Open Access Article
Open Access ArticleRecent advances in DNA-based electrogenerated chemiluminescence biosensors
Jingjing
Zhang
ab,
Jingfeng
Zhu
a and
Jie
Chao
 *ab
*ab
aKey Laboratory for Organic Electronics and Information Displays, Jiangsu Key Laboratory for Biosensors, Institute of Advanced Materials (IAM), Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts and Telecommunications, Nanjing 210023, China. E-mail: iamjchao@njupt.edu.cn
bSchool of Geographic and Biologic Information, Nanjing University of Posts and Telecommunications, Nanjing 210023, China
First published on 14th March 2023
Abstract
Electrogenerated chemiluminescence (also called electrochemiluminescence and abbreviated as ECL) has witnessed significant development in fields ranging from biological analysis to clinical diagnosis due to its outstanding advantages such as high sensitivity, wide detection range, and low background signal. DNA, a class of biopolymers with programmable self-assembly and molecular recognition functions, is a significant tool in the field of ECL bioanalysis. Benefiting from the unique structures and excellent properties of DNA, DNA-based ECL biosensors have aroused increasing attention. In this review, we summarize and classify the signal output mode of DNA-based ECL biosensors and introduce the different methods for the immobilization of DNA probes on electrodes. In addition, we discuss the recent progress and applications of DNA-based ECL biosensors, including the detection of nucleic acids, proteins, and small biomolecules. Finally, we present a brief summary and prospects in this field.
1. Introduction
In biosensing, the issue of selectivity, especially for low-abundant targets and in the presence of interfering substances is critical.1–3 Therefore, the development of biosensors with high selectivity and ease of operation can overcome this issue during sensing. In the 1960s, Clark and Lyons invented the first biosensor for the electrochemical measurement of glucose in biological samples and defined the basic concept of biosensors.4 Since then, biosensors have been rapidly applied in many fields, including analytical chemistry, medical diagnosis, environment monitoring, and food safety inspection.5–7 A biosensor is “a set of stand-alone integrated devices that provide specific quantitative and semi-quantitative analytical information by placing biometric elements in direct contact with transducers”.8,9 It is mainly composed of two parts, as follows: (i) bioactive materials, also known as molecular recognition elements, which determine the selectivity of the biosensor, and (ii) transducer, which can be physical or chemical and converts physical or chemical changes produced by various active substances into electrical signals.10–12 Thus, by matching the molecular recognition elements with appropriate transducers, the developed biosensors can show good analytical performances. With the assistance of the transducer, the relevant changes caused by the target are output as the related electrical signal (including capacitance, resistance, potential or current, etc.).13,14 The higher the concentration of the target, the higher/lower the electrical signal. Based on this principle, the quantitative analysis of the target can be realized. Biosensors combine the specificity and sensitivity of biological systems and the computational power of microprocessors, which have the characteristics of high specificity, fast speed, simple operation, and small sample preprocessing.15–17ECL biosensors are analytical technology that take biomolecules such as proteins, DNA/RNA, small molecules, and ions as targets and converts the signals generated during their specific recognition into optical signals for the quantitative detection of the targets.18 Compared with other biosensors, ECL biosensors have outstanding advantages, such as high sensitivity, low background signal, and high controllability.19–21 Thus, they are reliable platforms for detecting important disease markers with low abundance, which can be used for early disease diagnosis and prognostic monitoring.22–24 Since the 21st century, ECL biosensors have successfully entered the limelight and developed quickly. To build superior ECL biosensing systems, numerous types of materials have been intensively developed and applied.25
As one of the most attractive biological polymers, DNA has great potential in the field of biological analysis. In addition to being the carrier of genetic information and responsible for the conversion of genetic code into proteins, it can also be used as a functional material for bioanalysis, DNA nanotechnology, and nanomedicine.26–28 In recent years, DNA-based ECL biosensors have attracted significant attention for the ultra-sensitive detection of biomolecules.29–31 This type of biosensor typically uses DNA as a molecular recognition element to capture the target in the sample.32 By modifying single-stranded DNA (ssDNA) or DNA self-assembled nanostructured probe on the surface of the electrode, the DNA probe molecules can selectively hybridize with the target under the appropriate conditions and convert the generated biological signals into detectable chemical signals, thus achieving the purpose of target detection.33 With the advancement of nanotechnology, ingenious signal amplification strategies and nano-functional materials have been gradually developed and various DNA-based ECL biosensors manufactured with improved sensitivity and selectivity.
Herein, we review the latest research on DNA-based ECL biosensors in the past few years, focusing on their main signal output modes and the methods for the immobilization of DNA probes during the construction of ECL biosensors. In addition, the recent progress on DNA-based ECL biosensors in the field of biological analysis is highlighted according to the classification of the targets (Scheme 1). Due to the proliferation of related literature in recent years, this review only briefly introduces some of these studies as references as needed.
2. Signal output mode of DNA-based ECL biosensors
With the rapid development of DNA and the ECL technique, researchers have realized a variety of signal output modes for ECL biosensors, including single signal output mode and multi-signal switching mode.34 Generally, most ECL systems are based on a single signal output mode. The single signal output mode typically includes the “signal on” and “signal off” modes. In the “signal on” mode, the ECL signal usually increases with an increase in the analyte concentration, while the “signal off” mode is the opposite. The single signal output mode has certain disadvantages in ECL analysis, for example, it is not conducive to the stability, accuracy and efficiency. Alternatively, multi-signal mode ECL systems can avoid most of the defects of the single signal output mode, making ECL analysis more flexible and accurate. Generally, the multi-signal switching mode is used for the analysis of more than two analytes, and the ECL signal changes with the analytes concentration in real time, resulting in more complex signal changes. In this section, the different signal output modes of DNA-based ECL biosensors are categorized and summarized, and their applications in the field of bioanalysis are discussed and explained.2.1. “Signal on” mode
In DNA-based ECL biosensors, the “signal on” mode is a signal process from zero or weak to strong and is a widely researched ECL signal output mode, which has become an effective method in the field of biological analysis, disease diagnosis, etc.35–37 There are various conventional means to achieve the “signal on” strategy, including the introduction of luminescent substances, co-reaction reagents or co-reaction promoters and the application of ECL resonance energy transfer (ECL-RET) systems.Common luminescent materials such as ruthenium (Ru) complexes, luminol and its derivatives, and quantum dots have been used in biosensors. Recently, Peng et al.38 modified a luminous Ru complex on the electrode surface as an emitter for signal generation and achieved precise modulation of signal quenching to signal enhancement in a chain substitution reaction-based system by introducing DNA nanoscissors as a control structure (Fig. 1A). In the presence of the target miRNA, a strong ECL emission was obtained. Liu et al.39 successfully synthesized sulfur quantum dots (SQDs) as a signal emitter via the hydrothermal method. In combination with a three-stranded DNA (tsDNA) nanostructure and duplex-specific nuclease (DSN)-assisted target cycle amplification signal strategy, the ultra-sensitive detection of miRNA-21 was achieved (Fig. 1B). Compared with the quantum dots containing heavy metal elements, pure elemental quantum dot materials such as SQDs are suitable for the construction of ECL biosensors due to their advantages of low toxicity, good water solubility, high biocompatibility and outstanding ECL properties. This work extends the application of pure elemental quantum dots in ECL luminescent materials and provides a new direction for the construction of novel quantum dot-based ECL biosensors.
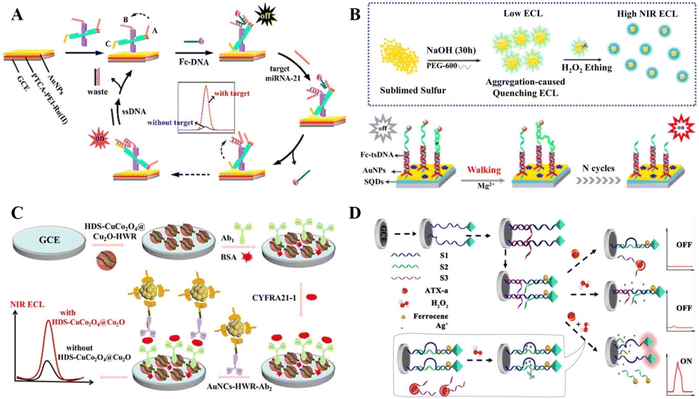 | ||
| Fig. 1 (A) Schematic illustration of a DNA nanoscissor-based ECL biosensor. Reprinted with permission from ref. 38. Copyright 2019, the American Chemical Society. (B) Schematic illustration of a novel ECL biosensor based on SQDs for the detection of miRNA-21. Reprinted with permission from ref. 39. Copyright 2020, the American Chemical Society. (C) Schematic illustration of an ECL biosensor based on HDS-CuCo2O4@Cu2O co-reactant accelerator. Reprinted with permission from ref. 40. Copyright 2022, the American Chemical Society. (D) Schematic illustration of a visual ECL biosensor for the detection of Pb2+. Reprinted with permission from ref. 41. Copyright 2018, the American Chemical Society. | ||
In the co-reactant ECL system, the co-reactant undergoes electrochemical oxidation or reduction to form a reducing or oxidizing intermediate, which can react with an oxidized or reduced ECL probe to generate excited states. The greater the generation of the intermediate of the co-reactant, the higher the ECL efficiency, thus producing a higher signal. The classical co-reactant systems such as Ru complex-TPrA and luminol–hydrogen peroxide have been widely used in the field of ECL analysis. Nowadays, new ECL luminescent materials and their co-reactant reagents have been developed with the deepening of research. Gold nanoclusters (AuNCs) are a class of ECL clusters, which exhibit the quantum size effect and excellent fluorescence properties, and thus attracted extensive attention from researchers in recent years. Jia et al.40 first prepared a hollow double-shell CuCo2O4@Cu2O (HDS-CuCo2O4@Cu2O) heterostructure, which can be used as a co-reactant accelerator to significantly enhance the luminescence of AuNCs in the ECL reaction (Fig. 1C). HDS-CuCo2O4@Cu2O is involved in the ECL signal generation process mainly by obtaining more electrons from TPIA through holes, which promotes the formation of Triisopropanolamine (TPIA)˙+. The possible mechanism of signal amplification is as follows:
| AuNCs − e− → AuNCs˙+ | (1) |
| TPIA + h+ → (HDS-CuCo2O4@Cu2O)TPIA˙+ | (2) |
| TPIA˙+ → TPIA˙ + H+ | (3) |
| AuNCs˙+ + TPIA˙ → AuNCs* | (4) |
| AuNCs* → AuNCs + hv (830 nm) | (5) |
In addition, the precise and effective introduction of the ECL-RET effect is also an effective strategy for the development of “signal on” mode ECL biosensors. The ECL-RET effect usually occurs when the emission spectrum of a donor molecule overlaps with the absorption spectrum of another acceptor molecule. The excitation of the donor molecule transfers energy to the acceptor molecule within a certain distance, inducing the absorption of energy by the acceptor molecule to emit light, while the intensity of the donor molecule itself decays. The conventional ECL-RET system usually faces the problem of low effective contact frequency between the donor and acceptor in the solution, resulting in a low quenching efficiency. Xia et al.41 designed a DNA-regulated ECL-RET probe, which can achieve dual quenching and dual stimulation response, with ultra-low background signal and highly strong signal output, to achieve the ultra-sensitive detection of anatoxin-a (Fig. 1D).
