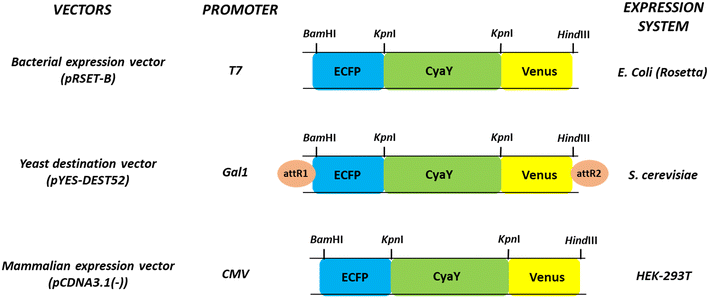 | ||
| Fig. 1 Schematic linear representation of FeOS constructs in various vectors for cellular expression in different systems for cell-based studies. | ||
Protein expression and purification
The recombinant protein was effectively purified using Ni–NTA His-tag affinity columns, and this purification process was carried out under light-protected conditions. To confirm the size and integrity of the sensor protein, the purified protein was analysed using a 10% SDS-PAGE gel, which exhibited the expected band.FRET emission spectra
FRET is a phenomenon that facilitates the transfer of energy from one fluorophore, known as the donor, to another, termed the acceptor, resulting in a measurable shift in fluorescence emission. This process is invaluable for monitoring dynamic interactions involving fluorescent nanoparticles, labeled proteins, or cells in real-time and non-invasively. Effective FRET relies on selecting donor–acceptor pairs with significant spectral overlap and utilizing bright donor fluorophores.20 In this study, in vitro spectral analysis of FeOS revealed changes in the fluorescence emission intensities of ECFP and Venus in response to varying iron concentrations. Specifically, the emission intensity of ECFP decreased, while that of Venus increased, indicating non-radiative energy transfer from the donor to the acceptor molecule, indicative of FRET (Fig. 2). FRET-based biosensors have become essential tools in various biological applications, including disease detection and drug screening. They offer several advantages, such as simplicity, uniform assay conditions, high sensitivity, specificity, and rapid response. These qualities make them valuable for conducting precise quantitative measurements, both in vitro and within living cells.21Evaluation of the sensor protein's stability
Minimal changes in the FRET ratio were observed when FeOS was evaluated in 20 mM PBS buffer, indicating its stability in this buffer (Fig. 3a). The variations in the FRET ratio observed across different buffer solutions when iron is not present are probably because of differences in buffer composition and solvent effects. Variations in pH, ionic strength, and solvent polarity across different buffers can influence the conformational dynamics and stability of the sensor, affecting energy transfer efficiency between fluorophores and leading to differences in FRET ratio. However, we chose the buffer in which the FRET ratio changes were least significant for further experiments which is PBS in our study. Consequently, 20 mM PBS buffer at pH 7.2 was chosen for further dilution of the sensor protein in subsequent experiments. FRET ratio measurements were recorded by diluting FeOS in 20 mM PBS buffer with a variable pH range of 5.0 to 8.0, both in the absence and presence of 0.1 μM iron. Significant ratiometric changes were observed under acidic conditions up to pH 7.0, while FeOS exhibited greater stability within the alkaline physiological range of 7.0 to 8.0 (Fig. 3b). Given that the cytosolic pH in S. cerevisiae typically falls within the range of approximately 7.2, FeOS is considered a suitable candidate for investigating iron uptake within yeast cells. It's crucial to rigorously assess the stability of the sensor protein, considering that environmental factors can influence the properties of fluorescent proteins. This ensures the specificity and reliability of the sensor protein.22Affinity mutants and their specificity evaluation
Site-directed mutagenesis was employed to introduce specific point mutations in the WT FeOS. The fidelity of these mutant variants was validated through sequencing, as shown in Fig. S3.