Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
QN-302 demonstrates opposing effects between i-motif and G-quadruplex DNA structures in the promoter of the S100P gene†
Effrosyni
Alexandrou
 ,
Dilek
Guneri
,
Dilek
Guneri
 ,
Stephen
Neidle
,
Stephen
Neidle
 * and
Zoë A. E.
Waller
* and
Zoë A. E.
Waller
 *
*
School of Pharmacy, University College London, 29-39 Brunswick Square, London, WC1N 1AX, UK. E-mail: s.neidle@ucl.ac.uk; z.waller@ucl.ac.uk
First published on 6th November 2023
Abstract
GC-rich sequences can fold into G-quadruplexes and i-motifs and are known to control gene expression in many organisms. The potent G-quadruplex experimental anticancer drug QN-302 down-regulates a number of cancer-related genes, in particular S100P. Here we show this ligand has strong opposing effects with i-motif DNA structures and is one of the most potent i-motif destabilising agents reported to date. QN-302 down-regulates the expression of numerous cancer-related genes by pan-quadruplex targeting. QN-302 exhibits exceptional combined synergistic effects compared to many other G-quadruplex and i-motif interacting compounds. This work further emphasises the importance of considering G-quadruplex and i-motif DNA structures as one dynamic system.
Promoter sequences within cancer-related genes frequently contain repeats of short G-tracts that can fold into higher-order quadruplex structures under appropriate conditions.1–5 Their complementary C-tract strands can also fold into i-motif arrangements.6,7 Stabilization of these structures by appropriate small-molecule compounds can result in transcriptional inhibition, and ultimately to cancer cell death.1,5 Several thousand such compounds have been described, and some show promise as potential drug candidates.8–12 We have developed several series of substituted naphthalene diimide derivatives,13–15 and the most recent, QN-302 (Fig. 1), shows high potency in cell growth inhibition assays, favourable pharmacological properties and antitumour activity in several in vivo cancer models.16 The transcriptional profile in cancer cells of genes down-regulated by QN-302 is in accordance with the hypothesis that it is a pan-quadruplex stabilising agent, affecting genes in several important cancer-related pathways.16 However, to date it cannot be excluded that QN-302 also stabilises i-motif structures formed on the complementary C-rich strand of G-quadruplex sequences. The present study addresses this issue with a major gene target as an exemplar.
QN-302 is currently in clinical development with Qualigen Therapeutics Inc. It has been granted Orphan Drug Designation status for the treatment of pancreatic cancer, clearance has been granted by the FDA in the USA to proceed to clinical trials for human cancers, which are now underway.
QN-302 down-regulates the expression of a number of significant cancer-related genes,16 including the S100P gene in cancer cells and in a xenograft model of pancreatic cancer.17 This gene codes for a small (10.4 kDa) calcium-binding protein and is highly upregulated in 70% of human pancreatic cancer patients, correlating with disease status.18,19 The S100P protein has been proposed as a plausible biomarker for diagnostic purposes and as a therapeutic target in pancreatic cancer.20,21 The S100P promoter22 contains a C-rich sequence containing four C-tracts on the coding strand, 48 nucleotides upstream from the transcription start site. The complementary four G-tract sequence on the template strand forms a stable G-quadruplex, which is further stabilised by QN-302.17 Here we report on the biophysical characterisation and comparison of both the G-rich and C-rich sequences from the promoter region of S100P.