As a typical single-signal output mode, the signal of the ECL biosensor in the “signal on” mode is gradually enhanced with an increase in the analyte concentration. This is the most commonly used signal output mode with a wide detection range and low background signal. In future research, achieving a stable and continuous signal output, avoiding rapid decay of the ECL signal, and improving the detection sensitivity are worthy of further study.
2.2. “Signal off” mode
The ECL emission is directly quenched by the ECL quencher, and thus the ECL signal decreases with an increase in the analyte concentration, which is the basic principle of “signal off” mode ECL biosensors.42–44By cleverly designing and amplifying the ECL response of the initial state, the “signal off” effect of the sensor can be effectively improved.45 This provides a new idea for the further development of ultra-sensitive “signal off” mode DNA-based ECL biosensors. Recently, Wang et al.46 constructed an “on–off” paper-based ECL sensing platform for the highly sensitive detection of miRNA-141. The “signal on” response of this biosensor is based on a dual signal amplification strategy (Fig. 2A). Firstly, 3D graphene with a large specific surface area was introduced on the surface of a paper-based platform, and a large amount of AuPd nanoparticles (NPs) was loaded to build an excellent conductive substrate, which can participate in the ECL reaction system as co-reactants to enhance the ECL emission. Secondly, a DNA walker based on Nt.BsmAI nucleic acid endonuclease was used to further amplify the ECL signal by hydrolyzing DNA through an enzymatic reaction to release the walker probe and excite the 3D DNA walker. The above-mentioned two enhancements caused the sensor to have a strong initial ECL signal. The quenching effect of the ECL biosensor was mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/cas12a-mediated trans-cleavage, and the CRISPR/Cas12a (LbCpf1) protein could indiscriminately digest any non-target ssDNA (called trans-cleavage), resulting in the weakest “signal off” response.
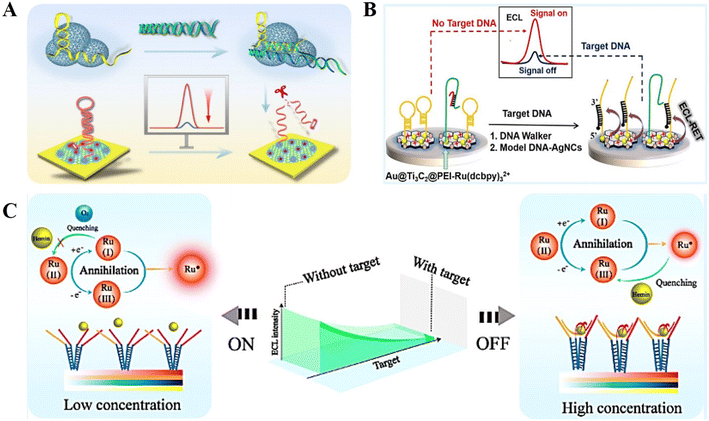 | ||
| Fig. 2 (A) Schematic illustration of an “on–off” paper-based ECL sensing platform for the highly sensitive detection of miRNA-141. Reprinted with permission from ref. 46. Copyright 2021, the American Chemical Society. (B) Schematic illustration of a “signal off” output mode ECL biosensor for the sensitive detection of SARS-CoV-2. Reprinted with permission from ref. 47. Copyright 2021, the American Chemical Society. (C) Schematic illustration of a “signal off” ECL biosensor based on DNA nanotweezer. Reprinted with permission from ref. 51. Copyright 2019, the American Chemical Society. | ||
To obtain higher sensitivity and specificity, various new nanomaterials have been introduced in ECL biosensors. Yao et al.47 successfully prepared an Au@Ti3C2@PEI–Ru(dcbpy)32+ nanocomposite, in which Ti3C2 as a class of two-dimensional transition metal compound nanomaterials with the advantages of high loading rate, good electrical conductivity, and polyethylenimine (PEI) was used as a co-reactant to enhance the signal (Fig. 2B). A DNA walker-based amplification strategy was employed to shear hairpin DNA under the action of nucleic acid endonucleases, and the designed model DNA-Ag nanoclusters (model DNA-AgNCs) were used as quenchers to quench the ECL. During the ECL reaction, Ti3C2–Ru(II) and PEI are firstly oxidized to Ti3C2–Ru(III) and PEI˙+. Then, PEI˙+ reacts with PEI˙ to reduce Ti3C2–Ru(III) to the excited-state Ti3C2–Ru(II)*. Finally, the excited Ti3C2–Ru(II)* radiates to the ground-state Ti3C2–Ru(II) and releases photons. The possible luminescence mechanism is as follows:
| Ti3C2–Ru(II) + PEI − 2e− → Ti3C2–Ru(III) + PEI˙+ | (6) |
| Ti3C2–Ru(III) + PEI˙+ → Ti3C2–Ru(III) + PEI˙ + H+ | (7) |
| Ti3C2–Ru(III) + PEI˙ → Ti3C2–Ru(II)* + PEI | (8) |
| Ti3C2–Ru(II)* → Ti3C2–Ru(II) + hv | (9) |
The rapid development of DNA nanotechnology has also laid a good foundation for the construction of novel ECL biosensors. The above-mentioned articles both involve the construction of a “signal off” output ECL biosensor with a DNA walker as the main component. In addition, DNA robots, DNA tetrahedrons, DNA dendrimers and other DNA self-assembled structures have been widely used in the design of ECL biosensors.48–50 Recently, Bian et al.51 designed a DNA nanotweezer-based ECL biosensor that enables intelligent control of hemin concentration for the ultra-sensitive detection of the early diagnostic biomarker fusion gene PML/RARα in leukemia (Fig. 2C). Depending on the principle of complementary base pairing, the switching status of the DNA nanotweezer is closely related to the presence or absence of the target. In the absence of the target, the DNA nanotweezer is open, which cannot capture the free hemin in solution, and the ECL response is in the “on” state. In the presence of the target, the DNA nanotweezer changes its configuration to capture hemin, while the high concentration of hemin on the electrode surface quenches the ECL signal and the biosensor is in the “off” state. Based on this highly intelligent and controlled DNA structure design, the proposed method exhibits excellent research value and application potential in bioanalysis.
As an effective signal output mode, the “signal-off” mode has been of increasing interest to researchers due to its high sensitivity, ease of operation, and reagent-free process. In the ongoing development of novel and efficient ECL quenchers, the design of effective steric hindrance strategy or nucleic acid-based cutting strategy will pave the way for the future “signal-off” mode ECL biosensors.
2.3. “Multiple signal switching” mode
Despite the advantages of “single on” or “single off” mode ECL biosensors, they suffer from the shortcoming of false-positive signals, which limits the application of single-switching mode ECL biosensors in practical sample detection. In recent years, a more complex output mode named “multiple signal switching” mode has attracted widespread attention. Generally, the “multiple signal switching” mode includes the “signal off–on–off” mode, “signal on–off–on” mode, and ratiometric mode.21Hu et al.52 proposed a “signal off–on–off” mode biosensing strategy for the detection of the antiviral drug amantadine (AMD) (Fig. 3A). In the construction of the biosensor, cyclic multiple changes in the ECL signal were achieved mainly based on the host–guest interaction among AMD, ferrocene (Fc), and cucurbit[7]uril (Q[7]). In the early stage, Fc quenches the Ru(bpy)32+ luminescence process on the electrode surface, which is in the “off” state with a low signal. Later, by introducing the intermediate Q[7], it will wrap Fc into its cavity, leading to the “on” state with a high signal. Finally, when AMD is added, Fc will be replaced by AMD and cause an obvious signal decrease due to the stronger binding ability between AMD and Q[7], thus achieving an “off–on–off” multiple signal response. Likewise, “signal on–off–on” mode ECL biosensors have been developed and used to identify targets with high specificity. Yang et al.53 researched a ternary ECL biosensing platform that achieved an initial high intensity ECL signal through material optimization (Fig. 3B). Subsequently, the Fc-labelled nucleic acid probe decreases the electron transfer rate at the electrode surface to obtain a low ECL response. Eventually, an enzyme-mediated strand displacement reaction is triggered in the presence of the target, resulting in signal enhancement and partial restoration of the “signal on” state. The possible ECL mechanism as described in the following eqn (10)–(17). In the solid-phase ternary ECL system, porous palladium nanosphere (pPdNS) with excellent catalytic activity was applied as the co-reaction accelerator to promote the ECL reaction of rubrene microblocks (RubMBs) as luminophores and dissolved O2 as an endogenous co-reactant.
| Rub − e− → Rub˙+ | (10) |
| O2 + e− → O2˙− | (11) |
| O2˙− + 2H2O + 2e− → ˙OH + 3OH− | (12) |
| O2 + 2H2O + 2e− → H2O2 + 2OH− | (13) |
| H2O2 + e− → OH− + ˙OH + O2 | (14) |
| O2˙− + Rub˙+ → 1R* + O2 | (15) |
| ˙OH + Rub˙+ → 1R* + H+ | (16) |
| 1R* → R + hv | (17) |
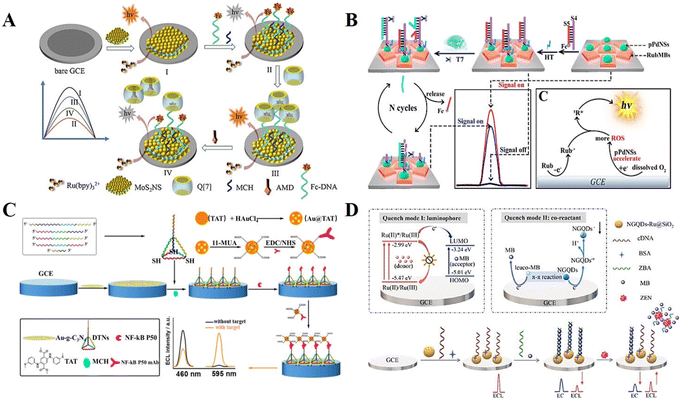 | ||
| Fig. 3 (A) Schematic illustration of a “signal off–on–off” mode ECL biosensor. Reprinted with permission from ref. 52. Copyright 2021, Elsevier. (B) Schematic illustration of a “signal on–off–on” mode ECL biosensor. Reprinted with permission from ref. 53. Copyright 2018, the American Chemical Society. (C) Schematic illustration of a dual-wavelength emission ECL-RET-based biosensor. Reprinted with permission from ref. 54. Copyright 2020, the American Chemical Society. (D) Schematic illustration of an ECL-EC ratiometric biosensor. Reprinted with permission from ref. 55. Copyright 2023, Elsevier. | ||
The “multiple signal switching” mode described above is mainly based on a single signal intensity change. During the detection process, false positive/negative errors may still occur due to instrument errors or environmental changes, such as degradation and dissociation of reagents, co-reactant concentrations, and pH value. The emergence of ratiometric ECL biosensors effectively avoids these interferences. The quantification principle of ratiometric ECL biosensors relies on the change in the ratio of two emission intensities rather than the absolute value, which is normalized by self-calibration, improving the accuracy of the detection. Fan et al.54 constructed a dual-wavelength emission ECL-RET-based biosensor, which achieved the ultrasensitive detection of nuclear factor-kappa B (NF-κB) based on the ratio change by comparing the signal emission intensity at 595 and 460 nm (Fig. 3C). Mutually independent, non-interfering dual-coupled ECL-electrochemical (EC) signal detection is also an effective method for constructing ratiometric ECL biosensors. Luo et al.55 firstly discovered the double quenching mechanism of methylene blue (MB) in nanocomposites (nitrogen-doped graphene quantum dots on Ru(bpy)32+-doped silica nanoparticles, NGQDs-Ru@SiO2) (Fig. 3D). Relying on this mechanism, an ECL-EC ratiometric biosensor was cleverly designed for zearalenone (ZEN) detection. Due to the MB-mediated quenching effect, the ECL biosensor initially possessed a high EC signal and a low ECL signal. After recognition binding with ZEN, the ECL signal was recovered and the EC signal was reduced, thus realizing the proportional detection.