† The WT-FeOS and the mutants, including I17C and D76H, exhibited relatively similar FRET ratio changes when exposed to different metal ions. This suggests that these metals could be competing with iron for the same binding site within the sensor. However, the double mutant A10D-I17C displayed increased specificity for Fe2+ despite the presence of other metal ions, as evidenced by the maximum change in the FRET ratio observed in this case (Fig. 4). These findings suggest that while other metals may indeed compete with iron for binding, the A10D-I17C mutant demonstrates a heightened affinity for iron, highlighting its potential as a selective iron sensor. In live cells, the levels of physiological trace metals like Cu2+ and Zn2+ can vary widely, influenced by factors such as cell type and their specific roles within the cell. Typically, copper concentrations range between 10 and 100 μM, while zinc concentrations are usually found within the 100–300 μM range. In this study, we observed that when the A10D-I17C sensor was exposed to Cu2+ and Zn2+ at concentrations in these ranges (10 μM, 50 μM), there was minimal change in the FRET ratio compared to the significant changes seen in the presence of iron, as depicted in Fig. 4c. This led us to conclude that the sensor exhibited specificity towards iron. The introduction of point mutations in the sensing domain has the potential to alter the substrate binding properties of the sensor, thus expanding its applicability for sensing iron concentrations.23Ligand binding isotherms of the sensor protein
The WT and affinity mutants were evaluated over a range of Fe2+ concentrations ranging from 10 nM to 300 μM. The FRET ratios exhibited an initial increase followed by stabilization, indicating the saturation of FeOS with Fe2+ ions, with no further significant change observed in the ratio (Fig. 5). This saturation curve provides evidence of the complete occupancy of the ligand-binding pocket of CyaY in FeOS with Fe2+. The calculated Kd values for WT-FeOS and mutants, including I17C, A10D-I17C, and D76H, were 1.02 μM, 0.98 μM, 0.90 μM, and 1.75 μM, respectively. The Kd values obtained for the sensor variants shed light on their affinities for iron, with lower Kd values indicating stronger binding affinity. This suggests that the sensor effectively binds to iron even at lower concentrations, enabling the detection of minute changes in the ratio. Moreover, the sensor demonstrates versatility by detecting iron concentrations across a broad range, from nanomolar to micromolar levels, as shown in Table 1. Iron concentrations can vary widely across different cell types ranging from around 1–20 μM in the cytosol of mammalian cells to 100–300 μM in mitochondria. This versatility underscores the sensor's utility for detecting iron within physiologically relevant concentration ranges. Table 1 provides the determined dynamic range of Fe2+ detection for both the WT and its variants. The mutant sensors exhibited altered Kd values compared to the wild type, consistent with the known phenomenon of point mutations in the sensing domain affecting the substrate binding properties of the sensor. Among all the developed sensors, A10D-I17C is considered the most suitable for investigating the dynamics of Fe2+ levels at the in vivo level, as it displayed the lowest binding constant and can measure Fe2+ levels across various physiological scales.| Sensor | K d (M) (±SDa) | Dynamic rangeb |
|---|---|---|
| Binding constants (Kd) determined in vitro.a Standard Deviation (SD).b Effective quantification range between 10 % and 90 % saturation of the nanosensor. | ||
| Wild type | 1.02 × 10−6 (±0.02) | 30 nM–100 μM |
| I17C | 9.83 × 10−7 (±0.02) | 10 nM–90 μM |
| A10D-I17C | 9.02 × 10−7 (±0.02) | 50 nM–200 μM |
| D76H | 1.75 × 10−6 (±0.02) | 10 nM–100 μM |
Intracellular monitoring of sensor A10D-I17C in E. coli
Live cell images confirmed the successful expression of the sensor in E. coli (Fig. S4†). For the in vivo bacterial study, sensor A10D-I17C and Fe2+ (1 μM) were mixed, with measurements taken over 60 minutes at 5-minute intervals. The recorded data showed a basal FRET ratio of 0.995 at the start (0 sec), which significantly increased over 50 minutes after the addition of 1 μM Fe2+, reaching saturation thereafter. The control experiment exhibited negligible FRET variations in the absence of Fe2+, confirming insignificant energy transfer from the donor to the acceptor without Fe2+. Negative control pRSET-B_ECFP_Venus was then expressed in E. coli and the FRET ratio was calculated. It was observed that upon adding Fe2+ the FRET ratio change was negligible, clearly indicating