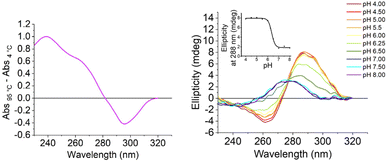
We initially characterised both the C-rich [5′-TCCCAACCCCACTGTCCCACCCT-3′] and G-rich [5′- AGGGTGGGACAGTGGGGTTGGGA-3′] sequences from the promoter region of the S100P gene. All experiments were performed in 10 mM lithium cacodylate and 100 mM KCl. The G-quadruplex forming sequence was examined at pH 7.0 and the i-motif at between pHs 4.0 and 8.0. UV melting and annealing experiments showed that the G-rich sequence had a Tm of 75.0 ± 0.2 °C and a Ta of 73.3 ± 0.7 °C at pH 7.0 (Fig. S1†). This is consistent with our previous CD experiments indicating that the G-quadruplex structure formed would be highly stable under physiological conditions.17 The complementary C-rich sequence had a Tm of 45.4 ± 0.6 °C and a Ta of 43.0 ± 0.0 °C at pH 5.5 (Fig. S2†). This Tm is similar to the melting temperature of other i-motifs of this length with three-cytosine long tracks at the same pH.23 UV thermal difference spectroscopy on the C-rich sequence showed positive peaks at 240 and 265 nm and a negative peak at 295 nm (Fig. 2, left), consistent with i-motif structure.24 Circular dichroism studies at acidic pH gave a spectrum with a positive peak at 288 nm and a negative peak at 260 nm, which is also consistent with an i-motif structure25 (Fig. 2, right). The i-motif forming sequence was found to have a transitional pH (pHT) of 6.4, which indicates that this C-rich sequence can form an i-motif at near-neutral pH.23,26
QN-302 is one of the most potent G-quadruplex binding ligands reported to date with a Kd of 4.9 nM for the G-quadruplex forming sequence from hTERT.15 It was previously shown to stabilise the G-quadruplex from the promoter region of S100P with ΔTm values of 7.4 ± 0.2 °C at 10 μM (1 eq.), 17.0 ± 0.1 °C at 20 μM (2 eq.) and 20.0 ± 1.3 °C at 50 μM (5 eq.) (Table 1).17 These data indicates that QN-302 has a strong stabilising effect on the G-quadruplex structure formed.
| [QN-302] μM | ΔTm (°C) | ΔTm (°C) |
|---|---|---|
| S100P G – quadruplex17 | S100P i – motif | |
| 10 | 7.4 ± 0.2 | −6.5 ± 1.7 |
| 20 | 17.0 ± 0.1 | −14.3 ± 0.1 |
| 50 | 20.0 ± 1.3 | −20.7 ± 1.1 |
We then focused in detail on the effects of QN-302 on the C-rich sequence from the S100P promoter. ΔTm values in the presence of QN-302 were determined in 10 mM lithium cacodylate, 100 mM KCl at pH 5.5, where the S100P sequence would be fully folded (Fig. 2). At 10 μM (1 eq.) of QN-302 the ΔTm values were found to be −6.5 ± 1.7 °C, −14.3 ± 0.1 °C at 20 μM (2 eq.) and −20.7 ± 1.1 °C at 50 μM (5 eq.), demonstrating a dose-dependent destabilisation of i-motif structure by QN-302 (Fig. 3, Table 1 and Fig. S4†). Other known G-quadruplex ligands such as berberine, BRACO-19, Phen-DC3, pyridostatin, RHPS4 and TmPyP4 have previously been shown to destabilise i-motifs, but to a lesser extent.27–29 For example, BRACO-19 has a ΔTm values of −7.3 ± 0.7 °C for the i-motif forming sequence from the promoter region of the DAP gene28 and −13.4 ± 0.5 °C for the i-motif from the human telomere. These ΔTm values are significantly smaller compared to our observations with QN-302. Di Porzio, Galli et al. have synthesised bis-triazolyl-pyridine derivatives that appear to have highly destabilising effects on the c-Myc and the hTelo i-motifs with ΔTm values of up to −29 ± 1 °C in one case. However, this destabilisation was achieved with double the number of ligand equivalents (10 molar equivalents) in phosphate buffer at pH 5.0.29 These ligands did not have the same high stabilising effect on their respective G-quadruplexes as we observe with QN-302. To the best of our knowledge QN-302 is one of the most potent destabilising agents for i-motifs reported to date. Highly destabilising activity was also observed when QN-302 was tested against the i-motif forming sequences from the human genome including the telomeric sequence (hTelo), the insulin linked polymorphic region (ILPR) and the promoter region of DAP (Fig. S5–S7 and Table S1†).