Compared with the single signal output mode, the “multiple signal switching” mode undoubtedly has a more sophisticated design concept and corresponding application prospects. This intelligent signal output mode not only has the characteristics of low background signal and high sensitivity, but also can effectively eliminate false negative or positive signals and improve the accuracy, thus promoting the application of ECL biosensors in real sample analysis. In the future, it is necessary to design more “multiple signal switching” strategies to meet different application requirements, and the controllability, reproducibility and stability of the “multiple signal switching” mode in the ECL biosensors also require more attention.
3. Immobilization methods of DNA on electrodes
In the construction of DNA-based ECL biosensors, the immobilization of DNA molecules on the electrode surface is a key step.56 To ensure that the DNA probes on the electrode surface possess high activity, stability and directionality, it is necessary to properly regulate the immobilization process of DNA probes and the coverage of the electrode surface.57 Whether as a signal or capture probe, the target recognition response in typical DNA-based ECL biosensors typically occurs at the interface.31 Therefore, new immobilization strategies need to be continuously developed to improve the accessibility, sensitivity, and specificity of DNA probes and the low thermodynamic and kinetic response at the biosensing interface. The method of immobilizing the probe usually depends on the electrode material and the substrate that modifies the electrode surface.58 The commonly used probe immobilization strategies generally include four types including chemical adsorption, physical adsorption (non-covalent bonding), covalent bonding and affinity method. In this section, we briefly introduce these four common types of DNA probe immobilization and illustrate their role in the construction of DNA-based ECL biosensors.3.1. Chemical adsorption method
The most common chemical adsorption method is the self-assembly adsorption of DNA probes labelled with sulfhydryl (or sulfhydryl derivatives) on the surface of gold (silver, platinum, palladium, etc.) electrodes through gold–sulfur bonds (Au–S).59 Given that there are no stable oxides under general environmental conditions, sulfhydryl groups can be spontaneously adsorbed on the gold surface through Au–S bonds, forming a dense and highly ordered self-assembled monolayer.60 Considering the simplicity and feasibility of this immobilization method, many reports in the literature fixed sulfur-containing probe DNA on the surface of gold electrodes for the construction of DNA-based ECL biosensors. However, this method of DNA immobilization method faces the problem of non-specific adsorption of non-sulfhydryl-functionalized DNA sequences. DNA probes tend to adsorb on metal surfaces such as gold, silver, platinum, and copper through base pairs non-specifically, making the hybridization efficiency based on pairs at the interface low.61 Fortunately, some interfacial engineering techniques have been used to position DNA probes vertically on the electrode surface, as well as using short-chain alkane thiols such as 6-mercaptohexanol (MCH) to replace and seal non-specific adsorption at the interface, or the direct adoption of sulfhydryl-functionalized DNA probes to form mixed monolayers for co-assembly, effectively reducing the non-specific adsorption of sulfhydryl-functionalized DNA.62–64 Thus, chemisorption allows the single-stranded DNA probe to be firmly and gently fixed to the gold electrode surface, and also preserves the DNA conformation.Recently, a wide variety of DNA-based ECL biosensors have been developed for clinical diagnostics, chemical research, pharmaceuticals and bioassays based on chemisorption-based DNA probe immobilization. Zhang et al.65 proposed an ECL strategy for miRNA detection. Hairpin DNA was immobilized on the electrode surface through Au–S bonds, and hybridized with target cyclic amplification products ssDNA, and thus the hairpin DNA opened and acted as a capture probe to achieve the capture of DNA nanoclaws loaded with ECL signal labels (Fig. 4A). The designed ECL biosensor could achieve the highly selective detection of miRNA-21 under the condition of complex biological samples. Similarly, Zhu et al.66 used Fe3O4@AuNP nanocomplexes with hairpin DNA fixed on their surface as the capture substrate. The target miRNA hybridized with the hairpin DNA to cause conformational changes, thus enabling the detection of miRNAs (Fig. 4B). Besides, Li et al.67 successfully achieved the capture hybridization of target DNA using sulfhydrylated capture DNA attached to the surface of a glassy carbon electrode (GCE) modified with AuNPs (Fig. 4C). Moreover, Ling et al.68 achieved early screening of group B streptococci (GBS), an important pathogen at the perinatal stage, based on enzyme-free nucleic acid amplification by assembling thiol-modified capture probes on the surface of gold-modified electrodes through gold thiol bonding (Fig. 4D).
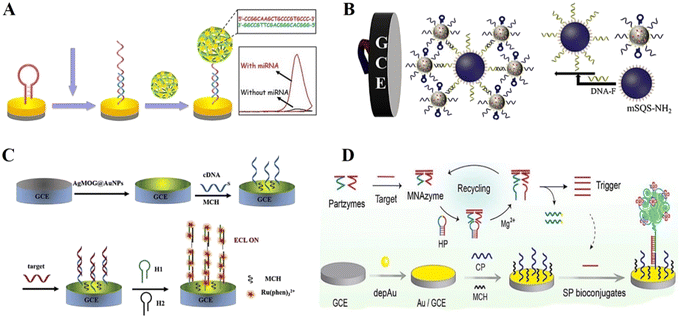 | ||
| Fig. 4 (A) Schematic illustration of an ECL biosensor in which hairpin DNA was immobilized on the electrode surface through Au–S bonds. Reprinted with permission from ref. 65. Copyright 2020, Elsevier. (B) Schematic illustration of an ECL biosensor based on Fe3O4@AuNP nanocomplexes with hairpin DNA fixed on the surface as the capture substrate by Au–S bonding. Reprinted with permission from ref. 66. Copyright 2019, Elsevier. (C) Schematic illustration of an ECL biosensor in which sulfhydrylated capture DNA probes are attached to the electrode (GCE) via Au–S bonds. Reprinted with permission from ref. 67. Copyright 2019, Elsevier. (D) Schematic illustration of an ECL biosensor based on enzyme-free nucleic acid amplification, in which thiol-modified capture probes are assembled on the electrode through gold thiol bonding. Reprinted with permission from ref. 68. Copyright 2019, Elsevier. | ||
In recent years, DNA immobilization by chemical adsorption, represented by the Au–S bond, has become the most commonly used method for probe immobilization because of its advantages of high structural order and surface binding stability. However, due to the high purity requirements of sulfhydryl DNA, the separation and purification operation are complicated, which may lead to an increase in the experimental cost. Generally, there are two methods for the chemisorption immobilization of sulfhydryl (or sulfhydryl derivatives) DNA probes on electrodes. One is to immobilize the modified probe directly on the surface of the gold electrode. The other is to immobilize the modified probe on gold nanoparticles (AuNPs) at the electrode, which can greatly increase the binding sites, thus improving the ECL response signal and reducing the detection limit.
3.2. Physical adsorption method (non-covalent bonding)
Physical adsorption (non-covalent bonding) is also a classical immobilization method in DNA-based ECL biosensors. Physical adsorption of DNA monolayers usually depends on the utilization and combination of certain features and factors, including the charge on the electrode surface, the negatively charged phosphate skeleton, the structure of the DNA, and the electrostatic/hydrophobic interactions of the bases.69Graphene, as a new material with sp2-hybridized connected carbon atoms tightly stacked into a monolayer two-dimensional honeycomb lattice structure, has excellent optical, electrical, and mechanical properties. It has important application prospects in materials science, micro and nano processing, energy, biomedicine, and drug delivery, and is considered a revolutionary material of the future.70 Nowadays, the majority of DNA-based ECL biosensor designs are limited to two-dimensional (2D) graphene, whereas 3D graphene has a larger specific surface area and higher catalytic activity, which can play a more significant role in ECL detection.71 Wu et al.72 developed a functionalized ECL sensing platform based on 3D graphene, where Ru(bpy)32+-modified DNA probes can self-assemble on the surface of the platform by non-covalent attraction and act as hybridization probes emitting strong ECL signals (Fig. 5A). When the target is present, spontaneous hybridization and complementary pairing will occur to reduce the signal, thus enabling the detection of the target DNA. Song et al.73 employed a silver orthophosphate and zirconium-based metal–organic framework (Ag3PO4-UiO-66) as an ECL donor and graphene nanosheets as an ECL acceptor for the fabrication of an ECL-RET biosensor for the highly sensitive detection of diethylstilbestrol (DES) (Fig. 5B). In the sensing platform construction, aptamer and graphene nanosheets are fixedly attached by electrostatic adsorption, which is used to control the distance between the ECL donor and acceptor to obtain the desired ECL-RET effect.