that energy transfer is not occurring when the Fe2+ sensing domain CyaY is absent from the construct (Fig. 6a). No significant change in the FRET ratio was obtained by the addition of metal ions (Cu2+ and Zn2+), confirming the in vivo specificity of the sensor (Fig. 6b). All measurements were performed in triplicate. The intracellular results displayed a time-dependent response curve of FeOS to Fe2+, illustrating its suitability for real-time monitoring of Fe2+ in live bacterial cells. This study contributes to the development of genetically encoded FRET-based nanosensors, offering non-invasive and real-time monitoring of specific metabolite concentrations in vivo with high spatio-temporal resolution.24,25In vivo monitoring and cell imaging of A10D-I17C in S. cerevisiae and HEK-293 T cells
Yeast cells, due to their highly conserved protein nature, are ideal for studying metal ion trafficking and nutrient imbalance-related diseases in humans.26 Real-time in vivo imaging revealed significant fluorescence in the cytosol, indicating successful A10D-I17C expression (Fig. 7a). Fe2+ transport into the cytosol was observed, generating FRET ratio signals, enabling dynamic Fe2+ concentration monitoring post 1 μM Fe2+ addition. The recorded data showed an initial rapid ratio increase from 0.99 to 1.20 at 8 minutes, followed by saturation. Conversely, no ratio change was observed in the control group where no Fe2+ was added to the sensor. No change in the ratio was also acquired with the ECFP-Venus only indicating that the FRET is the result of conformational changes in CyaY upon binding of Fe2+. These ratiometric changes were visualized in a plotted graph (Fig. 7b). Although we have not directly calculated the signal-to-noise ratio of the sensor, we conducted a control experiment where no analyte (iron) was added, as shown in Fig. 7b. Under the control conditions, the observed FRET signal was almost negligible, serving as the baseline signal in the absence of iron. Any additional FRET signal detected would be attributed to the presence of iron when the sensor is incubated within the cell. This demonstration underscores the sensor's ability to distinguish between background noise and the actual signal, thus contributing to its effectiveness in detecting intense fluorescence, which is a crucial criterion for a superior sensor. Yeast cells, when incubated with Cu2+ and Zn2+, showed no significant change in the FRET ratio, confirming the in vivo specificity of the sensor, as these metal ions do not impact the FRET ratio (Fig. S5†).Confocal imaging of the transfected cells confirmed the effective expression of sensor A10D-I17C within the cells (Fig. 8a). The recorded FRET changes exhibited a continuous increase upon Fe2+ addition, up to 5 minutes, followed by saturation. Conversely, the HEK-293 T cell line transfected with sensor A10D-I17C in the absence of Fe2+ displayed no significant changes in the FRET ratio, indicating negligible energy transfer. The HEK-293 T cell line transfected with pcDNA3.1(−) containing the ECFP-Venus construct as the negative control did not yield significant changes in the FRET ratio (Fig. 8b). In a similar manner, when HEK cells were exposed to Cu2+ and Zn2+, there was no notable alteration in the FRET ratio (Fig. S6†). The cell-based assessments of yeast and mammalian cells collectively demonstrated that sensor A10D-I17C possesses remarkable potential for monitoring real-time dynamics of Fe2+ levels in eukaryotic cells under in vivo conditions. Genetically encoded nanosensors utilizing FRET offer rapid, sensitive, precise, and real-time metabolite dynamics analysis, potentially overcoming current methodological constraints.27,28
Conclusions