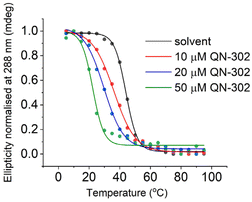 | ||
| Fig. 3 Representative CD melting experiments with 10 μM S100P i-motif in 10 mM lithium cacodylate 100 mM KCl buffer (pH 5.5), and 0, 10, 20 or 50 μM QN-302, as indicated. | ||
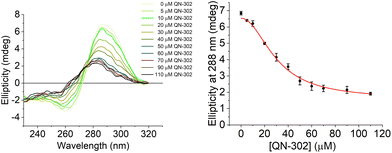
To further investigate the destabilising effects of the S100P i-motif by QN-302 CD titrations were performed (Fig. 4). Upon addition of QN-302 at a concentration range from 0 to 110 μM, the CD signal intensity at 288 nm was found to decrease in a dose-dependent fashion until a point at ∼50 μM beyond which no further reduction in the ellipticity was observed. The decrease in the CD signal suggested ligand-dependent disruption of the S100P i-motif, consistent with unfolding of the structure to a single strand. This agrees with the CD melting experiments showing destabilisation. A plot of ellipticity against QN-302 concentration gave a sigmoidal-shaped curve (Fig. 4), indicative of a cooperative unfolding effect. By fitting the sigmoidal-shaped curves to the Hill 1 equation using Origin software, we obtained Hill coefficients (n) of 2.3 ± 0.2. This reveals that the binding of QN-302 exhibits positive cooperativity (n > 1) for the S100P i-motif. Additionally, the concentration of QN-302 that is required to reach 50% reduction of the molar ellipticity was determined to be 31 ± 4 μM. Analogous CD titration experiments with the G-quadruplex forming sequence showed no significant changes in topology (Fig. S3†). Indicating that the ligand is G-quadruplex stabilising and has both i-motif destabilising and unfolding properties. Taken together, the biophysical data illustrate the dynamic interplay of the two higher order DNA structures.
To further compare the affinity of QN-302 for the S100P i-motif and G-quadruplexes, UV titrations were performed (Fig. S8–S13 and Table S2†) and the dissociation constants (Kd) were determined. The Kd for the G-quadruplex (Kd = 2.0 ± 0.3 μM) was found to be about six times lower than for the i-motif (Kd = 11.7 ± 2.9 μM), indicating that QN-302 has higher affinity for G-quadruplex compared to i-motif. This was not unexpected, given QN-302 was designed to target G-quadruplex structures.
Numerous studies have shown that high expression of the S100P gene is correlated with pancreatic cancer progression in humans.18–21 The proposed mode of action involves the stabilisation of the G-quadruplex sequence in the promoter.17,22 This stabilisation would inhibit transcription factor binding and the progression of RNA polymerase, resulting in direct downregulation of S100P gene expression at the transcriptional level analogous to other ligands such as pyridostatin.16,17,30 In this study we have further examined the mechanistic details of QN-302 interacting with the higher-order structures that can be formed in this promoter region of the S100P gene and in particular have examined the potential role of the i-motif formed by the C-rich strand. QN-302 has a strong destabilising effect on the S100P i-motif as it is illustrated by CD melting and titration experiments. Therefore, QN-302 by stabilizing the G-quadruplex structure and destabilizing the i-motif structure, has a dual role and may exert a synergistic effect on the inhibition of transcription of the S100P gene. Ligand-induced G-quadruplex stabilization inhibits gene expression whereas stabilization of i-motifs could activate transcription.31,32 This highlights the importance of evaluating the effects of a compound on both the i-motifs and the G-quadruplexes potentially formed from a duplex region of appropriate sequence. This is particularly important given the fact that G-quadruplex and i-motif formation in cells are interdependent.33 In the case of S100P, there is biological evidence that QN-302 can switch off gene expression17 which may be a consequence of both the stabilisation of the G-quadruplex and the destabilisation of the i-motif. This suggests how these two alternative structures operate together in the S100P promoter.