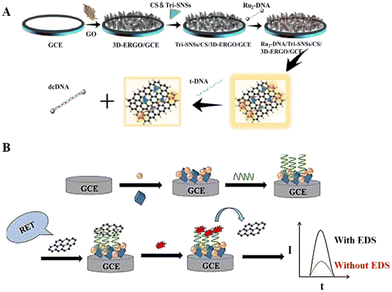 | ||
| Fig. 5 (A) Schematic illustration of a functionalized ECL biosensor based on 3D graphene in which DNA probes can self-assemble on the surface of the platform by non-covalent attraction. Reprinted with permission from ref. 72. Copyright 2018, Elsevier. (B) Schematic illustration of an ECL-RET-based biosensor relying on physical adsorption binding. Reprinted with permission from ref. 73. Copyright 2023, Elsevier. | ||
As one of the simplest methods of DNA immobilization, physical adsorption mainly depends on electrostatic interactions. Given that electrostatic adsorption does not require any modification, the effect on the DNA molecule is weaker. However, the direct non-covalent fixation of DNA makes the DNA skeleton close to the electrode or material surface, causing damage to the DNA probe and the target molecule. This is also a pressing issue for DNA probe immobilization by physical adsorption in the construction of DNA-based ECL biosensors.
3.3. Covalent bonding method
Covalent bonding is another commonly used method to immobilize probes in DNA-based ECL biosensors, which is based on the formation of covalent bonds between chemically modified DNA probe ends and activated substrate surfaces.74–76 Schiff base reactions have been widely used for the construction of this type of sensor.77 For example, indium tin oxide (ITO) electrodes were silanized by 3-aminopropyl trethoxy silanes (APTS), and acetaldehyde-modified trapped DNA was subjected to a Schiff base reaction to obtain electrodes that immobilized the capture probe. The silanized ITO electrode was cross-linked with a glutaraldehyde cross-linker, and could also be used to immobilize amino-modified DNA or aptamer.78 Another common covalent bonding method is carbodiimide bonding, which is formed by immobilizing chemically modified DNA on the surface of an activated sensor with oxygen groups.79 For instance, DNA attached to an amino group can be bound to the modified electrode with carboxyl group through the classical amide reaction. Besides, click reaction also provides a promising means for DNA immobilization.80 However, compared with chemical adsorption and physical adsorption, the covalent binding reaction conditions are more complex and demanding, and some covalent binding reaction paths will even damage the base and the recognition ability of the DNA probe. Therefore, once covalent binding is selected, the conditions under which bonding occurs are limited, and it is difficult to ensure that the reaction occurs under mild conditions without affecting the subsequent hybridization reactions. However, a significant advantage of covalent binding is that the bond force is very strong, which not only ensures the fixed amount of DNA probe, but also allows easy removal of small molecules with weak binding force and non-specific adsorption.81Feng et al.82 constructed an ECL sensing platform based on a DNA tetrahedral structure (TDN), which acts as a capture probe with a strand extending from the top and three vertices at the bottom modified with amino functional groups, thus immobilizing multilayer silica nanoparticles on the electrode surface (Fig. 6A). The constructed ECL biosensor achieved the accurate determination of glucose oxidase (GOD) with a detection limit as low as 40 aM. Liu et al.83 investigated the effect of the distance between MoS2 nanosheets and sulfur-doped boron nitrogen QDs (S-BN QDs) on the enhancement of the MoS2 surface plasma response and developed a surface plasmon coupling (SPC) ECL biosensor based on this (Fig. 6B). Hairpin DNA was covalently coupled to S-BN QDs via amino and carboxyl groups and served to modulate the DNA strand distance, thus exploring the signal effects of different lengths of DNA strands on plasmonic nanostructures. As a proof of concept, the constructed ECL sensor could achieve the effective detection of the hepatitis C virus (HCV) gene. Xu et al.84 developed a Zn2+-driven DNA rolling machine by borrowing the design concept of DNA walker, which was successfully used in the ultrasensitive detection of miRNA (Fig. 6C). During the construction of the sensing platform, the amino-modified ssDNA is covalently bound to the carboxyl-modified CdS:Mn QDs incubated on the electrode surface, and the ssDNA serves as a track path, which enables the DNA rolling machine to walk, and finally generate the ECL signal. Liu et al.85 developed an efficient ECL biosensor for miRNA detection based on the signal amplification strategy of nicking enzymes Nb.BbvCI (Fig. 6D). The amino-functionalized hairpin DNA and QDs with carboxyl groups form covalent coupling through amide bonds. The combination of hairpin probes endowed the biosensor with excellent selectivity (Fig. 6).
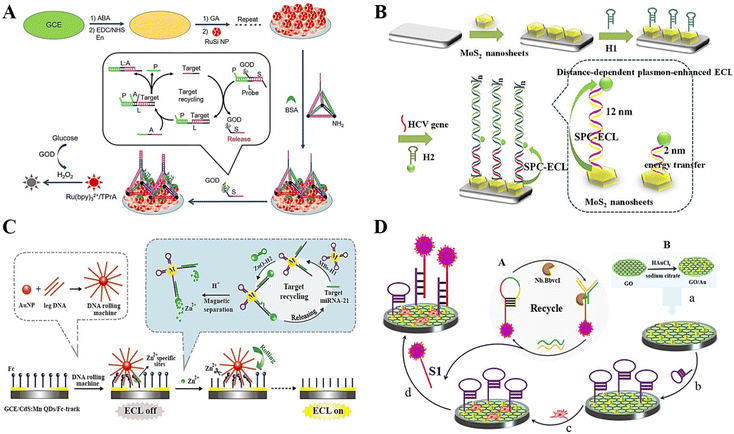 | ||
| Fig. 6 (A) Schematic illustration of an ECL biosensor based on DNA tetrahedral structure. Reprinted with permission from ref. 82. Copyright 2017, Elsevier. (B) Schematic illustration of an ECL sensor platform, showing the distance relation between MoS2 nanosheet plasmonic effects and the ECL luminescence of S-BN QDs. Reprinted with permission from ref. 83. Copyright 2020, Elsevier. (C) Schematic illustration of a Zn2+-driven DNA rolling machine-based biosensor. Reprinted with permission from ref. 84. Copyright 2019, the American Chemical Society. (D) Schematic illustration of an efficient ECL biosensor for miRNA detection based on the signal amplification strategy of nicking enzymes Nb.BbvCI. Reprinted with permission from ref. 85. Copyright 2017, Elsevier. | ||
3.4. Affinity method
The affinity method represents another effective immobilization method, which has been widely used in DNA-based ECL biosensors. Although there are a variety of affinity methods, the biotin–streptavidin system is one of the most commonly used.86 Because streptavidin has a tetramer conformation, it can bind four biotin molecules with high affinity and selectivity. This diversity makes it possible to amplify weak signals and improve the detection sensitivity. As a promising immobilization method, the assembly of DNA probes by the affinity method will be further applied in future biosensing applications.Cui et al.87 proposed a dual-signal amplification ECL sensing strategy based on isothermal strand-displacement polymerase reaction (ISDPR) and bridge DNA-AuNP nanocomposites for the highly sensitive detection of miRNA-21, in which a streptavidin-modified Ru(bpy)32+ complex (SA-Ru) can be immobilized with bridge DNA-AuNPs by the specific binding between biotin and streptavidin, resulting in an ECL emission (Fig. 7A). Li et al.88 developed a highly specific ECL biosensor based on catalytic hairpin assembly and nonmetallic surface plasmon resonance (SPR) effect, which can be used for the highly sensitive detection of miRNA-21. Hairpin DNA is bound to the surface of SnS2 nanoplates via the specific interactions between biotin and streptavidin, enabling the immobilization of the biosensor signal probe (Fig. 7B).
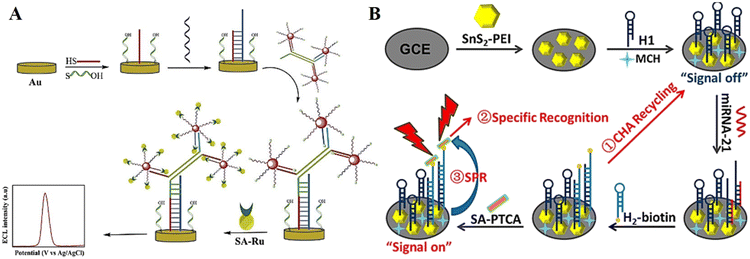 | ||
| Fig. 7 (A) Schematic illustration of a dual-signal amplification ECL sensing platform. Reprinted with permission from ref. 87. Copyright 2019, Elsevier. (B) Schematic illustration of a highly specific ECL biosensor based on the catalytic hairpin assembly cascade nonmetallic SPR effect. Reprinted with permission from ref. 88. Copyright 2022, Elsevier. | ||
The biotin–streptavidin binding method is fast, specific and has high affinity and can withstand the extreme effects of pH, temperature, organic solvents and denaturants. Considering the unique advantages of this affinity method, it can realize powerful streamlined bioassay applications. Moreover, owing to the versatility of the biotin–streptavidin system, streptavidin can be coupled to a variety of molecular markers and participate in a variety of immunoassay processes. Therefore, this affinity method is gradually gaining popularity among researchers.
4. Applications of DNA-based ECL biosensors
Nucleic acids, proteins, and small biological molecules in organisms play an essential role in many physiological activities.89,90 Their expression levels are also closely related to the occurrence of various human diseases (e.g., malignant tumors, cardiovascular and cerebrovascular diseases), and thus they are widely used as relevant markers for disease diagnosis and prediction.91,92 However, due to the low expression level of these disease markers at the early stage, monitoring abnormal changes at their low abundance levels, and thus judging the occurrence of diseases, have become a huge challenge for early diagnosis.In recent years, ECL biosensing has been developing rapidly. By combining DNA nanotechnology, electronics and optical imaging technology, it has attracted much attention in the fields of food safety and environmental monitoring. Accordingly, achieving efficient and sensitive detection has become one of the research hotspots in the field of ECL bioanalysis. It is capable of target conversion and signal amplification by recognizing targets through a target conversion strategy that converts a small number of targets into a large number of analog targets through chemical reactions or intermolecular specific recognition interactions.93,94 With the help of enzyme-free nucleic acid chain reaction, catalytic hairpin assembly (CHA), hybridization chain reaction (HCR), rolling circle amplification (RCA), DNAzyme-induced chain reaction and other signal amplification strategies, the emergence of DNA-based ECL biosensors provides highly sensitive detection tools for disease marker analysis, which is mainly reflected in the detection of nucleic acids, proteins, small molecules, and other types of targets.95–97 In this section, we focus on the developments and practical applications of DNA-based ECL biosensors in recent years.