In summary, we have successfully developed a genetically encoded sensor for the detection and characterization of intracellular Fe2+ levels both in vitro and in living cells. The affinity variant A10D-I17C engineered through mutagenesis identified the physiological range of Fe2+ measurements and exhibited sensitivity to variations in Fe2+ concentrations establishing it to be one of the most effective nanosensors built. A10D-I17C was successfully expressed in live cells and proved capable of tracking real-time flux rates of Fe2+ across various cell types, even at low concentrations. This underscores the sensor's versatility and effectiveness in both prokaryotic and eukaryotic systems. The sensor's design is straightforward and efficient, making it a valuable tool for studies, enabling the monitoring of changes in cellular Fe2+ concentrations with high spatial and temporal resolution. Compared to chemical and protein-based methods, this genetically encoded sensor offers advantages in terms of resilience, reusability, and ease of regeneration in cellular contexts. What sets this sensing tool apart is that it is genetically encoded and uses fluorescence sensing assay technology to monitor changes continuously and rapidly in real-time, presenting the results as fluorescence signals from living cells. The sensor can facilitate investigations into the complex mechanisms through which frataxin influences iron metabolism and how changes in iron concentrations correlate with increased oxidative stress—a hallmark of FRDA. Furthermore, the sensor holds potential for drug development and diagnostic purposes, advancing our understanding of FRDA and potentially revealing novel therapeutic avenues.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
N. S. and M. M. designed the study and prepared the original manuscript. N. S. & M. M. conducted all in vitro and in vivo experiments and analyzed the data. N. S. and M. M. did the live cell imaging of yeast and HEK cells and analyzed the data. All authors were engaged in commenting on the manuscript. All authors read and approved the final manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The first author (N. S.) would like to express gratitude to the Indian Council of Medical Research, Government of India, for funding this study through a grant provided as part of their scheme (grant number 2020-7063/CMB-BMS) to support the research associate position.References
- D. W. Domaille, E. L. Que and C. J. Chang, Synthetic fluorescent sensors for studying the cell biology of metals, Nat. Chem. Biol., 2008, 4(3), 168–175 CrossRef CAS PubMed.
- K. Pantopoulos, S. K. Porwal, A. Tartakoff and L. Devireddy, Mechanisms of mammalian iron homeostasis, Biochemistry, 2012, 51(29), 5705–5724 CrossRef CAS PubMed.
- J. Wang and K. Pantopoulos, Regulation of cellular iron metabolism, Biochem. J., 2011, 434(3), 365–381 CrossRef CAS PubMed.
- F. Bou-Abdallah, S. Adinolfi, A. Pastore, T. M. Laue and N. D. Chasteen, Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli, J. Mol. Biol., 2004, 341(2), 605–615 CrossRef CAS PubMed.
- C. Pastore, M. Franzese, F. Sica, P. Temussi and A. Pastore, Understanding the binding properties of an unusual metal-binding protein− a study of bacterial frataxin, FEBS J., 2007, 274(16), 4199–4210 CrossRef CAS PubMed.
- D. R. Richardson, D. J. Lane, E. M. Becker, M. L. H. Huang, M. Whitnall, Y. S. Rahmanto, A. D. Sheftel and P. Ponka, Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(24), 10775–10782 CrossRef CAS PubMed.
- J. Gao, Y. Chen, Z. Guo and W. He, Recent endeavors on molecular imaging for mapping metals in biology, Biophys. Rep., 2020, 6, 159–178 CrossRef PubMed.
- Y. Lu, G. Ruan, W. Du, J. Li, N. Yang, Q. Wu, L. Lu, C. Zhang and L. Li, Recent progress in rational design of fluorescent probes for Fe2+ and bioapplication, Dyes Pigm., 2021, 190, 109337 CrossRef CAS.
- S. Sundararajan and H. Rabe, Prevention of iron deficiency anemia in infants and toddlers, Pediatr. Res., 2021, 89(1), 63–73 CrossRef PubMed.
- I. C. Macdougall, Iron supplementation in nephrology and oncology: what do we have in common?, Oncologist, 2011, 16(3), 25–34 CrossRef PubMed.
- R. P. van Swelm, J. F. Wetzels and D. W. Swinkels, The multifaceted role of iron in renal health and disease, Nat. Rev. Nephrol., 2020, 16(2), 77–98 CrossRef CAS PubMed.
- M. G. Lee, S. J. Cho, J. K. Yang, H. K. Song and S. W. Suh, Crystallization and preliminary X-ray crystallographic analysis of Escherichia coli CyaY, a structural homologue of human frataxin, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2000, 56(7), 920–921 CrossRef CAS PubMed.