The findings reported here demonstrate that QN-302 both strongly stabilizes the S100P promoter G-quadruplex and strongly destabilizes the complementary i-motif in vitro. These data are consistent with and supportive of previous conclusions16 that QN-302 down-regulates the expression of numerous cancer-related genes by pan-quadruplex targeting. This is particularly important given the recent analysis of TCGA PanCancer Atlas PDAC datasets that indicate poor prognosis in patients with high S100P expression.34 QN-302 exhibits exceptional combined synergistic effects compared to many other G-quadruplex and i-motif interacting compounds. Overall, this work further emphasises the importance of considering these two alternative DNA structures as one dynamic system and as one target.
Data availability
Data is available on Figshare: 10.6084/m9.figshare.24476551.Author contributions
S. N. and Z. A. E. W. conceived the study, E. A, D. G. and Z. A. E. W. designed the experiments E. A. and D. G. performed the experiments E. A., D. G., S. N. and Z. A. E. W. wrote the paper, contributed to the manuscript revision, and approved the final version.Conflicts of interest
S. Neidle is a paid consultant and Advisory Board member of Qualigen Inc.Acknowledgements
D. G. is supported by the Biotechnology and Biological Sciences Research Council grant BB/W001616/1.References
- A. Siddiqui-Jain, C. L. Grand, D. J. Bearss and L. H. Hurley, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11593–11598 CrossRef CAS.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2007, 35, 406–413 CrossRef CAS PubMed.
- S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261–275 CrossRef CAS PubMed.
- K. Shin, N. A. Chapman, M. Sarker, C. Kenward, S. K. Huang, N. Weatherbee-Martin, A. Pandey, D. J. Dupré and J. K. Rainey, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 1901–1912 CrossRef CAS PubMed.
- S. Lago, M. Nadai, F. M. Cernilogar, M. Kazerani, H. Domíniguez Moreno, G. Schotta and S. N. Richter, Nat. Commun., 2021, 12, 3885 CrossRef CAS PubMed.
- S. L. Brown and S. Kendrick, Pharmaceuticals, 2021, 14, 96 CrossRef CAS PubMed.
- K. L. Irving, J. J. King, Z. A. E. Waller, C. W. Evans and N. M. Smith, Biochimie, 2022, 198, 33–47 CrossRef CAS.
- S. Neidle, J. Med. Chem., 2016, 59, 5987–6011 CrossRef CAS PubMed.
- H. Xu and L. H. Hurley, Bioorg. Med. Chem. Lett., 2022, 77, 129016 CrossRef CAS PubMed.
- E. Mendes, I. M. Aljnadi, B. Bahls, B. L. Victor and A. Paulo, Pharmaceuticals, 2022, 15, 300 CrossRef CAS PubMed.
- E. Ruggiero and S. N. Richter, Bioorg. Med. Chem. Lett., 2023, 79, 129085 CrossRef CAS PubMed.
- A. Heine, S. Juranek and P. Brossart, Mol. Cancer, 2021, 20, 52 CrossRef CAS.
- M. Micco, G. W. Collie, A. G. Dale, S. A. Ohnmacht, I. Pazitna, M. Gunaratnam, A. P. Reszka and S. Neidle, J. Med. Chem., 2013, 56, 2959–2974 CrossRef CAS PubMed.
- C. Marchetti, K. G. Zyner, S. A. Ohnmacht, M. Robson, S. M. Haider, J. P. Morton, G. Marsico, T. Vo, S. Laughlin-Toth, A. A. Ahmed, G. Di Vita, I. Pazitna, M. Gunaratnam, R. J. Besser, A. C. G. Andrade, S. Diocou, J. A. Pike, D. Tannahill, R. B. Pedley, T. R. J. Evans, W. D. Wilson, S. Balasubramanian and S. Neidle, J. Med. Chem., 2018, 61, 2500–2517 CrossRef CAS.
- T. Vo, S. Oxenford, R. Angell, C. Marchetti, S. A. Ohnmacht, W. D. Wilson and S. Neidle, ACS Med. Chem. Lett., 2020, 11, 991–999 CrossRef CAS.
- A. A. Ahmed, R. Angell, S. Oxenford, J. Worthington, N. Williams, N. Barton, T. G. Fowler, D. E. O'Flynn, M. Sunose, M. McConville, T. Vo, W. D. Wilson, S. A. Karim, J. P. Morton and S. Neidle, ACS Med. Chem. Lett., 2020, 11, 1634–1644 CrossRef CAS PubMed.