4.1. Nucleic acid detection
Nucleic acid is an essential constituent of all known life forms, which formed by the polymerization of many nucleotide monomers. Depending on their chemical composition, nucleic acids can be mainly classified as deoxyribonucleic acid (DNA) and ribonucleic acid (RNA).98,99 The detection of specific nucleic acid sequence targets, including DNA, RNA, miRNA and siRNA, is important for the early diagnosis and treatment of genetic diseases and medical analysis.100,101 Currently, the main approach for the detection of nucleic acids is based on the principle of complementary base pairing. In the presence of a target nucleic acid, it undergoes base recognition with the corresponding nucleic acid to induce nucleic acid amplification or DNA self-assembly, after which a small number of target nucleic acid molecules is converted into a mock target associated with an ECL signal, thus enabling detection. In the past few years, many high-performance DNA-based ECL biosensors have been developed for the sensitive detection of specific nucleic acid sequences, and significant progress has been made in the development of strategies for the construction of biosensors.To further improve the ECL efficiency, various materials have been developed to enhance the signal. Among them, silver-based materials can be used as a co-reactant of peroxydisulfate to promote the production of radicals and excited states during electrochemical reactions, which is an effective ECL-enhanced emitter.102 Xiao et al.103 developed a novel silver metal organic framework (AgMOF) that can catalyze the production of SO4˙− in the ECL reaction system, enabling self-enhanced ECL emission for the detection of miRNA-107 (Fig. 8A). In addition, the proposed biosensor exhibited a good ECL performance with a linear range of 20 to 120 fM and a detection limit as low as 4.33 fM by combining double-specific nuclease (DSN) for cyclic amplification of the target and resonance energy transfer effects between the donor and acceptor.
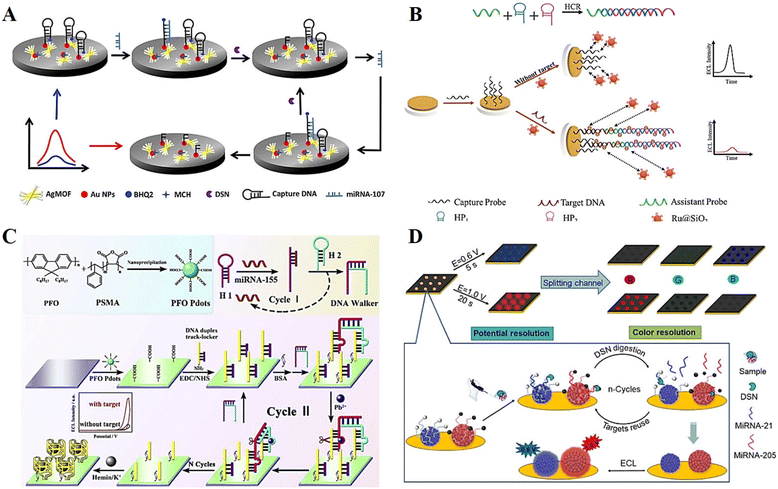 | ||
| Fig. 8 (A) Schematic illustration of a novel ECL biosensor based on silver metal organic framework for the detection of miRNA-107. Reprinted with permission from ref. 103. Copyright 2022, the American Chemical Society. (B) Schematic illustration of a novel homogeneous ECL biosensor based on Ru@SiO2 NPs for the sensitive detection of HPV16. Reprinted with permission from ref. 104. Copyright 2022, the American Chemical Society. (C) Schematic illustration of a dual-wavelength emission ECL-RET-based biosensor. Reprinted with permission from ref. 107. Copyright 2020, Elsevier. (D) Schematic illustration of an ECL-EC ratiometric biosensor. Reprinted with permission from ref. 108. Copyright 2021, the American Chemical Society. | ||
Most conventional DNA-based ECL biosensors require the DNA probe and the ECL reporter to be immobilized on the electrode surface, which is usually a tedious and low reproducible process. In contrast, homogeneous ECL biosensors based on sequence length differences are simple and reproducible by using the electrostatic interaction between the DNA strand length variation and the electrode surface to achieve ECL signal changes. Lu et al.104 constructed a novel homogeneous ECL biosensor based on tris(2,2′-bipyridyl) ruthenium(II) chloride hexahydrate-doped SiO2 nanoparticles (Ru@SiO2 NPs) for the highly sensitive detection of human papillomavirus 16 (HPV16) (Fig. 8B). The strength of the ECL signal was varied by the electrostatic interaction between the negative charge carried by the Ru@SiO2 NPs and the negatively charged electrode surface, which enabled the detection of the target HPV concentration by using long and short strand DNA.
miRNA is a type of important endogenous RNA in the human body. It plays an important regulatory role in the immune response and cellular activity, and can be used as an important marker in the diagnosis and treatment of cancer and other major diseases.105 Currently, polymer dots (Pdots), as an emerging nanomaterial with excellent luminescence and carrier mobility, have attracted wide interest in the fields of optoelectronic materials research and electrochemical analysis.106 Furthermore, novel ECL sensing platforms based on Pdots have become an emerging research hotspot in miRNA detection. Herein, Liu et al.107 developed an ECL sensor based on poly(9,9-di-n-octylfluorenyl-2,7-diyl) polymer dots (PFO Pdots) and CHA-triggered DNA walker for the ultrasensitive detection of miRNA-155 (Fig. 8C). At +1.25 V, PFO Pdots have strong ECL emission, which can be inhibited and quenched by adding H2O2. The DNA walker introduced by the target-triggered CHA reaction can consume the H2O2 in the solution, allowing the recovery of the ECL signal, and thus enabling the accurate determination of intracellular miRNAs.
In addition to single-component detection, multi-component detection is also a hot research topic in the field of ECL biosensors. In this case, Wang et al.108 synthesized luminol-doped Pdots (L-Pdots) and diethylamine-coupled Pdots (N-Pdots) and successfully implemented a potential and color dual-localization ECL sensing strategy for the simultaneous detection of multi-pathway miRNAs (Fig. 8D). L-Pdots and N-Pdots emit the strongest blue and red ECL emission at +0.6 V and +1.0 V, respectively, without interfering with each other. This provides a high-throughput sensing means for dual-potential and dual-color identification, further expanding the application of ECL bioanalysis and clinical diagnosis.
4.2. Protein detection.
Protein, a polypeptide chain composed of amino acids, is coiled and folded into a spatially structured substance during the “dehydration condensation” process, which is the material basis of life.109 Thus, the detection of proteins plays a key role in early clinical diagnosis and disease prevention. Currently, the strategies employed for protein detection are mainly specific biological recognition (including antigen–antibody reactions and nucleic acid aptamer binding, etc.) and chemical reactions between substances.110–112The selection of suitable high-performing ECL luminophores for efficient emission is one of the important issues in the field of ECL biosensors. Nanomaterials have been used to improve the sensitivity and selectivity of DNA-based ECL biosensors for protein detection due to their better biocompatibility, easy modification of aptamers and bioenzyme activity. Chen et al.113 developed a polymeric nanomaterial luminescence cluster induced by β-cyclodextrin (β-CD)-mediated enrichment of co-reactants (Fig. 9A). Due to the good stability and excellent ECL performance of the nanocomposite material, the SARS-CoV-2 nucleocapsid protein (ncovNP) could be sensitively detected by a sandwich-type immune system with detection limits down to the femtomolar level.
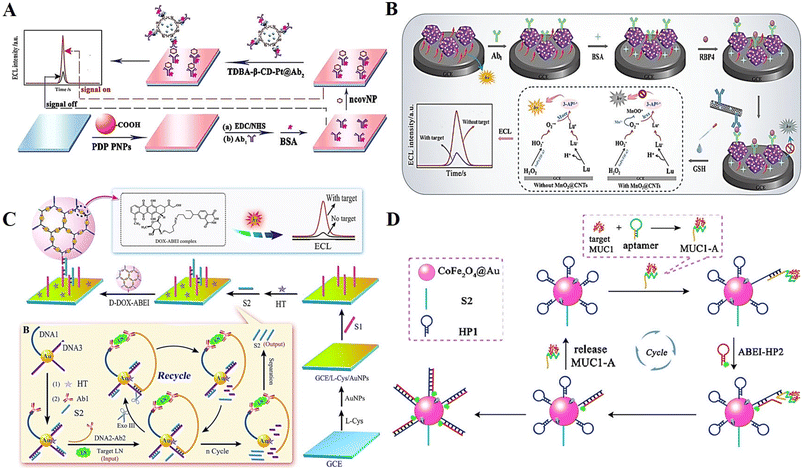 | ||
| Fig. 9 (A) Schematic illustration of an ECL biosensor based on polymeric nanomaterial luminescence cluster induced by β-CD-mediated enrichment of co-reactants for ncovNP determination. Reprinted with permission from ref. 113. Copyright 2022, the American Chemical Society. (B) Schematic illustration of a double-quenched ECL-RET immunosensor for the detection of typical diabetic proteins. Reprinted with permission from ref. 116. Copyright 2021, Springer Nature. (C) Schematic illustration of an ECL biosensor based on dendritic macromolecular structure for LN detection. Reprinted with permission from ref. 117. Copyright 2018, Elsevier. (D) Schematic illustration of an ECL biosensor relying on an aptamer–protein binding-initiated DNA nanomachine for ultra-sensitive detection of MUC1. Reprinted with permission from ref. 120. Copyright 2017, the American Chemical Society. | ||
Metal organic frameworks (MOFs), as a type of new nanomaterials with high porosity, low density and large specific surface area, have attracted wide attention in the field of electrochemical analysis in recent years.114 Among them, zeolitic imidazolate frameworks (ZIFs) are often used to fabricate composites with good ECL performances through nano-doping due to their excellent catalytic and loading performance.115 Gong et al.116 firstly synthesized an Au and Pt bimetallic-anchored and luminol-loaded ZIF material and used it to construct a double-quenched ECL-RET immunosensor for the detection of typical diabetic proteins (Fig. 9B).
DNA nanomaterials with non-toxicity and high biocompatibility, are an excellent and efficient nanoluminescence carriers with potential. Recently, Li et al.117 constructed an ECL biosensor based on a DNA dendrimer-containing luminophore system for the ultrasensitive detection of laminin. The DNA dendrimer with abundant double-stranded DNA was synthesized via hybridization, which could be loaded with a large amount of ECL luminophore (Fig. 9C). A DNA nanomachine was also involved, which could achieve the ultra-sensitive detection of target proteins by an enzyme-induced cyclic amplification system. This ECL biosensor may hold promising potential in biological analysis and clinical application.
Additionally, the strategy for protein detection of aptamer-based target conversion is the mainstream way of building DNA-based ECL biosensors. In most reported biosensors, a common way to achieve the detection of the target protein is to convert the protein into output ssDNA through transformation by enzymatic cleavage, which is then used to participate in the fabrication of a biosensor.118,119 However, this approach suffers from a series of drawbacks such as complicated operation, time-consuming process and high cost. Fortunately, with the rapid development of DNA nanotechnology, a variety of DNA nanoprobes, nanostructures and nanomachines has been developed, thus enabling direct protein recovery assays under laboratory conditions. As a typical design, Jiang et al.120 developed an ECL biosensor based on a protein–aptamer binding-initiated DNA nanomachine and successfully used it for the ultra-sensitive detection of mucin 1 (MUC1) (Fig. 9D). Due to its excellent sensitivity, this strategy may provide an efficient method for clinical application, especially in trace protein determination.