- V. K. Gupta, A. K. Jain, S. Agarwal and G. Maheshwari, An iron (III) ion-selective sensor based on a μ-bis (tridentate) ligand, Talanta, 2007, 71(5), 1964–1968 CrossRef CAS PubMed.
- M. A. Farrukh, N. Siraj and I. I. Naqvi, Comparative study of spectroscopic techniques for the estimation of iron in apple and vegetables, J. Saudi Chem. Soc., 2010, 14(2), 209–212 CrossRef CAS.
- J. L. Vinkenborg, S. M. van Duijnhoven and M. Merkx, Reengineering of a fluorescent zinc sensor protein yields the first genetically encoded cadmium probe, Chem. Commun., 2011, 47(43), 11879–11881 RSC.
- S. I. Bidichandani and M. B. Delatycki, Friedreich Ataxia. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20301458, 2017.
- A. B. Uceda, J. Donoso, J. Frau, B. Vilanova and M. Adrover, Frataxins emerge as new players of the intracellular antioxidant machinery, Antioxidants, 2021, 10(2), 315 CrossRef CAS PubMed.
- R. Evstatiev and C. Gasche, Iron sensing and signalling, Gut, 2011, gut-2010 Search PubMed.
- M. Nair, S. Adinolfi, C. Pastore, G. Kelly, P. Temussi and A. Pastore, Solution structure of the bacterial frataxin ortholog, CyaY: mapping the iron binding sites, Structure, 2004, 12(11), 2037–2048 CrossRef CAS PubMed.
- D. M. Charron and G. Zheng, Nanomedicine development guided by FRET imaging, Nano Today, 2018, 18, 124–136 CrossRef CAS.
- X. Zhang, Y. Hu, X. Yang, Y. Tang, S. Han, A. Kang, H. Deng, Y. Chi, D. Zhu and Y. Lu, FÖrster resonance energy transfer (FRET)-based biosensors for biological applications, Biosens. Bioelectron., 2019, 138, 111314 CrossRef CAS PubMed.
- J. Zhang, R. E. Campbell, A. Y. Ting and R. Y. Tsien, Creating new fluorescent probes for cell biology, Nat. Rev. Mol. Cell Biol., 2002, 3(12), 906–918 CrossRef CAS PubMed.
- J. C. Ewald, S. Reich, S. Baumann, W. B. Frommer and N. Zamboni, Engineering genetically encoded nanosensors for real-time in vivo measurements of citrate concentrations, PLoS One, 2011, 6(12), e28245 CrossRef CAS PubMed.
- E. S. Potekhina, D. Y. Bass, I. V. Kelmanson, E. S. Fetisova, A. V. Ivanenko, V. V. Belousov and D. S. Bilan, Drug screening with genetically encoded fluorescent sensors: Today and tomorrow, Int. J. Mol. Sci., 2020, 22(1), 148 CrossRef PubMed.
- R. Bano, N. Soleja and M. Mohsin, Genetically Encoded FRET-Based Nanosensor for Real-Time Monitoring of A549 Exosomes: Early Diagnosis of Cancer, Anal. Chem., 2023, 95(13), 5738–5746 CrossRef CAS PubMed.
- M. S. Cyert and C. C. Philpott, Regulation of cation balance in Saccharomyces cerevisiae, Genetics, 2013, 193(3), 677–713 CrossRef CAS PubMed.
- Z. Zhang, X. Cheng, Y. Zhao and Y. Yang, Lighting up live-cell and in vivo central carbon metabolism with genetically encoded fluorescent sensors, Annu. Rev. Anal. Chem., 2020, 13, 293–314 CrossRef CAS PubMed.
- S. John, G. Calmettes, S. Xu and B. Ribalet, Real-time resolution studies of the regulation of pyruvate-dependent lactate metabolism by hexokinases in single cells, PLoS One, 2023, 18(11), e0286660 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00091a |
| This journal is © The Royal Society of Chemistry 2024 |