- A. A. Ahmed, W. Greenhalf, D. H. Palmer, N. Williams, J. Worthington, T. Arshad, S. Haider, E. Alexandrou, D. Guneri, Z. A. E. Waller and S. Neidle, Molecules, 2023, 28, 2452 CrossRef CAS PubMed.
- F. Prica, T. Radon, Y. Cheng and T. Crnogorac-Jurcevic, Am. J. Cancer Res., 2016, 6, 562–576 CAS.
- Y. Wu, Q. Zhou, F. Guo, M. Chen, X. Tao and D. Dong, Front. Oncol., 2021, 11, 711180 CrossRef CAS.
- N. Ideno, Y. Mori, M. Nakamura and T. Ohtsuka, Diagnostics, 2020, 10, 1056 CrossRef CAS PubMed.
- W. Zou, L. Li, Z. Wang, N. Jiang, F. Wang, M. Hu and R. Liu, Bioengineered, 2021, 12, 9006–9020 CrossRef CAS PubMed.
- A. Gibadulinova, I. Oveckova, S. Parkkila, S. Pastorekova and J. Pastorek, Oncol. Rep., 2008, 20, 391–396 CAS.
- E. P. Wright, J. L. Huppert and Z. A. E. Waller, Nucleic Acids Res., 2017, 45, 2951–2959 CrossRef CAS PubMed.
- J.-L. Mergny, J. Li, L. Lacroix, S. Amrane and J. B. Chaires, Nucleic Acids Res., 2005, 33, e138–e138 CrossRef.
- J. Kypr, I. Kejnovská, D. Renciuk and M. Vorlícková, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- J. Zhou, C. Wei, G. Jia, X. Wang, Z. Feng and C. Li, Mol. BioSyst., 2010, 6, 580–586 RSC.
- A. Pagano, N. Iaccarino, M. A. S. Abdelhamid, D. Brancaccio, E. U. Garzarella, A. Di Porzio, E. Novellino, Z. A. E. Waller, B. Pagano, J. Amato and A. Randazzo, Front. Chem., 2018, 6, 281 CrossRef.
- M. A. S. Abdelhamid, A. J. Gates and Z. A. E. Waller, Biochemistry, 2019, 58, 245–249 CrossRef CAS.
- A. Di Porzio, U. Galli, J. Amato, P. Zizza, S. Iachettini, N. Iaccarino, S. Marzano, F. Santoro, D. Brancaccio, A. Carotenuto, S. De Tito, A. Biroccio, B. Pagano, G. C. Tron and A. Randazzo, Int. J. Mol. Sci., 2021, 22, 11959 CrossRef CAS PubMed.
- J. Spiegel, S. M. Cuesta, S. Adhikari, R. Hänsel-Hertsch, D. Tannahill and S. Balasubramanian, Genome Biol., 2021, 22, 117 CrossRef CAS PubMed.
- H. J. Kang, S. Kendrick, S. M. Hecht and L. H. Hurley, J. Am. Chem. Soc., 2014, 136, 4172–4185 CrossRef CAS PubMed.
- S. Kendrick, H. J. Kang, M. P. Alam, M. M. Madathil, P. Agrawal, V. Gokhale, D. Yang, S. M. Hecht and L. H. Hurley, J. Am. Chem. Soc., 2014, 136, 4161–4171 CrossRef CAS.
- J. J. King, K. L. Irving, C. W. Evans, R. V. Chikhale, R. Becker, C. J. Morris, C. D. Peña Martinez, P. Schofield, D. Christ, L. H. Hurley, Z. A. E. Waller, K. S. Iyer and N. M. Smith, J. Am. Chem. Soc., 2020, 142, 20600–20604 CrossRef CAS.
- K. Srivastava, K. E. Lines, D. Jach and T. Crnogorac-Jurcevic, Oncongene, 2023 DOI:10.1038/s41388-023-02851-y.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, supporting UV melting, CD melting and CD titrations. See DOI: https://doi.org/10.1039/d3ob01464a |
| This journal is © The Royal Society of Chemistry 2024 |