4.3. Small biomolecule detection
In the fields of molecular biology and pharmacology, small biomolecules refer to organic compounds with a low molecular weight (usually less than 1000 Daltons), which are directly involved in biological reactions as substrates or products, such as glucose, amino acids and cholesterol, as well as secondary metabolites such as lipids, glycosides, alkaloids and natural phenols.121 Small biomolecules can combine with specific large biomolecules and act as effectors to change the target activity or function.122 Similarly, given that small biomolecules cannot be detected directly by the principle of complementary base pairing, indirect binding to DNA through intercalation or electrostatic interaction, and the ability to bind small molecules to aptamers also provide new ideas for small molecule detection.Small biomolecules such as food additives, pesticide residues, drugs and other chemicals are closely related to daily life. Carbaryl is a broad-spectrum insecticide that can protect trees and crops from pests.123 However, due to its high stability at room temperature, it may be residual and accumulate in agricultural crops and related foods. Therefore, it is very important to establish a fast and sensitive method to detect carbaryl residues in food. Liu et al.124 developed an ECL biosensor based on the dual identification of Eu@MOF253, and successfully achieved highly sensitive and selective carbaryl detection in actual samples (Fig. 10A).
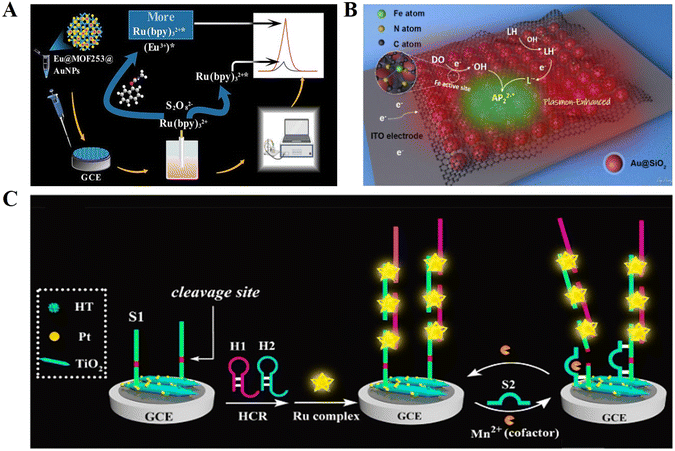 | ||
| Fig. 10 (A) Schematic illustration of an ECL biosensor based on the dual identification of Eu@MOF253 for carbaryl detection. Reprinted with permission from ref. 124. Copyright 2022, the American Chemical Society. (B) Schematic illustration of a novel ECL biosensor based on two-dimensional layered Au@SiO2 nanomaterial for the detection of dopamine. Reprinted with permission from ref. 125. Copyright 2021, the American Chemical Society. (C) Schematic illustration of a sensitive ECL biosensor for GSH detection. Reprinted with permission from ref. 127. Copyright 2019, the American Chemical Society. | ||
In addition, some biomolecules (e.g., the neurotransmitter dopamine and antioxidant glutathione) are also important markers indicating life activities. Bushira et al.125 synthesised a 2D Au@SiO2 nanomaterial coupled with Fe-SACs and further employed it to boost luminol-DO ECL (Fig. 10B). The developed ECL biosensor successfully achieved the rapid and sensitive detection of dopamine and broadened the application of plasmon effect-based ECL signal enhancement strategy in the field of biosensors. Glutathione (GSH) is widely found in animals and plants, and its expression in living organisms plays an important role in maintaining normal immune system function.126 Unfortunately, the aptamer of glutathione has not been successfully screened thus far, and it is not feasible to achieve the detection of glutathione using the target-aptamer target transformation strategy. Zhang et al.127 proposed a new GSH detection strategy by using the properties of glutathione to reduce MnO2 nanosheets to Mn2+, and then by detecting Mn2+ to achieve the conversion of the target (Fig. 10C). TiO2 acts as a co-reactant in the ECL ternary reaction system to enhance the ECL luminescence, and the helical space groove in the middle of the DNA phosphate backbone provides an insertion site for Ru complexes. SsDNA is modified on the electrode surface, which initially has a high ECL signal and suppresses the signal generation when Mn2+ is present, thus achieving the highly sensitive detection of GSH.
4.4. Others
In addition to the above-mentioned biosensors, DNA-based ECL biosensors are also widely used for cellular analysis, metal ions, etc. Considering the important role of cells in life sciences and human health, cell-related bioassays have become a popular research topic in recent years. By introducing different types of functional nanomaterials and aptamers, novel ECL cell sensors are being developed rapidly.128Caspase-3 plays an irreplaceable role in apoptosis, and thus is an important biomarker for monitoring apoptosis.129 Liang et al.130 prepared a high-efficiency ECL emitter by doping carbon dots (CDots) and AuNPs in ZIF-8. CDots@ZIF-8/AuNPs not only acted as a carrier for the ECL luminophore, but also a co-reactant to enhance the signal. As a proof of concept, CDots@ZIF-8/AuNPs were successfully applied in the detection of caspase-3 activity during cell apoptosis (Fig. 11A). Exosomes are a class of small membrane vesicles containing complex RNA and proteins, which can be used as effective immune markers for the diagnosis and treatment of tumor-like diseases.131 Zhang et al.132 proposed a multiplex ECL signal amplification strategy based on NiFe–Ru(bpy)32+ nanocomposites and DSN-mediated target cycle for cellular exosome detection, exemplifying the great potential of DNA-based ECL analysis technology in the early diagnosis of diseases (Fig. 11B).
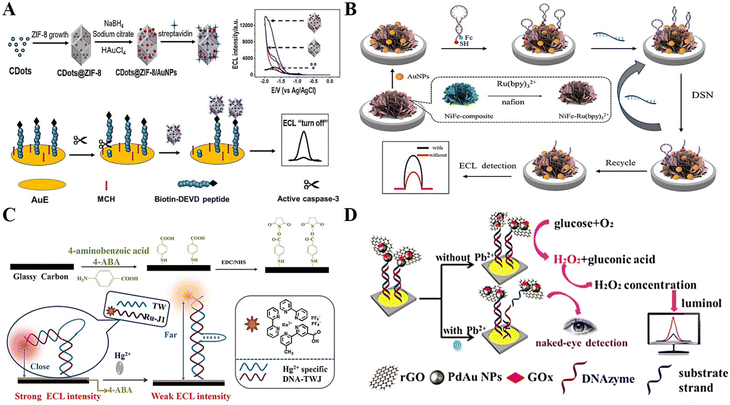 | ||
| Fig. 11 (A) Schematic illustration of a ZIF-based ECL sensing platform for detection of caspase-3. Reprinted with permission from ref. 130. Copyright 2021, Elsevier. (B) Schematic illustration of an ultrasensitive ECL biosensor based on multiple signal amplification for the detection of cellular exosome. Reprinted with permission from ref. 132. Copyright 2022, Elsevier. (C) Schematic illustration of a highly selective and stable ECL biosensor for the detection of Hg2+. Reprinted with permission from ref. 133. Copyright 2019, Elsevier. (D) Schematic illustration of a visual ECL biosensor for the detection of Pb2+. Reprinted with permission from ref. 134. Copyright 2018, the American Chemical Society. | ||
An excess or lack of metal ions in the human body will cause an imbalance, leading to serious diseases. Ma et al.133 assembled a unique structure of DNA three-way junction (DNA-TWJ) and applied it for the preparation of a highly selective and stable ECL biosensor (Fig. 11C). Finally, they successfully realized the sensitive detection of the metal ion Hg2+ with a detection limit as low as 0.04 pM. Xu et al.134 designed a visual ECL biosensor based on a cross-like all-in-one lab-on-paper analytical device to realize the on-site visible monitoring of Pb2+ in tap water and river water, providing a new scheme for the detection of heavy metal ions in the technical field of environmental monitoring (Fig. 11D).
5. Summary and prospect
In the last few years, DNA-based ECL biosensors have been flourishing based on different signal output modes and immobilization methods, combined with various types of signal amplification strategies. Has accordingly, they have become an excellent analytical tool and widely used in many fields such as medical diagnostics, biopharmaceuticals, food safety, and environmental monitoring. Currently, there are two main paths for the development and innovation of DNA-based ECL biosensors, as follows: (i) novel materials. Continuously developing new materials with high luminescence efficiency, high stability and electrocatalytic properties or co-reactant materials with the ability to accelerate electron transfer and improve the reaction rate of ECL systems. In this regard, benefiting from the rapid development of nanotechnology, various nanomaterials, such as carbon nanomaterials, noble metal nanomaterials, semiconductor nanomaterials and conductive polymer nanomaterials in ECL biosensing are also increasing. (ii) Signal amplification strategy. Efficient ECL emission can be achieved by combining multiple signal amplification reactions through clever strategy design. In addition, the in-depth research on DNA nanotechnology has overcome the limitation of using DNA as genetic material, and thus DNA is no longer limited to being a capture probe or signal hybridization probe for DNA-based ECL biosensors. Through precise structural design based on the principle of complementary base pairing, it is used for the construction of biosensors in various forms and means (e.g., DNA molecular motors, DNA walkers, and DNA robots). This bringing a new perspective to the field of biosensors.Looking ahead, we believe that there are still many problems that need to be overcome and improved. For example, the current ECL biosensors are still mainly focused on single-component detection, and most of the existing studies for two-component detection are based on two luminescent reagents to achieve common detection, with cross-reaction. A few sensing devices that implement one luminescent reagent require the building of complex mathematical models, which are laborious and time-consuming. Therefore, one of the problems to be solved in the future is the crossing of the direct influence between two luminescent reagents in ECL two-component detection and realizing the purpose of using one luminescent reagent to achieve the two-component detection. In addition, building a universal ECL biosensing platform, solving the problem that traditional biosensors are difficult to regenerate, minimizing the regeneration steps, and reducing experimental costs should also be the focus of future research. Furthermore, achieving the miniaturization of ECL biosensing technology will be the focus of future application in commercial products. By combining microelectrode array technology and microfluidic platforms, the migration from laboratory biosensing detection tools to commercial biological medical devices will receive increasing attention.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Nature Science Foundation of China (62101281, 21922408, and 22274081), Natural Science Foundation of Jiangsu Province (BK20210584), Natural Science Research Start-up Foundation of Recruiting Talents of Nanjing University of Posts and Telecommunications (NY221032).Notes and references
- M. Oliverio, S. Perotto, G. C. Messina, L. Lovato and F. De Angelis, ACS Appl. Mater. Interfaces, 2017, 9, 29394–29411 CrossRef CAS PubMed.
- S. Lakard, I.-A. Pavel and B. Lakard, Biosensors, 2021, 11, 179 CrossRef CAS PubMed.
- E. A. Songa and J. O. Okonkwo, Talanta, 2016, 155, 289–304 CrossRef CAS PubMed.
- L. C. Clark Jr and C. Lyons, Ann. N. Y. Acad. Sci., 1962, 102, 29–45 CrossRef CAS PubMed.
- M. Soltani, B. R. Davis, H. Ford, J. A. D. Nelson and B. C. Bundy, Biochem. Eng. J., 2018, 138, 165–171 CrossRef CAS.
- A. Salari and M. Thompson, Sens. Actuators, B, 2018, 255, 3601–3615 CrossRef CAS.
- P. Mehrotra, J. Oral. Biol. Craniofac. Res., 2016, 6, 153–159 CrossRef PubMed.
- D. R. Thévenot, K. Toth, R. A. Durst and G. S. Wilson, Anal. Lett., 2001, 34, 635–659 CrossRef.
- D. R. Thevenot, K. Toth, R. A. Durst and G. S. Wilson, Pure Appl. Chem., 1999, 71, 2333–2348 CrossRef CAS.
- S. A. Lim and M. U. Ahmed, RSC Adv., 2016, 6, 24995–25014 RSC.
- R. R. Breaker, Curr. Opin. Biotechnol., 2002, 13, 31–39 CrossRef CAS PubMed.
- F. S. Ligler, K. E. Sapsford, J. P. Golden, L. C. Shriver-Lake, C. R. Taitt, M. A. Dyer, S. Barone and C. J. Myatt, Anal. Sci., 2007, 23, 5–10 CrossRef PubMed.
- R. S. Sethi, Biosens. Bioelectron., 1994, 9, 243–264 CrossRef CAS.
- S. P. Mohanty and E. Kougianos, IEEE Potentials, 2006, 25, 35–40 Search PubMed.
- C. Ziegler and W. Göpel, Curr. Opin. Chem. Biol., 1998, 2, 585–591 CrossRef CAS PubMed.
- M. N. Velasco-Garcia and T. Mottram, Biosyst. Eng., 2003, 84, 1–12 CrossRef.
- S. Vigneshvar, C. Sudhakumari, B. Senthilkumaran and H. Prakash, Front. Bioeng. Biotechnol., 2016, 4, 11 CAS.
- V. Gaudin, in Electrochemical Biosensors, Elsevier, 2019, pp. 307–365 Search PubMed.
- H. Qi and C. Zhang, Anal. Chem., 2019, 92, 524–534 CrossRef PubMed.
- Z. Xu, L. Liao, Y. Chai, H. Wang and R. Yuan, Anal. Chem., 2017, 89, 8282–8287 CrossRef CAS PubMed.
- Y. Zhuo, H.-J. Wang, Y.-M. Lei, P. Zhang, J.-L. Liu, Y.-Q. Chai and R. Yuan, Analyst, 2018, 143, 3230–3248 RSC.
- J. E. Ardill and T. M. O'Dorisio, Endocrinol. Metab. Clin., 2010, 39, 777–790 CrossRef CAS PubMed.
- P. Steinacker, A. Huss, B. Mayer, T. Grehl, J. Grosskreutz, G. Borck, J. Kuhle, D. Lulé, T. Meyer and P. Oeckl, Amyotrophic Lateral Scler. Frontotemporal Degener., 2017, 18, 112–119 CrossRef CAS PubMed.
- M. Khalil, C. E. Teunissen, M. Otto, F. Piehl, M. P. Sormani, T. Gattringer, C. Barro, L. Kappos, M. Comabella and F. Fazekas, Nat. Rev. Neurol., 2018, 14, 577–589 CrossRef CAS PubMed.
- M. A. Arugula, Y. Zhang and A. L. Simonian, Anal. Chem., 2014, 86, 119–129 CrossRef CAS PubMed.
- J. Kim and C. J. Easley, Bioanalysis, 2011, 3, 227–239 CrossRef CAS PubMed.
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2017, 3, 1–23 Search PubMed.
- R. Chhabra, J. Sharma, Y. Liu, S. Rinker and H. Yan, Adv. Drug Delivery Rev., 2010, 62, 617–625 CrossRef CAS PubMed.
- J. Zhang, H. Qi, Y. Li, J. Yang, Q. Gao and C. Zhang, Anal. Chem., 2008, 80, 2888–2894 CrossRef CAS PubMed.
- M.-L. Zhao, W.-J. Zeng, Y.-Q. Chai, R. Yuan and Y. Zhuo, Anal. Chem., 2020, 92, 11044–11052 CrossRef CAS PubMed.
- L. D. Zhao, X. Yang, X. Zhong and Y. Zhuo, ChemPlusChem, 2022, 87, e202200070 CAS.
- Z. Ning, M. Chen, G. Wu, Y. Zhang and Y. Shen, Biosens. Bioelectron., 2021, 191, 113462 CrossRef CAS PubMed.
- D. Li, S. Song and C. Fan, Acc. Chem. Res., 2010, 43, 631–641 CrossRef CAS PubMed.
- H. Jin, R. Gui, J. Yu, W. Lv and Z. Wang, Biosens. Bioelectron., 2017, 91, 523–537 CrossRef CAS PubMed.
- J. Zhang, P. Chen, X. Wu, J. Chen, L. Xu, G. Chen and F. Fu, Biosens. Bioelectron., 2011, 26, 2645–2650 CrossRef CAS PubMed.
- A. Cui, J. Zhang, W. Bai, H. Sun, L. Bao, F. Ma and Y. Li, Biosens. Bioelectron., 2019, 144, 111664 CrossRef PubMed.
- X. Ou, X. Tan, X. Liu, Q. Lu, S. Chen and S. Wei, Biosens. Bioelectron., 2015, 70, 89–97 CrossRef CAS PubMed.
- L. Peng, Y. Yuan, X. Fu, A. Fu, P. Zhang, Y. Chai, X. Gan and R. Yuan, Anal. Chem., 2019, 91, 3239–3245 CrossRef CAS PubMed.
- L. Liu, Y. Zhang, R. Yuan and H. Wang, Anal. Chem., 2020, 92, 15112–15119 CrossRef CAS PubMed.
- H. Jia, J. Li, L. Yang, D. Fan, X. Kuang, X. Sun, Q. Wei and H. Ju, Anal. Chem., 2022, 94, 7132–7139 CrossRef CAS PubMed.
- M. Xia, F. Zhou, X. Feng, J. Sun, L. Wang, N. Li, X. Wang and G. Wang, Anal. Chem., 2021, 93, 11284–11290 CrossRef CAS PubMed.
- L. Liu, Q. Ma, Y. Li, Z. Liu and X. Su, Biosens. Bioelectron., 2015, 63, 519–524 CrossRef CAS PubMed.
- W. Bai, A. Cui, M. Liu, X. Qiao, Y. Li and T. Wang, Anal. Chem., 2019, 91, 11840–11847 CrossRef CAS PubMed.
- Y. Zhang, Y. Li, Y. Wei, H. Sun and H. Wang, Talanta, 2017, 170, 546–551 CrossRef CAS PubMed.
- M. H. Jiang, S. K. Li, X. Zhong, W. B. Liang, Y. Q. Chai, Y. Zhuo and R. Yuan, Anal. Chem., 2019, 91, 3710–3716 CrossRef CAS PubMed.
- Q. Wang, Y. Liu, J. Yan, Y. Liu, C. Gao, S. Ge and J. Yu, Anal. Chem., 2021, 93, 13373–13381 CrossRef CAS PubMed.
- B. Yao, J. Zhang, Z. Fan, Y. Ding, B. Zhou, R. Yang, J. Zhao and K. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 19816–19824 CrossRef CAS PubMed.
- K. Zhang, Z. Fan, Y. Huang, Y. Ding and M. Xie, Talanta, 2022, 236, 122868 CrossRef CAS PubMed.
- K. Zhang, Z. Fan, Y. Ding and M. Xie, Chem. Eng. J., 2022, 429, 132472 CrossRef CAS PubMed.
- L. Li, C. Niu, T. Li, Y. Wan, Y. Zhou, H. Wang, R. Yuan and P. Liao, Biosens. Bioelectron., 2018, 101, 206–212 CrossRef CAS PubMed.
- X. Bian, B. Guo, M. Zhao, D. Han, W. Cheng, F. Song and S. Ding, ACS Appl. Mater. Interfaces, 2019, 11, 3715–3721 CrossRef CAS PubMed.
- L. Hu, M. Lei, J. Zhang and Y. Dong, Sens. Actuators, B, 2021, 344, 130328 CrossRef CAS.
- S. S. Yang, M. H. Jiang, Y. Q. Chai, R. Yuan and Y. Zhuo, ACS Appl. Mater. Interfaces, 2018, 10, 38648–38655 CrossRef CAS PubMed.
- Z. Fan, Z. Lin, Z. Wang, J. Wang, M. Xie, J. Zhao, K. Zhang and W. Huang, ACS Appl. Mater. Interfaces, 2020, 12, 11409–11418 CrossRef CAS PubMed.
- L. Luo, X. Liu, X. Bi, L. Li and T. You, Biosens. Bioelectron., 2023, 222, 114991 CrossRef CAS PubMed.
- X.-m. Chen, B.-y. Su, X.-h. Song, Q.-a. Chen, X. Chen and X.-R. Wang, TrAC, Trends Anal. Chem., 2011, 30, 665–676 CrossRef CAS.
- E. Sobhanie, F. Salehnia, G. Xu, Y. Hamidi, S. Arshian, A. Firoozbakhtian, M. Hosseini, M. R. Ganjali and S. Hanif, TrAC, Trends Anal. Chem., 2022, 116727 CrossRef CAS PubMed.
- C. Fang, H. Li, J. Yan, H. Guo and T. Yifeng, ChemElectroChem, 2017, 4, 1587–1593 CrossRef CAS.
- W.-W. Zhao, J.-J. Xu and H.-Y. Chen, Chem. Rev., 2014, 114, 7421–7441 CrossRef CAS PubMed.
- C. Vericat, M. Vela, G. Benitez, P. Carro and R. Salvarezza, Chem. Soc. Rev., 2010, 39, 1805–1834 RSC.
- P. Kingshott and H. J. Griesser, Curr. Opin. Solid State Mater. Sci., 1999, 4, 403–412 CrossRef CAS.
- K. Thapa, W. Liu and R. Wang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1765 CAS.
- J. J. Gooding, Electroynalysis, 2002, 14, 1149–1156 CrossRef CAS.
- S. Campuzano, F. Kuralay and J. Wang, Electroanalysis, 2012, 24, 483–493 CrossRef CAS.
- Y. Zhang, G. Xu, G. Lian, F. Luo, Q. Xie, Z. Lin and G. Chen, Biosens. Bioelectron., 2020, 147, 111789 CrossRef CAS PubMed.
- H.-Y. Zhu and S.-N. Ding, Biosens. Bioelectron., 2019, 134, 109–116 CrossRef CAS PubMed.
- Y. Li, L. He, C. Z. Huang and Y. F. Li, Biosens. Bioelectron., 2019, 134, 29–35 CrossRef CAS PubMed.
- J. Ling, M. Zhao, F. Chen, X. Zhou, X. Li, S. Ding and H. Tang, Biosens. Bioelectron., 2019, 127, 167–173 CrossRef CAS PubMed.
- T.-S. Wong, B. Brough and C.-M. Ho, MCB Mol. Cell. Biomech., 2009, 6, 1 Search PubMed.
- A. K. Geim, Science, 2009, 324, 1530–1534 CrossRef CAS PubMed.
- S. Nardecchia, D. Carriazo, M. L. Ferrer, M. C. Gutiérrez and F. del Monte, Chem. Soc. Rev., 2013, 42, 794–830 RSC.
- Y. Wu, J. Yang, Z. Zheng, Z. Li, F. Lu, Y. Chen and W. Gao, Talanta, 2018, 188, 58–65 CrossRef CAS PubMed.
- Q. Song, X. Shan, L. Bu, A. Dai, D. Jiang, W. Wang, H. Shiigi and Z. Chen, J. Electroanal. Chem., 2023, 928, 116987 CrossRef CAS.
- Z. Hu, Z. Suo, W. Liu, B. Zhao, F. Xing, Y. Zhang and L. Feng, Biosens. Bioelectron., 2019, 131, 237–249 CrossRef CAS PubMed.
- A. Sassolas, B. D. Leca-Bouvier and L. J. Blum, Chem. Rev., 2008, 108, 109–139 CrossRef CAS PubMed.
- Y. Du and S. Dong, Anal. Chem., 2017, 89, 189–215 CrossRef CAS PubMed.
- Y. Jia and J. Li, Chem. Rev., 2015, 115, 1597–1621 CrossRef CAS PubMed.
- D. Liu and M. La, Int. J. Electrochem. Sci., 2020, 15, 11371–11386 CrossRef CAS.
- X.-B. Yin, TrAC, Trends Anal. Chem., 2012, 33, 81–94 CrossRef CAS.
- M. Tarnowska and T. Krawczyk, Biosens. Bioelectron., 2020, 169, 112614 CrossRef CAS PubMed.
- I. Rahmawati, Y. Einaga, T. A. Ivandini and A. Fiorani, ChemElectroChem, 2022, 9, e202200175 CrossRef CAS.
- Q. M. Feng, Y. H. Guo, J. J. Xu and H. Y. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 17637–17644 CrossRef CAS PubMed.
- Y. Liu, Y. Nie, M. Wang, Q. Zhang and Q. Ma, Biosens. Bioelectron., 2020, 148, 111823 CrossRef CAS PubMed.
- Z. Xu, Y. Chang, Y. Chai, H. Wang and R. Yuan, Anal. Chem., 2019, 91, 4883–4888 CrossRef CAS PubMed.
- Q. Liu, C. Ma, X. P. Liu, Y. P. Wei, C. J. Mao and J. J. Zhu, Biosens. Bioelectron., 2017, 92, 273–279 CrossRef CAS PubMed.
- A. Sassolas, L. J. Blum and B. D. Leca-Bouvier, Biotechnol. Adv., 2012, 30, 489–511 CrossRef CAS PubMed.
- A. Cui, J. Zhang, W. Bai, H. Sun, L. Bao, F. Ma and Y. Li, Biosens. Bioelectron., 2019, 144, 111664 CrossRef PubMed.
- J. Li, R. Cai and W. Tan, Anal. Chem., 2022, 94, 12280–12285 CrossRef CAS PubMed.
- Y. Zhao, X. Zuo, Q. Li, F. Chen, Y.-R. Chen, J. Deng, D. Han, C. Hao, F. Huang and Y. Huang, Sci. China: Chem., 2021, 64, 171–203 CrossRef CAS PubMed.
- S. L. Kuan and M. Raabe, ChemMedChem, 2021, 16, 94–104 CrossRef CAS PubMed.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi, X. Lou, J. Shi and C. Fan, Nat. Protoc., 2016, 11, 1244–1263 CrossRef CAS PubMed.
- E. E. Ferapontova, E. M. Olsen and K. V. Gothelf, J. Am. Chem. Soc., 2008, 130, 4256–4258 CrossRef CAS PubMed.
- S. Goggins and C. G. Frost, Analyst, 2016, 141, 3157–3218 RSC.
- P. Scrimin and L. J. Prins, Chem. Soc. Rev., 2011, 40, 4488–4505 RSC.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- I. Willner, B. Shlyahovsky, M. Zayats and B. Willner, Chem. Soc. Rev., 2008, 37, 1153–1165 RSC.
- J. D. Watson and F. H. Crick, Nature, 1953, 171, 737–738 CrossRef CAS PubMed.
- J. Liu, Z. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- N. Risch and K. Merikangas, Science, 1996, 273, 1516–1517 CrossRef CAS PubMed.
- J. J. McCarthy and R. Hilfiker, Nat. Biotechnol., 2000, 18, 505–508 CrossRef CAS PubMed.
- S. Y. Xiao, Y. Li, S. J. Zhen, C. Z. Huang and Y. F. Li, Electrochim. Acta, 2020, 357, 136842 CrossRef CAS.
- S. Xiao, X. Wang, C. Yang, Y. Jiang, S. Zhen, C. Huang and Y. Li, Anal. Chem., 2022, 94, 1178–1186 CrossRef CAS PubMed.
- Y. Lu, X. Rong, P. Wu, J. Shou, L. Chen, F. Luo, C. Lin, J. Wang, B. Qiu and Z. Lin, Anal. Chem., 2022, 94, 5823–5829 CrossRef CAS PubMed.
- Y. Cai, X. Yu, S. Hu and J. Yu, Genomics, Proteomics Bioinf., 2009, 7, 147–154 CrossRef CAS PubMed.
- C. Wu and D. T. Chiu, Angew. Chem., Int. Ed., 2013, 52, 3086–3109 CrossRef CAS PubMed.
- D. Liu, X. Zhang, J. Zhao, S. Chen and R. Yuan, Biosens. Bioelectron., 2020, 150, 111872 CrossRef CAS PubMed.
- N. Wang, L. Chen, W. Chen and H. Ju, Anal. Chem., 2021, 93, 5327–5333 CrossRef CAS PubMed.
- S. Jones and J. M. Thornton, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13–20 CrossRef CAS PubMed.
- X. Cheng, Y. Li, K. Jun, D. Liao, W. Zhang, L. Yin, S. Man and L. Ma, Biosens. Bioelectron., 2022, 215, 114559 CrossRef CAS PubMed.
- H. Xiong, J. Yan, S. Cai, Q. He, D. Peng, Z. Liu and Y. Liu, Int. J. Biol. Macromol., 2019, 132, 190–202 CrossRef CAS PubMed.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2008, 47, 6330–6337 CrossRef CAS PubMed.
- Y. Chen, Y. He, J. Zhao, J. Zhang, R. Yuan and S. Chen, Anal. Chem., 2022, 94, 4446–4454 CrossRef CAS PubMed.
- J. Zhou, Y. Li, W. Wang, X. Tan, Z. Lu and H. Han, Biosens. Bioelectron., 2020, 164, 112332 CrossRef CAS PubMed.
- J. Zhang, Y. Tan and W.-J. Song, Microchim. Acta, 2020, 187, 1–23 CrossRef PubMed.
- W. Gong, S. Yang, F. Zhang, F. Tian, J. Chen, Z. Yin, S. Ding, W. Yang and R. Luo, J. Nanobiotechnol., 2021, 19, 272 CrossRef CAS PubMed.
- L. Li, C. Niu, T. Li, Y. Wan, Y. Zhou, H. Wang, R. Yuan and P. Liao, Biosens. Bioelectron., 2018, 101, 206–212 CrossRef CAS PubMed.
- Z.-H. Yang, Y. Zhuo, R. Yuan and Y.-Q. Chai, Anal. Chem., 2016, 88, 5189–5196 CrossRef CAS PubMed.
- S. Fredriksson, M. Gullberg, J. Jarvius, C. Olsson, K. Pietras, S. M. Gústafsdóttir, A. Östman and U. Landegren, Nat. Biotechnol., 2002, 20, 473–477 CrossRef CAS PubMed.
- X. Jiang, H. Wang, H. Wang, Y. Zhuo, R. Yuan and Y. Chai, Anal. Chem., 2017, 89, 4280–4286 CrossRef CAS PubMed.
- C. Feng, S. Dai and L. Wang, Biosens. Bioelectron., 2014, 59, 64–74 CrossRef CAS PubMed.
- D. D. Ryu and D. H. Nam, Biotechnol. Prog., 2000, 16, 2–16 CrossRef CAS PubMed.
- W. R. Melchert and F. R. Rocha, Talanta, 2010, 81, 327–333 CrossRef CAS PubMed.
- C. Liu, L. Cai, Y. Wang, H. Wang, G. Fang and S. Wang, J. Agric. Food Chem., 2022, 70, 6264–6271 CrossRef CAS PubMed.
- F. A. Bushira, S. A. Kitte, C. Xu, H. Li, L. Zheng, P. Wang and Y. Jin, Anal. Chem., 2021, 93, 9949–9957 CrossRef CAS PubMed.
- J. Yin, Y. Kwon, D. Kim, D. Lee, G. Kim, Y. Hu, J.-H. Ryu and J. Yoon, Nat. Protoc., 2015, 10, 1742–1754 CrossRef CAS PubMed.
- R. Zhang, X. Zhong, A. Y. Chen, J. L. Liu, S. K. Li, Y. Q. Chai, Y. Zhuo and R. Yuan, Anal. Chem., 2019, 91, 3681–3686 CrossRef CAS PubMed.
- J. Shen, Y. Li, H. Gu, F. Xia and X. Zuo, Chem. Rev., 2014, 114, 7631–7677 CrossRef CAS PubMed.
- A. G. Porter and R. U. Jänicke, Cell Death Differ., 1999, 6, 99–104 CrossRef CAS PubMed.
- G.-X. Liang, K.-R. Zhao, Y.-S. He, Z.-J. Liu, S.-Y. Ye and L. Wang, Microchem. J., 2021, 171, 106787 CrossRef CAS.
- P. Li, M. Kaslan, S. H. Lee, J. Yao and Z. Gao, Theranostics, 2017, 7, 789 CrossRef CAS PubMed.
- J. Zhang, L. Hao, Z. Zhao, D. Jiang and J. Chao, Sens. Actuators, B, 2022, 369, 132332 CrossRef CAS.
- F. Ma, Y. Chen, Y. Zhu and J. Liu, Talanta, 2019, 194, 114–118 CrossRef CAS PubMed.
- J. Xu, Y. Zhang, L. Li, Q. Kong, L. Zhang, S. Ge and J. Yu, ACS Appl. Mater. Interfaces, 2018, 10, 3431–3440 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |