Open Access Article
Open Access ArticlePlasmonic and metamaterial biosensors: a game-changer for virus detection
Junfei
Wang
 a,
Zhenyu
Xu
a,
Zhenyu
Xu
 a and
Domna G.
Kotsifaki
a and
Domna G.
Kotsifaki
 *ab
*ab
aPhotonics Lab, Division of Natural and Applied Sciences, Duke Kunshan University, Kunshan, 215316, Jiangsu Province, China. E-mail: domna.kotsifaki@dukekunshan.edu.cn; dk310@duke.edu
bData Science Research Center, Duke Kunshan University, Kunshan, 215316, Jiangsu Province, China
First published on 24th March 2023
Abstract
One of the most important processes in the fight against current and future pandemics is the rapid diagnosis and initiation of treatment of viruses in humans. Currently available viral diagnostic methods detect only known pathogens, which comprise a small number of virus strains. In addition, identifying viral genomes is challenging due to low viral abundance and possible contamination by host nucleic acids. To ensure the distinction between the infected and non-infected people and predict the outbreak of disease, alternative approaches should be considered. In the ongoing hunt for new developing tests and diagnostic kits with high selectivity and sensitivity, plasmonic platforms, which control light in subwavelength volumes, have opened up exciting prospects for biosensing applications. They can identify particular viruses in a cost-effective, sensitive, label-free, rapid, and reproducible way due to their tunable plasmonic properties. In particular, plasmonic-assisted virus detection platforms can be achieved by various approaches, including propagating surface and localized plasmon resonances, as well as surface-enhanced Raman spectroscopy. In this review, we discuss both the fundamental principles governing a plasmonic biosensor and prospects for achieving improved sensor performance. We highlight several nanostructure schemes to combat virus-related diseases. We also examine the technological limitations and challenges of plasmonic-based biosensing, such as reducing the overall cost and handling of complex biological samples. Finally, we provide a future perspective for opportunities to improve plasmonic-based approaches to increase their impact on global health issues.
1 Introduction
At the dawn of the twenty-first century, humanity faces multiple health challenges with substantial global economic and social impacts.1–6 The monitoring and early detection of biological entities necessitate platforms that are able to analyze extremely low concentrations of analytes in real samples near the point of care (PoC) and sometimes at the place of patient care. The early detection and timely treatment of diseases can improve cure rates and reduce treatment costs. Commonly used analytical methods7–9 rely upon culture-based methods, serological tests, or nucleic acid-based amplification techniques such as polymerase chain reaction (PCR), gene sequencing, virus isolation, hemagglutination assay, and enzyme-linked immunosorbent assay (ELISA). In spite of their inherent advantages, these techniques are time-consuming and involve sophisticated instrumentation that requires skilled operators. In addition, time-consuming predeveloped protocols are typically limited to specific strains or types of viruses and may have high false-negative rates, which limit their effectiveness to lower the risk of new infections.10 Viruses are structures of several sizes (from 20 to 900 nm) and morphological shapes, composed of genetic material covered in proteins, glycoproteins, or lipids.11 Approximately one-third of all infectious disease-related deaths are caused by viral infections.12 One fundamental issue in considering viral diagnostics sensitivity is the uncertainty about the range of viral loads that constitute a transmission risk.13 Those infected with SARS-CoV-2, for example, are most infectious around the time of system onset when viral loads in the upper respiratory tract are highest. Similar to pre-symptomatic individuals, asymptotic individuals contribute to viral spread as well.13 Hence, it is crucial to diagnose viruses during their incubation period in order to maximize healing rates and reduce the risk of pandemics.12 It is equally important to detect viruses in order to monitor the environment because they can also infect plants and animals.14 Although PCR can provide accurate and sensitive viral diagnostics by amplification of specific DNA/RNA sequences, it is more expensive than plasmon-based methods.7–9 Consequently, the need for new diagnostic approaches that are fast and cost-effective has brought into focus the development of real-time PoC testing devices,15 based on plasmonic nanostructures that offer new sensing capabilities for rapid diagnosis of virus particles and could be a game changer for disease management.With the growing need for new PoC diagnostic platforms, the World Health Organization has created the ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end users) framework, outlining directions and guidelines for their development.16 Current PoC tests, such as paper-based devices,17 succeed in providing rapid, cost-effective, and facile results but are held back by inadequate sensitivity, selectivity, and overall reliability, highlighting the challenges faced by PoC diagnostics.18 Early diagnosis is essential for a wide range of conditions, including infectious diseases, auto-immune disorders, and inflammatory diseases, for which timing is important to maximize the efficacy of therapy. In addition, continuous monitoring of viral biomarkers or therapeutic drug levels at the bedside can provide valuable feedback to physicians and allow them to tailor the treatment options for individual patients.19–21 In this aspect, nanostructure-based PoC approaches that can rapidly provide the molecular profile of a patient could become instrumental in paving the way towards precision diagnosis.22,23
Plasmonic-based biosensing (Fig. 1) has embraced the challenge of offering on-site strategies to complement traditional diagnostic methods and has attracted significant attention owing to its versatility and ability to achieve label-free monitoring with low response times.5,22,27–33 These characteristics, achieved by exploiting the properties of nanomaterials,34–39 have allowed for the design of ultrasensitive nanobiosensors, which could be implemented in diagnostic tools to alleviate the burden of infectious diseases in the developing world. By patterning metal films into nanostructures even tighter electromagnetic field confinement is possible, allowing for the detection of single virus particles.40 Therefore, plasmonic-based biosensing utilizes the interaction of electromagnetic fields to detect virus particles, antigens, or nucleic acids from clinical specimens (such as blood, serum, saliva, etc.) with high selectivity and sensitivity. Furthermore, this method offers the advantages of easy operation, minimal sample pretreatment, and simple cost-effective instrumentation.41 Likewise, as light sources, detectors, and optical components are abundant in the visible-to-near infrared electromagnetic spectrum range, the design of plasmonic biosensors in this range is particularly advantageous.24 Such biosensors require structural dimensions on the few-nanometer scale and can be fabricated using nanolithography techniques.18,24,41,42 In addition, plasmonic biosensors enable direct detection of analytes from heterogeneous biological media without the need for exogenous labels.43,44 This is a key factor in plasmonic-based biosensor design since it facilitates bio-assay procedures by eliminating tedious washing, amplifying, and labeling steps.1,10,45 For the abovementioned reasons, plasmonic-based biosensors are seen as promising candidates for the essential elements of future biosensor PoC platforms.
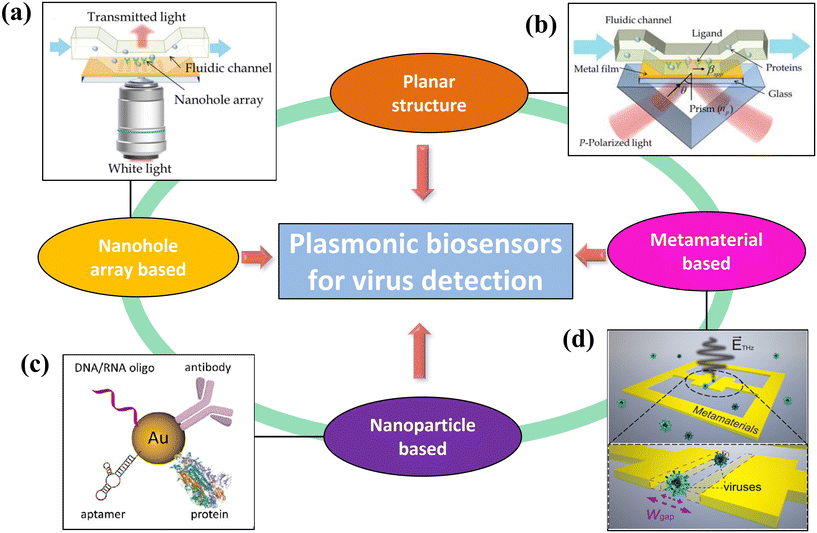 | ||
| Fig. 1 Schematic illustration of various plasmonic-based virus sensing platforms. (a) An array of nanoholes can increase the binding potential for flowing virus antigens and enhance the sensitivity through the extraordinary transmission effect (reproduced with permission from ref. 24). (b) Planar structure in which surface plasmon is generated in between a dielectric and a metal (reproduced with permission from ref. 24). (c) Localized surface plasmon around nanoparticles can increase sensitivity (reproduced with permission from ref. 25). (d) Metamaterials can efficiently enhance the electromagnetic fields of light, leading to ultrasensitive biosensing (reproduced with permission from ref. 26). | ||
In this tutorial review, we present the advances in plasmonic-based biosensing for virus detection and highlight the scope of future work in this research field. We address the fundamental physical principles of plasmonic effects and biosensing strategies. The integration of metallic nanostructures into commercial microfluidic platforms for future devices that can alert the public to biological threats is also discussed. Because of the ongoing coronavirus disease 2019 (COVID-19) pandemic, slight emphasis is given to coronavirus detection techniques. Finally, we discuss the challenges that need to be overcome for the future development of plasmonic-based biosensors and note how such biosensors are already impacting the diagnosis of infectious diseases in the developing world. We believe that this comprehensive review will be a useful resource for researchers, physicians, and students interested in constructing ultra-dense and high-throughput clinical screening plasmonic devices.
2 Physical considerations
2.1 A brief historical introduction
The interaction of light with plasmonic nanostructures has long been a subject of interest in the classical and quantum worlds.46 A key feature of plasmon resonances is that they are excited by electromagnetic waves, either evanescent or localized.46 Their first observation dates back to Wood,47 who reported anomalous reflective patterns when polarized light was shone on a metalized diffraction grating. A few years later, Rayleigh48 provided a phenomenological explanation for these patterns, but the underlying physical mechanism remained a mystery. In 1957, significant advances in our understanding of surface plasmon resonance (SPR) were made when Ritchie49 confirmed the presence of metal surface plasma excitations, while Powell et al. determined that the excitation of surface plasmons involved electrons at metal interfaces.50 In 1968, Otto used an attenuated total reflection prism-coupled method to enable the coupling of an electromagnetic field with surface plasmon waves.51 Similarly, Kretschmann and Raether reported the excitation of SPR by utilizing a 10–100 nm thin gold film on the surface of a prism.52 The potential exploitation of SPR for biosensing first appeared in 1974 with observations made by Fleischmann and colleagues,54 who noted an enhancement of Raman scattering near a roughened metal surface; this enhancement was later found to be associated with an electromagnetic effect.55 Ten years later, Liedberg et al. observed refractive index (RI) changes on the surface of a metallic film after the absorption of biomolecules.56 Since then, the label-free nature of SPR biosensing has become an important tool in biophysics, molecular biology, and pharmaceutical research.41,42,57–59 Today, several companies, such as Biocore, PhotonicSys, and Plasmetric, manufacture devices used to evaluate the performance of biosensor chips for PoC applications.2.2 Fundamental mechanism of plasmon resonance biosensors
A simple technique of generating surface plasmons from a metal–dielectric interface is the Kretschmann configuration (Fig. 2(a)). The underlying physics of SPR sensors based on an evanescent field has been reviewed extensively in the literature.44,64,65 Briefly, when light, which implies an electromagnetic wave, strikes the metal, the electric field of the light interacts with conducting electrons. The coupling of the incident electromagnetic wave to the collective oscillations of the conduction electrons forms an evanescent wave, which is known as SPR. To achieve this, the momentum of incident photons should match the momentum of the conduction band of electrons. This momentum matching condition depends on the refractive index (RI) of the dielectric medium at the surface of the metal layer and is given by the following expression:64
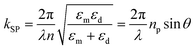 | (1) |
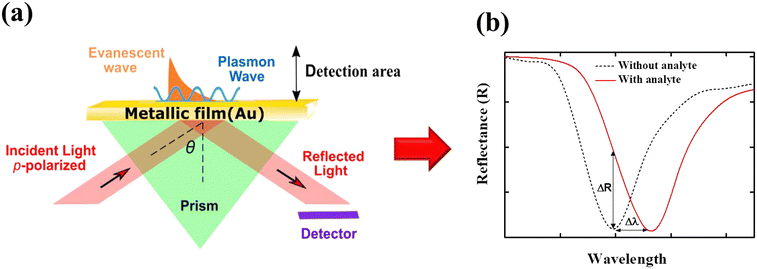 | ||
| Fig. 2 (a) Schematic illustration of the Kretschmann configuration.53 The surface plasmon polariton can be excited when momentum mismatching is satisfied. The surface plasmon decays exponentially from the surface and propagates to a distance of a few tens of microns. (b) Typical sensor readouts: spectrum of reflected light before and after the binding of the analyte, which leads to refractive index changes. The momentum mismatching condition exists at certain incident angles θ. Δλ is the resonance shift and ΔR indicates the intensity changes due to analyte binding. | ||
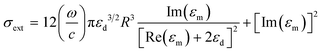 | (2) |
For the detection mechanisms in both SPR and LSPR, the sensitivity to changes within their associated plasmon decays with length.65 LSPR changes can be detected within tens of nanometers in the visible range, whereas SPR changes, which occur along the propagation surface, can be detected within a few hundred nanometers.67 In biosensing, LSPRs are usually utilized through surface-enhanced techniques such as surface-enhanced Raman scattering spectroscopy73 (SERS), surface-enhanced infrared absorption spectroscopy,74 surface-enhanced fluorescence,75 and through resonance shifts induced by nearby analytes.76
2.3 Plasmonic metamaterials
Plasmonic metamaterials have been utilized to further control collective plasmonic modes and electromagnetic field enhancement.77–80 The concept of these materials was first introduced in 1968 by Veselago, who observed the unusual behavior of light refracted by a left-handed material.81 A few years later, Pendry et al. noted that microstructures, fabricated from nonmagnetic conducting sheets smaller than the excitation wavelength, could be tuned to show varying magnetic permeability, including imaginary components.82 Based on these observations,81,82 a practical way to manufacture a left-handed material that does not follow the conventional right-hand was determined. In 2000, Smith et al. demonstrated the first left-handed metamaterial, which simultaneously exhibited negative permeability and permittivity at microwave frequencies.83 Since then, metamaterials have been explored extensively for a variety of applications in optics,84 photonics,85 energy harvesting,86 sensing,87 imaging,88 and spectroscopy.89 Compared with conventional SPR-based methods, metamaterials can be more easily fabricated through nanolithography techniques.78 For periodic arrays of metamolecules, near- and far-field coupling is utilized to generate resonance with a high-quality factor (Q-factor). This breaks the damping limit of a single metamolecule in the dipole approximation,90 thus making such arrays promising candidates for biosensing applications.782.4 Surface enhanced Raman spectroscopy (SERS) mechanism
SERS is a highly analytical tool91,92 that has many applications in the field of diagnostics.93 It can be used to enhance weak Raman signals of analytes through the use of plasmonic nanostructures.94–96 Raman spectroscopy evaluates the vibrational and rotation modes of biomolecules through the analysis of inelastic Raman scattering of a laser beam.94 Specifically, metallic nanostructures possess a localized electromagnetic field as a result of LSPR, which affects the Raman signal of an active analyte in close proximity to the nanostructure by enhancing the Raman scattering cross-section.95 Overall, SERS shows a broad range of benefits, such as high selectivity due to the unique fingerprint signatures of analytes, easy sample preparation, high possibility of single-entity detection, high throughput, and PoC applicability by using available Raman probes.91,922.5 Surface plasmon resonance imaging (SPRi)
Surface plasmon resonance imaging (SPRi) is a real-time optical detection technique that monitors and analyzes biomolecular interactions without using any labels. While SPR and SPRi share similar detection principles, the latter provides high-throughput biosensing or screening capabilities.97–100 The most commonly used experimental setup for SPRi is based on Kretschmann geometry.97,100 By using, for instance, the changes in reflectivity of a thin gold film that occur upon analyte absorption, SPRi measurements allow for monitoring tens, hundreds, or even thousands of interactions simultaneously.101 The combination of LSPRs that are obtained by nanoparticles close to the metallic surface and SPRs has been shown to enhance SPRi sensitivity in viral diagnostics.99,100 SPRi biosensors have been demonstrated so far based on angles, wavelengths, phases, and polarization interrogations.97–100 Angle-resolved interrogation mode, for example, continuously scans the incident angle with a fixed wavelength, while wavelength interrogation mode fixes the incident angle, whereas the SPR spectral profile and dip can be obtained by scanning the incident wavelength.97–100 Consequently, SPRi configurations provide a variety of applications for molecular sensing, healthcare testing and environmental screening with high throughput characteristics.97–1002.6 Plasmonic optical fibers
Generally, convection optical fibers are made of silica glass and have a solid core surrounded by a slightly lower refractive index cladding.102–105 By using the total internal reflection effect, they can guide light within the core. On the other hand, plasmonic fiber-optic biosensors rely on a thin metallic film or nanostructure along the length of the sensing area to generate SPR or LSPR.106 Therefore, part or all of the fiber cladding can be removed via chemical etching or by side-polishing methods, and nanoparticles or nanofilms can be deposited. Several optical fiber configurations such as unclad fibers, side-polished (or D-shaped) fibers, tapered, and U-shaped fibers have been demonstrated for sensing applications.106–109 As a label-free method, these plasmonic biosensors detect biomolecular interactions with high sensitivity and low levels of detection (LOD). Their broadband operation, along with their structural flexibility and nanomaterial functionalization, makes plasmonic optical fiber-based biosensors ideal for real-time and in situ biosensing and healthcare applications.106,107,1092.7 General characteristics of plasmonic biosensors
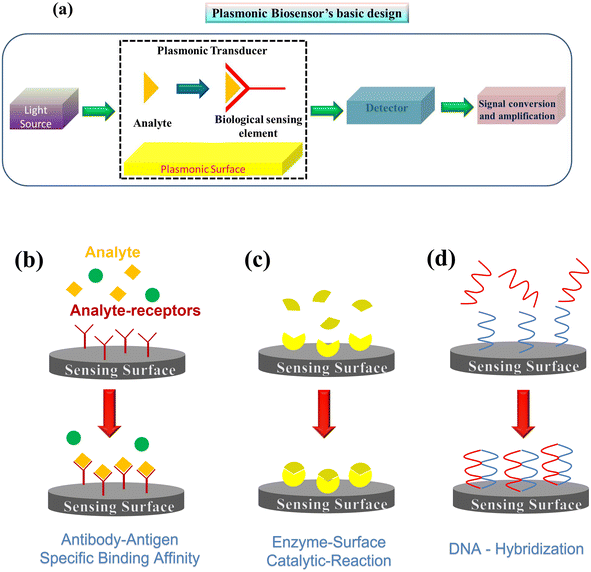
The basic components of a biosensor are illustrated in Fig. 3(a) and consist of the target analyte bound to the bioreceptor, the transducer, which converts the signal into a measurable quantity, and the reader device, which generates the results110,111 (detailed descriptions of these components are available elsewhere and are beyond the scope of this work110,111). In addition, chemical activation of the surface is crucial to improve the sensing efficiency of single virus particles. Some important features of analyte–receptor coupling on the plasmonic surface are shown in Fig. 3(b). Typically, in affinity-based plasmonic biosensors, surfaces are activated by biological receptors, such as antibodies, nucleic acids, cell membrane receptors, specifically designed peptides, aptamers, or molecularly imprinted polymers (MIPs).112 These biological entities show great affinity and specificity for certain analytes, allowing for the selective capture of the target with high sensitivity from complex biological samples. In the following section, we will focus on several parameters used to assess the performance of a biosensor.110 In the context of biosensing, the most important feature of a sensor is its sensitivity, S,110 it is described by eqn (3) and can be determined by the slope of the analytical calibration curve,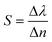 | (3) |
Another key parameter is the limit of detection (LOD) or sensor resolution, which is defined as the smallest amount of analyte that can be reliably detected by a specific measurement process. It is determined by the concentration of the analyte that produces a biosensor response corresponding to the standard deviation, σblank, of the biosensor response measured with no analyte and is given by:113,115
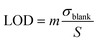 | (4) |
The performance of a plasmonic biosensor is strongly influenced by the spectral shape and background noise of the readout system. For the spectral shape, the Q-factor is an important parameter since it is a reliable indicator of the resolution of the detector for certain analytes and is given by:118
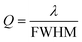 | (5) |
Another important characteristic of a biosensor is the figure of merit (FOM), which is the ratio between the sensitivity and full width at half maximum of the resonance spectra.114
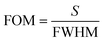 | (6) |
3 Plasmonic biosensors
Plasmonic optical biosensor technology has emerged as a powerful diagnostic tool.138–141 By selecting the appropriate biorecognition element, the technology can be applied to virtually any type of target molecule, from proteins, nucleic acids, bacteria, and drugs, and up to human cells,1,43,119,142,143 while many studies have demonstrated its utility in the biomedicine and environmental fields.30,37,89,95,115 In medicine, the accurate diagnosis of specific diseases is key for the timely and appropriate treatment and clinical management of a patient. Moreover, the rapid and early identification of certain viral diseases before the appearance of external symptomatology can also be important. This is the case with COVID-19; the availability of plasmonic biosensors for the rapid and accurate detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) may be useful for massive population screening, early detection of infected patients, and more efficient management of the pandemic.1,2,4,144 Owing to the versatility of plasmonic biosensors, the detection process can be modified. For example, the use of genomic RNA sequences of the virus target, instead of viral antigens, has enabled the rapid development of specific reverse transcription RT-PCR-based genomic assays.1 Hence, plasmonic biosensors can be applied to the direct and label-free detection of viral RNA by designing and immobilizing single-stranded DNA probes, as receptors, with complementary sequences to specific SARS-CoV-2 gene fragments on the sensor surface.1 Moreover, the sensitivity and specificity for SARS-CoV-2 could be increased with the combination of several probes targeting genes of the same virus.1 Henceforth, in this review, we will discuss recent plasmonic biosensor platforms for virus detection, with an emphasis on SARS-CoV-2 (Table 1).| Structure | Virus detected | Detection format | LOD sensitivity | Ref. |
|---|---|---|---|---|
| Ag/Au (35 nm/10 nm) chips | Avian influenza H7N9 | Monoclonal antibody (IgM) | 144 copies per mL | 122 |
| Cr/Ag/Au (3 nm/40 nm/10 nm) chips | Human enterovirus 71 | Enterovirus antibody | 67 virus particles (vp) per mL | 123 |
| Cr/Au (2.5 nm/47 nm) chips | H1N1, RSV, adenovirus, SARS | PCR amplified viral bodies | 0.5 nM for adenovirus/2 nM for SARS | 124 |
| Au SPR chip | Ebola virus | Monoclonal antibodies | 0.5 pg mL−1 | 125 |
| Biacore X bare gold chip | HIV | Hairpin DNA, capture probes | 48 fM | 126 |
| Array of Au nanoprism | Rotavirus | Rotavirus capsid (2B4) antibody | 126 ± 3 PFU mL−1 | 127 |
| Array of Au nanodisks and nanodots | Ebola virus | A/C protein | 220 fg mL−1 | 18 |
| Planar toroidal gold metamaterial | Zika virus | Immobilized antibody | 5.81 GHz log(pg mL−1)−1 | 128 |
| Au toroidal metasensor | SARS CoV-2 | SARS antibody | 4.2 fmol | 129 |
| Au nanospikes | SARS CoV-2 | SARS antibody | 0.08 ng mL−1 | 13 |
| Au nano-island layer | SARS CoV-2 | Thiol cDNA receptor | 0.22 pM | 130 |
| Hetero-assembled Au nanoparticles layer | Hepatitis B virus | Hepatitis antibody | 100 fg mL−1 | 131 |
| Au spike-like nanoparticles | Avian influenza virus | DNA–hemagglutinin binding aptamer | 1 pg mL−1 | 132 |
| Au (∼20 nm) particles | Norovirus | Norovirus recognizing affinity peptide | 9.9 copies per mL | 133 |
| Bioconjugated Au nanoparticle (10–15 nm) | Dengue and West Nile viruses | Antiflavivirus 4G2 antibody | 10 plaque-forming units (PFU) per mL | 134 |
| SiO2/Au particles (4 nm/100 nm) | Zika virus | Anti-Zika (NS1) antibody | 10 ng mL−1 | 135 |
| Ag particles (20–80 nm) | Dengue virus | NS1 antibody | 0.06 μg mL−1 | 136 |
| Au particles (40 nm) | SARS CoV-2 | Nucleocapsid (N) protein | 150 ng mL−1 | 137 |
3.1 Biosensing using plasmonic nanostructures
The first implementation of plasmonic label-free biosensors for influenza virus detection was reported 25 years ago.145 Since then, researchers have developed a variety of biosensor assays for rapid virus detection and quantification based on plasmonic technologies.146 Chang et al. reported a simple strategy for avian influenza A (H7N9) virus detection using an intensity-modulated SPR biosensor integrated with a monoclonal antibody (Fig. 4(a)).122 Specifically, the authors employed a Kretschmann configuration using an Ag/Au (35 nm/10 nm) chip to increase the selectivity for the virus. They noted an LOD of 144 copies per mL, which indicated a sensitivity 20-fold higher than with target-captured ELISA using antibodies, and better than conventional RT-PCR tests.122 Furthermore, they evaluated their configuration using mimic clinical specimens containing the H7N9 virus mixed with nasal mucosa from patients with flu-like symptoms and noted a detection limit of 402 copies per mL, which was far superior to conventional influenza detection assays, and a rapid testing time of under 10 min.122 Likewise, Prabowo et al.123 demonstrated a portable SPR biosensor for the quantification of enterovirus antibodies, which showed a detection limit of 67 copies per mL. In another study, an SPR-based biosensor was used to detect nine common respiratory viruses with an LOD of 2 nM for SARS,124 while an SPR chip developed to detect the Ebola virus showed a sensitivity of 0.5 pg mL−1.125 The authors modified a gold chip with 4-mercaptobenzoic acid and used three monoclonal antibodies of Ebola virus to study the efficiency based on the affinity constant.125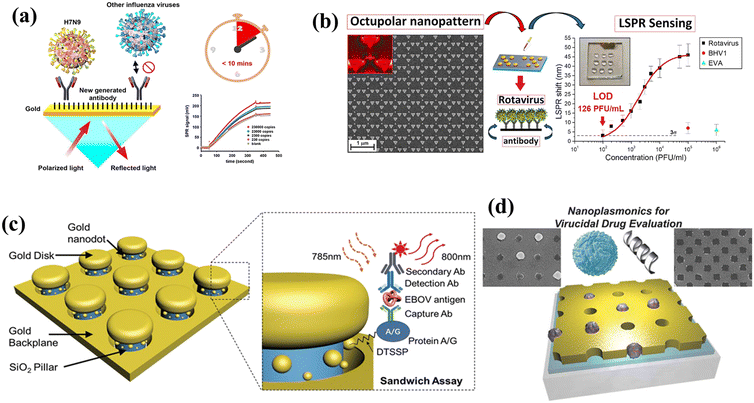 | ||
| Fig. 4 (a) Schematic of a plasmonic biosensor used to identify avian influenza H7N9. A bare Ag/Au chip is cleaned before surface functionalization with self-assembled monolayers. The capture antibody, at a concentration of 10 μg mL−1, is covalently immobilized to the reaction spot of the chip (reproduced with permission from ref. 122). (b) Left: Scanning electron microscopy images of the octupolar geometry-based Au nanostructure used for rotavirus detection. The minimum interparticle distance between two unit cells is 25 nm. Right: Average LSPR peak shift (black square) and Langmuir isotherm fitting (red line) for various concentrations of rotavirus in distilled water (reproduced with permission from ref. 127). (c) Schematic of a nano-antenna array for Ebola virus detection. The gold nanodisks and backplane are separated by SiO2 nanopillars, forming nanocavities. Gold nanoparticles are present on the SiO2 pillar surfaces, where the localized electromagnetic field around the nanostructure is highest (reproduced with permission from ref. 147). (d) Schematic of a periodic gold nanohole array that was designed in order to selectively capture lipid vesicles and virus particles inside the nanoholes. The 10 × 10 mm gold nanohole array was formed on a glass substrate by the template-stripping method. An optical adhesive layer is present between the gold and glass (reproduced with permission from ref. 148). | ||
A biosensing platform developed by Diao et al.126 based on the Biacore X analytical system was able to obtain 48 fM of HIV-1-related DNA using entropy-driven strand displacement reactions (ESDRs) as an isothermal, label-free nucleic acid amplification technique. The authors developed a sensitive SPR biosensing strategy for enzyme and label-free detection based on DNA nanotechnology.126 The whole detection process was accomplished in 60 min with high accuracy and reproducibility.126 The authors noted that the observed biosensing performance could be attributed to the perfect combination of a hairpin probe, ESDR circuit, and DNA tetrahedra on the SPR biosensing chip.126 Another SPR device has been demonstrated to investigate antigen–antibody interactions of chicken infectious bronchitis coronavirus.149 The authors utilized a prism configuration using a 50 nm Au film to increase the specificity of the virus.149 Compared with the expected response rate, the SPR sensor had much smaller differences (up to 6.3 times) between specific and non-specific interactions.149 The effect of buffer acidity changes was also investigated, showing a six-fold reduction in non-specific interactions.149
A two-dimensional octupolar geometry-based gold nanostructure was fabricated by Rippa et al.127 to detect ultrasmall concentrations of rotavirus, which is the main cause of childhood viral gastroenteritis in humans (Fig. 4(b)). Specifically, the authors designed an array of units comprising three large identical triangular gold nanoprisms (side length, 200 nm) and one smaller inner prism (side length, 80 nm), with a 25 nm separation between adjacent units.127 An LOD of 126 ± 3 PFU mL−1 using a very low sample volume (2 μL) was estimated. In addition, the authors evaluated their plasmonic biosensor with two more viruses (bovine herpesvirus [BHV1] and equine viral arteritis [EVA]) to confirm its sensitivity and specificity. A maximum LSPR peak shift of 7 nm from a concentration of 1 × 105 PFU mL−1 for BHV1 was measured, while a maximum shift of 6 nm was observed for EVA.127 A microfluidic polymerase chain reaction with a gold nanoslit-based SPR sensor was fabricated to detect the DNA sequence of latent membrane protein 1 (LMP1) from Epstein–Barr virus (EBV)-positive cells.150 The device was divided into the PCR microchannel and gold nanoslit of 80 nm width.150 The microfluidic PCR was integrated with the nanoslit SPR chip, using heat-resistant double-sided tape.150 Finally, the sensor was evaluated using samples from nasopharyngeal cancer patients and completed the analytical procedure in 36 min, with an LOD of 10−11 g mL−1.150
Recently, an array of gold nano-antennas that uses a sandwich immunoassay format has been fabricated for single-molecule detection of Ebola virus antigens (Fig. 4(c)).147 The nano-antenna consists of SiO2 nanopillars bound to gold nanodisks and nanodots, which enhance the fluorescence signal through the formation of nanocavities.147 The authors used a thiol–gold link and a protein A/C layer to simultaneously functionalize the surface of the nanopillars and to prevent signal losses on the gold surfaces.147 They noted a detection sensitivity for the Ebola virus soluble glycoprotein in human plasma of 220 fg mL−1; this was a significant improvement over the recommended immunoassay test for Ebola virus antigens.147 In addition, the interaction of light with the periodic array of nanoholes enabled the extraordinary optical transmission effect,151 which enhanced the transmission of light at specific wavelengths. These spectral characteristics have facilitated the development of high-sensitivity plasmonic biosensors that can be integrated with microfluidics. A metallic nanohole-based assay was developed148 to capture single virus-like particles (Fig. 4(d)). The diameter of the nanoholes was chosen to fit the size distribution of virus particles that had been treated with a virucidal drug candidate.148 The sensing performance of the platform was evaluated by monitoring resonance shifts for the virucidal-induced capture of single virus-like particles, showing a minimum RI resolution of 5.5 × 10−5 RIU.148 The authors noted high RI sensitivity in the functionalized nanoholes with a low surface coverage when compared with non-functionalized nanoholes.148
Since terahertz waves are non-ionizing and harmless to organic tissues and biomolecules, they may become increasingly attractive for biomedical applications.156 A terahertz gold metasensor was designed for Zika virus envelope protein detection.128 Based on toroidal metamaterial properties, these devices support resonances that possess much higher sensitivity to RI perturbations in the surrounding media.82,83 The toroidal metamaterial consisted of an array of mirroring asymmetric split resonators and had the ability to support a Q-factor128 around 30. By measuring the shift of the toroidal dipolar momentum, the authors determined the LOD and sensitivity of the metasensor to be 560 pg mL−1 and 5.81 GHz log(pg mL−1)−1, respectively, for a variety of Zika virus concentrations.128
A plasmonic nanohole array with S protein antibodies immobilized on the surface was fabricated152 to detect a broad range of pathogens in a typical biology laboratory setting (Fig. 5(a)). By capturing the S proteins, whole virus particles could be suspended in the nanohole array, which resulted in a red-shift of the resonance.152 A plasmonic microfluidic biosensing platform was developed by Funari et al. who demonstrated the utility of electrodeposition-based gold nanospikes combined with optical probes.13 Based on local RI changes caused by the interaction of the SARS CoV-2 S protein and antibodies in the diluted human serum, a shift of the LSPR resonance peak was detected, with a detection concentration of 0.08 ng mL−1.13 The authors noted that the proposed platform could complement existing serological assays and improve COVID-19 diagnosis.13 A dual functional plasmonic detection platform that combines the plasmonic photothermal33 and LSPR effects has been reported for SARS-CoV-2 detection130,153 (Fig. 5(b)). Two-dimensional gold nano-islands functionalized with RdRp-COVID cDNA (RdRp-COVID-C) receptors permit the selective detection of RdRp-COVID-C through DNA hybridization, giving an LOD for the cDNA of 0.22 pM. This provides a new approach for SARS-CoV-2 detection.130,153
 | ||
| Fig. 5 (a) Schematic of the thermoplasmonic-assisted dual-mode approach. Amplification-free-based direct viral RNA detection and amplification-based cyclic fluorescence probe cleavage detection are combined to provide SARS-CoV-2 detection within 30 min (reproduced with permission from ref. 152). (b) A label-free optofluidic nanoplasmonic sensor that can detect vesicular stomatitis virus and pseudotyped Ebola virus from biological media with little to no sample preparation (reproduced with permission from ref. 153). (c) Schematic of a biosensor based on a plasmonic plastic optical fiber coupled with a novel type of synthetic molecularly imprinted polymer (MIP) receptor for the specific recognition of subunit 1 of the SARS-CoV-2 spike (S) protein (reproduced with permission from ref. 154). (d) Biosensing configuration based on an SPR D-shaped plastic optical fiber integrated with an aptamer for the recognition of the receptor-binding domain (RBD) of the S glycoprotein of SARS-CoV-2 (reproduced with permission from ref. 155). | ||
A label-free detection assay scheme based on an antifouling polymer brush biointerface prepared on a gold-coated piezoelectric quartz crystal microbalance chip was fabricated to enable the quantitative analysis of N-protein of SARS-CoV-2.158 This device improved the bioassay sensitivity to a clinically relevant LOD of 1.3 × 104 PFU mL−1 within a detection time of only 20 min.159 A customized SPR serological biosensor based on the Kretschmann configuration incorporated with microfluidic components in a compact and user-friendly platform was demonstrated for identification and quantification of SARS-CoV-2 antibodies in clinical samples.160 By using polyclonal antibodies, this portable plasmonic device has diagnostic sensitivity (99%) and specificity (100%) for clinical COVID-19 positive and negative samples.160 Ansah et al. introduced a methodology for the synthesis of interior hotspots templated with protein for label-free and on-site SERS detection of the virus.161 Specifically, the structure consisted of Au nanocavity electrode (AuNC) with a large surface area, high aspect ratio, and negligible background noise fabricated on a Si substrate.161 A supporting electrolyte solution of NaCl, protein and Au precursor was used during electrochemical deposition to synthesize the core–shell structure that encapsulated the lysate protein of SARS-CoV-2 (SLs) protein.161 The whole process was carried out under an applied potential of 0.3 V and illumination of a laser beam at 785 nm.161 They authors detected SLs with an LOD value of 10−1 PFU mL−1.161 An inverted gold pyramidal metasurface was designed, fabricated, and evaluated for label-free hepatitis A virus (HAV) detection by employing SERS.162 The authors fabricated 300 × 300 μm2 Au nanostructures based on periodic arrays of inverted pyramidal nanoholes (PNHs) by using an EBL method.162 The PNHs are equilateral triangular based (side size of 390 nm) and arranged in a triangular geometry.162 For this study, the 785 nm excitation wavelength was used to test the SERS performances of the PNHs.162 The HAV was detected at a concentration of 103 PFU mL−1, corresponding to 13 pg mL−1.162
3.2 Biosensing using plasmonic two-dimensional materials
A number of studies on two-dimensional (2D) layered materials for biochemical sensing applications have exploded since the synthesis of graphene.163 In a variety of healthcare applications, these materials (e.g. graphene, transition metal dichalcogenides, graphitic and transition-metal carbides) can serve as active sensing elements or supporting substrates due to their electrical, optical, electrochemical, and physical properties, which are often tunable.164–167 By reducing their geometrical dimensions, 2D-materials enhance plasmonic field confinement and enable the excitation of plasmons, excitons, and phonons to demonstrate new biosensing functionalities.166–168 In the literature, several review papers164,165,169,170 have addressed the role of 2D-materials in developing detection platforms with high sensitivity and selectivity towards the target analyte.For example, graphene oxide (GO)-based fluorogenic peptide probes were designed to allow the differentiation of Ebola virus from Marburg virus which has a similar capsid protein composition as well receptor-extensive vesicular stomatitis virus.171 The authors used an array of three fluorescent peptide fragments with modest affinity and selectivity for these three viruses.171 By increasing the concentration of the peptide probes and applying statistical analysis, the authors determined the characteristic patterns for each virus in a simple way without the need for specific peptide ligands.171 A reduced-GO-based SPR sensor functionalized with specific antibodies was developed for the detection of dengue virus for concentrations of 0.1 pM.172 Peng et al. demonstrated a sensing platform based on ultrathin 2D-MXene Ti3C2 nanosheets for the detection of PVR-amplified HPV-18 DNA from cervical scrape samples obtained from human papillomavirus infected patients.173 The authors noted high sensitivity and selectivity for HPV-18 determination, with an LOD of 100 pM.173 Molybdenum disulfide (MoS2) nanostructures166,167,174,175 were synthesized for nanomedicine and biosensing applications.176,177 For instance, a MoS2-based biosensor was developed to quantify hepatitis C virus gene (HCV).175 The authors employed an isothermal enzyme-free hybridization chain reaction for DNA amplification to form long dsDNA to explore the plasmon effect of MoS2 nanosheets.175 The proposed biosensor detected HCV gene from 0.5 pmol L−1 to 1 nmol L−1 with an LOD of 0.17 pmol L−1.175
3.3 Two-dimensional (2D) materials for SARS-CoV-2 detection
A 2D heterostructure based on PtSe2/graphene was attached to the gold film of the SPR biosensor for the rapid detection of the coronavirus.178 In this work, the performance of the biosensor was investigated with three different ligand–analytes: (i) the monoclonal antibodies as the ligand and the coronavirus spike receptor-binding domain (RBD) as the analyte, (ii) the virus spike RBD as the ligand and the virus anti-spike protein (IgM, IgG) as the analyte, and (iii) the specific RNA probe as the ligand and the virus single-stranded RNA as the analyte.178 The heterostructure PtSe2/graphene provided an increased surface area for better absorption of the target analyte and enhanced the sensitivity of the biosensor. The authors noted a sensitivity of 833 THz RIU−1 in SPR frequency for COVID-19 virus spike RBD detection.178 One year later, the same research group proposed a graphene-coated BK7/WS2/Au/BaTiO3 sensor for detecting SARS-CoV-2 virus at an early stage.179 The authors noted a sensor angular sensitivity of 230.7 deg RIU−1 for detecting the SARS-CoV-2 whole virus and an angular sensitivity of 227.6 deg RIU−1 for the detection of monoclonal antibodies against the SARS-CoV-2 virus.179 In another study, selective detection of COVID-19 virus is demonstrated by using an antibody functionalized graphene material as a Raman transducer platform.180 The phononic energy of graphene is strongly influenced by the change in its doping level when an analyte molecule is attached.180 The authors noted that when negatively charged COVID-19 spike RBD proteins are bonded to CoV-2 spike RBD antibody functionalized graphene, a blue shift in the phonon vibration mode peak results from the p-doping of the p-type graphene.180 The achieved LOD with this graphene-based phononic biosensor was 3.75 fg mL−1 and 1 fg mL−1 of spike virus protein in artificial saliva and buffer solution, respectively.1803.4 Biosensing using plasmon-based optical fibers
In the past few decades, optical fibers have evolved from an optical transmission waveguide to important components of applications ranging from small particle manipulation102–105,181 to medical imaging.182 In the past decades, a new class of optical fiber sensors based on SPR has been added to the family of PoC devices.183 Plasmonic fiber-optic biosensors offer an interesting alternative to classical prism-based configurations and are advantageous in terms of flexibility and cost. Plasmonic optical fiber platforms have provided miniaturized sensing approaches for the determination of clinical biomarkers.184An SPR-based optical fiber device has been developed for the analysis of avian influenza virus subtype H6.185 The SPR-based optical fiber consists of a 40 nm thin gold film and a side-polished structure.185 To optimize the self-assembled monolayers and subsequent antibody functionalization, the detection surface of the SPR-based optical fiber was modified with plasma at low temperature, which rendered better results than chemical modification.185 The binding interaction between immobilized antibodies and antigens on the cell surface was evaluated with 104 to 108 embryo infectious dose (EID)50 per 0.1 mL of virus, leading to an LOD of 5.14 × 105 EID50 per 0.1 mL and an average response time of 10 min.185 By combining the optical properties of LSPR nanostructures with the total internal reflection of optical fiber configurations, better spatial sensitivity can be achieved. For example, the integration of gold nanorods into a fiber-optic platform permitted the development of an immunosensor for the determination of Cymbidium mosaic virus and Odontoglossum rings spot virus186 in plants. To achieve direct sensing of the analytes, gold nanorods were employed to generate a near-infrared sensing window to solve the color interference issue of sample matrices.186 The optical fiber LSPR-based platform provided an LOD of 48 pg mL−1, while the RI resolution was 8 × 10−6 RIU.186 The authors noted that the improvement in sensitivity in comparison with ELISA was attributed to the properties of nanorods, which simultaneously prevented the color interference of similar-sized nanospheres.186 A tilted fiber grating surface coated with gold nanoparticles has been demonstrated for the detection of Newcastle disease virus (NDV).187 Modification of the fiber cladding with gold nanoparticles (with an average diameter of 80 nm) enhanced the sensitivity as a result of the LSPR field, while activation of the nanoparticles with staphylococcal protein A improved the bioactivity of anti-NDV monoclonal antibodies by up to ten times compared with that of a tilted fiber grating without gold nanoparticles.187 Monitoring of resonance wavelength red-shifts showed a minimum detectable amount for virus of approximately 5 pg; this is slightly better than that achievable by RT-PCR (10 pg).
3.5 Biosensing using plasmonic nanoparticles
The characteristics of metal nanoparticles have found the greatest use in LSPR biosensing, with several applications utilizing this technique.144,192–194 For example, a sandwich immunoassay with gold nanoparticles was demonstrated to detect the hepatitis B virus (HBV) surface antigen (HBsAg).131 For this purpose, a glass substrate was fabricated with synthesized gold nanoparticles (AuNPs) of three different sizes (15, 30, and 50 nm) and conjugated with an anti-HBsAg antibody to detect the target antigen.131 After 10 min, a second layer of AuNPs conjugated with the anti-HBsAg antibody was added to obtain a hetero-assembled chip, the LOD of which was 100 fg mL−1.131 Takemura et al. used the LSPR signal from Ab-conjugated thiol-capped AuNPs to amplify the fluorescence intensity signal of quantum dots for the detection of nonstructural protein 1 (NS1) of the Zika virus.195 Their biosensor had a wide detection range of 10–107 RNA copies per mL and maintained its specificity with human serum.195 Chowdhury et al. also developed a biosensor using AuNPs and CdSeTeS quantum dots to identify the serotypes of dengue virus.196 The biosensor had LOD at the femtomolar level and was successfully applied to RNA extracted from dengue virus culture fluids.196 Lee et al. constructed a label-free biosensor for avian influenza virus (H5N1) using hollow spike-like AuNPs and a three-way DNA junction.132 To achieve multifunctionality, each piece of DNA was tailored to aptamers specific for the hemagglutinin (HA) protein of the virus, fluorescence dye, and thiol group. The sensor detected the HA protein of H5N1 in phosphate-buffered saline and chick serum with an LOD of 1 pM.132 Heo et al. fabricated gold nanoparticles with an approximate average size of 20 nm to detect human norovirus.133 The authors' novel sensing approach utilized noroviral protein-recognizing affinity peptides, which are relatively cost-effective compared with antibodies, to bind noroviral proteins.133 They noted an LOD of the capsid protein of 9.9 copies per mL.133AuNPs or roughened gold surfaces are also widely used in SERS spectroscopy because of their LSPR properties.144 Paul et al. developed an antibody-conjugated AuNP-based SERS probe for the identification of mosquito-borne viruses.134 They successfully detected dengue virus type-2 and West Nile virus at a low concentration of 10 PFU mL−1.134 Camacho et al. designed SERS nanoprobes using gold shell-isolated nanoparticles, which contained 100 nm gold nanoparticles and 4 nm silica shells. The silica shells modified with Nile blue functioned as the Raman reporter.135 This configuration was irradiated with a 633 nm wavelength laser beam, and the Raman signal was recorded by a mapping process. The nanoprobes successfully detected Zika virus at a very low concentration (around 10 ng mL−1) and without any cross-reactivity with dengue virus.135 Luan et al. developed a stable and bright fluorescent plasmonic nanoscale construct that consisted of a bovine serum albumin (BSA) scaffold with approximately 210 IRDye 800CW fluorophores, a polysiloxane-coated gold nanorod acting as a plasmonic antenna, and biotin as a high-affinity biorecognition element.197 This configuration was able to improve the LOD of fluorescence-linked immunosorbent assays by up to 4750-fold, shorten the overall assay time (to 20 min), and used lower sample volumes.197 The authors attributed this improvement in sensitivity to the BSA blocking method, in which BSA acts as a blocking agent to minimize non-specific binding of the plasmonic fluorophore to arbitrary surfaces and biomolecules. In another study, an ATR-LSPR-based optical platform was employed to demonstrate the detection of ChiLCVa plant virus using its complementary DNA sequence as a receptor.198 Gold nanoparticles of 50 nm diameter were immobilized on a functionalized coverslip surface by the ATR-configured evanescent wave-based LSPR absorption method.198 The sensor's LOD was 1.0 μg ml−1 for the plant viral DNA sample, while unique binding dynamics were observed compared to non-specific DNA.198
Compared with gold nanoparticles, silver nanoparticles display a higher efficiency of LSPR excitation and a wider wavelength range.199 Moreover, silver nanoparticles have sharper LSPR bands, are less dissipative, and perform better in SERS.193 However, there are fewer studies on AgNPs than AuNPs as plasmonic biosensors. One reason behind this may be that AgNPs display toxicity200,201 and antiviral effects.202,203 Another reason is that bare AgNPs are not as stable as AuNPs because of quicker oxidation.204 To deal with these drawbacks, AgNPs are usually coated with different materials.205 The thickness and the type of coating material greatly influence the optical properties of AgNPs. A thermally annealed thin silver film deposited onto a silicon substrate was used to detect the NS1 antigen of dengue virus in whole blood.136 After the annealing process, silver nanoparticles with diameters from 20 to 80 nm were generated and with inter-structural spacing ranging from a few tens to about 100 nm.136 The authors determined the system to have an RI sensitivity of 10−3, while an increase in absorption and a red-shift of 108 nm of the peak absorption wavelength were observed with antigen binding.136 The sensitivity of this configuration was found to be 9 nm μg−1 mL−1, and the LOD was 0.06 μg mL−1. Hong et al. developed hybrid slot antenna structures with silver nanowires in the terahertz frequency range to detect bacteriophage PRD1 and obtained an enhancement factor of 2.5 for a slot antenna width of 3 μm.206
A SERS platform has been used to detect HBV.207 The authors used a standard, label-free Ag nanoparticle solution as the SERS-active substrate to test blood serum samples from HBV patients and healthy volunteers.207 The SERS spectra of the serum samples from both the infected patients and healthy volunteers were compared by employing linear discriminant analysis.207 Using this approach, a SERS spectrum was produced in 10 min for each sample and with a diagnostic sensitivity of 91.4%, indicating the great potential of this platform for a quick, non-invasive, label-free diagnostic method through the implementation of principal component analysis (PCA).207
The detection of spike proteins of alpha, beta, and gamma variants of the COVID-19 virus was achieved by specific nanostructured molecularly imprinted polymers (nanoMIPs).211 The nanoMIP-functionalised LSPR sensor detected all 3 protein variants with a limit of detection of 9.71 fM, 7.32 fM and 8.81 pM using wavelength shifts for alpha, beta and gamma spike protein variants, respectively.211 The LSPR sensing scheme of this device is based on Ag nanoparticles with an average diameter of 22.47 nm and an aspect ratio of 1.15 on a glass substrate.211 A three-dimensional porous microplasma-engineered nanoassembly (AgMEN) was fabricated to provide a high sensitivity to SARS-CoV-2 S variants, including wild-type, alpha, delta, and omicron detection via remarkable SERS signal collection.212 An LOD of 1 fg mL−1 and 0.1 pg mL−1 was noted for the S and N spike proteins, respectively.212
Behrouzi and Lin applied LSPR of antigen-coated AuNPs to detect the N protein of SARS-CoV-2.137 This detection method gave naked-eye results in 5 min and an LOD of 150 ng mL−1.137 Park et al. used self-assembled AuNP arrays for the detection of the SARS-CoV-2 S protein.213 Their biosensor gave quick results with high sensitivity in just 10 min and without any purification steps.213 Both aforementioned sensors could be used for the PoC detection of SARS-CoV-2. In addition, Das et al. achieved an LOD of the S protein of 111.11 deg RIU−1 using a gold nanorod with a Kretschmann prism configuration.214 Zavyalov et al. built a SERS aptasensor based on hydroxylamine-reduced AgNP substrates and successfully detected SARS-CoV-2 in about 7 min with an LOD of 5.5 × 104 median tissue culture infectious dose per mL.215 Tripathi et al. deposited AgNPs over glass coverslips and used them as SERS substrates.216 The sensor was used to detect the Japanese encephalitis virus and demonstrated ultrasensitive detection, with a detection limit of 7.6 ng mL−1 and a linear response from 5 to 80 ng mL−1.216 A colorimetric assay based on AgNPs with diameters less than 60 nm and capped with thiol-modified antisense oligonucleotides specific for the N protein of SARS-CoV-2 has been demonstrated as being capable of diagnosing positive COVID-19 cases from isolated RNA samples within approximately 10 min.217 A naked-eye (equipment-free), label-free, and RNA extraction-free method for SARS-CoV-2 detection was developed by employing anisotropic Au nanoparticles.218 In this study, a specific sequence in the N-gene of SARS-CoV-2 was selected as a target to design antisense oligonucleotides (ASOs) with an extra strand polyguanine (G12).218 Inactivated virus samples were added to anisotropic AuNPs synthesized on four ASOs, and after the annealing process, the color of the solution changed from red to purple.218 The authors noted that in microfluidic paper-based analytical devices, this conjugation method allowed hybridization and annealing without a denaturation step, and its corresponding color change could be observed by the naked eye in the detection zone.218
4 Perspectives and outlook
Progress in material science and fundamental optics will continue to provide advantages to biosensing research. An important facilitator of this progress is the use of numerical analysis tools, such as COMSOL Multiphysics software,38 to explore geometrical and material parameters for the optimization of the biosensor's performance. The combination of various optical and non-optical, for example electrochemical, detection configurations on a single platform could also enable multifunctional biosensors to extract information from a given sample. For example, plasmon-enhanced electrochemiluminescence (ECL)-based sensors are a promising option that deserves further attention.219 The ECL process uses an oxidized luminescent substrate to stimulate a fluorescent acceptor.219 Therefore, non-specific signals that are caused by external light can be minimized. In this regard, Fan et al. developed an ECL-based biosensor that amplifies the signal by employing a DNA walker strategy for the detection of the SARS CoV-2 RdRp gene.220 Despite their simplicity and high sensitivity, plasmon-enhanced ECL-based biosensors present many challenges when used in point of care tests;220 more efforts are needed to overcome them. In light of this, plasmonic colorimetric biosensing based on etching or growth of metal nanomaterials presents excellent prospects due to their simplicity and ease of use as test strips.221 In order to detect SARS-CoV-2, colorimetric tests were developed using plasmonic biosensors with Au nanoparticles functionalized with polyclonal antibodies (f-AuNPs).222 The authors observed intense color changes with the naked eye when f-AuNPs accumulated on the virus and noted a detection limit of 0.28 PFU mL−1 in human saliva.222 In addition, the integration of SERS with etching-based plasmonic colorimetric sensors could lead to novel, extremely sensitive detection devices.221 Thus, it is possible to significantly improve detection accuracy and reproducibility, as well as detect targets in complex samples and serum, by combining several detection methods.Likewise, two-dimensional materials, such as graphene, can provide dynamic control of plasmonic resonances, which is needed for small molecule detection.223 Moreover, we envision that hybrid substrate integrating polar materials, van der Waals heterostructures, metal antennas, and metasurfaces will open new avenues for future biosensing innovation. Furthermore, integration with birefringent and chiral metamaterials for example chiral plasmon224 layers for measuring chiro-optic response could advance enantioselective biochemical sensing applications.
Another alternative technique to be considered is nanopore technology, which allows for the precise detection of subunits as well as the sequencing of pathogen DNA and RNA in an effective and versatile way; this technology will be at the forefront of future state-of-the-art approaches.225 Nanopore-based sequencing systems, such as the one developed by Oxford Nanopore Technologies, were successfully applied to SARS-CoV-2 strains at the early stages of the pandemic.226,227 The rapid and real-time detection of mutagenized virus is a key benefit of this technique, providing important data for further epidemiological analysis.
In parallel, the availability of a variety of metal nanoparticle synthesis protocols, as well as an increase in the number of commercial nanoparticle providers, may contribute to the development of novel biosensors with high specificity and selectivity.1,5,43,144 To develop high-sensitivity tests for SARS-CoV-2, the selection of metallic nanoparticles with appropriate sizes and shapes is a key point since their physical and optical properties can greatly influence the performance of a nanoparticle-based diagnostic system. Although spherical nanoparticles have been studied most extensively because of their ease of synthesis, other shapes are worth investigating when a higher sensitivity or a different sensing strategy is desired but not achievable with nanospheres. Regarding nanoparticle size, large metallic nanoparticles have large absorption cross-sections and may result in systems with higher sensitivities than those utilizing small nanoparticles. However, all of these parameters need to be addressed and evaluated on a project-by-project basis as many other factors could influence the LOD.228
Despite the excellent biosensing performance of plasmonic diagnostic tools, several technological aspects still require considerable improvement before fully operative devices for clinical diagnosis and real-world applications can be realized. Factors that need to be addressed include cost, sensitivity, specificity, and reproducibility, as well as user interfaces and connectivity that allow for real-time monitoring of data collection. In terms of cost, inexpensive disposal chips are necessary to avoid cross-contamination issues and complicated cleaning procedures while handling biological samples or body fluids. In this regard, an integrated approach that allows for single-use cartridges and a stand-alone reader is desirable. Microfluidic technology can also play a key role in providing disposable, stable over time, and easy to manipulate cartridges through the incorporation of biochips with specific biofunctionalities for each detection assay. For airborne respiratory viruses, it will be essential to integrate such cartridges with additional safety steps, including sample preprocessing, before final detection.229 Another possible advancement is the merging of plasmonic devices with smartphones; their light sources, cameras, and image processing and communication capabilities can reduce costs and facilitate large scale distribution.230–233 Therefore, the development of portable and wireless nanobiosensors is essential for diverse applications. Sample collection and processing is an additional consideration for on-site biosensing. The large diversity of analytes and the matrix composition of specimens such as body fluids still remain a challenge. For example, virus detection from clinical specimens is still limited owing to the lack of suitable methods to prevent the interference of biomolecules in body fluids. In this sense, the design of antifouling coatings that can take into account either the composition of the media or the biological receptor's characteristics may help to bridge the gap between common analytical methods and plasmonic biosensing applications.
In summary, this tutorial review highlighted the physics underpinning the mechanics of plasmonic-based biosensors, the current progress of biosensor research, and the ability of such devices to detect virus particles viruses. It is worth noting that, although high sensitivity is always the main goal of any biosensor, for better clinical and commercial translation, it is essential to balance the trade-off between the sensitivity, cost-effectiveness, portability, and stability of these plasmonic-based systems. Against the backdrop of the COVID-19 global pandemic, continued biosensor development is crucial for the realization of more portable and affordable platforms that can meet global healthcare needs.
Conflicts of interest
The authors declare that they don't have any competing financial interests or personal relationships that could have appeared to influence the work reported in this tutorial review.Acknowledgements
This work was supported by funding from Duke Kunshan University and Synear and Wang-Cai donation labs (DKU).Notes and references
- M. Soler, M. C. Estevez, M. Cardenosa-Rubio, A. Astua and L. M. Lechuga, ACS Sens., 2020, 5, 2663–2678 CrossRef CAS PubMed.
- G. Giovannini, H. Haick and D. Garoli, ACS Sens., 2021, 6, 1408–1417 CrossRef CAS PubMed.
- M. M. Hassan, F. S. Sium, F. Islam and S. M. Choudhury, Sens. Bio-Sens. Res., 2021, 33, 100429 CrossRef.
- A. Asghari, C. Wang, K. M. Yoo, A. Rostamian, X. Xu, J.-D. Shin, H. Dalir and R. T. Chen, Appl. Phys. Rev., 2021, 8, 031313 CAS.
- H. Altug, S.-H. Oh, S. A. Maier and J. Homola, Nat. Nanotechnol., 2022, 17, 5–16 CrossRef CAS PubMed.
- A. A. Serafetinides, M. Makropoulou, D. G. Kotsifaki and G. Tsigaridas, Society of Photo-Optical Instrumentation Engineers (SPIE) Conference Series, 2016, p. 1022613 Search PubMed.
- A. M. Caliendo, Clin. Infect. Dis., 2011, 52, S326–S330 CrossRef PubMed.
- N. Boonham, J. Kreuze, S. Winter, R. van der Vlugt, J. Bergervoet, J. Tomlinson and R. Mumford, Virus Res., 2014, 186, 20–31 CrossRef CAS PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. Chu, T. Bleicker, S. Brünink, J. Schneider, M. L. Schmidt, D. G. Mulders, B. L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J.-L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M. P. Koopmans and C. Drosten, Eurosurveillance, 2020, 25, 1–8 Search PubMed.
- F. Cui and H. S. Zhou, Biosens. Bioelectron., 2020, 165, 112349 CrossRef CAS PubMed.
- J. Louten, Essential Human Virology, Academic Press, Boston, 2016, pp. 19–29 Search PubMed.
- C. M. Michaud, Encyclopedia of Microbiology (Third Edition), Academic Press, Oxford, 3rd edn, 2009, pp. 444–454 Search PubMed.
- R. Funari, K.-Y. Chu and A. Q. Shen, Biosens. Bioelectron., 2020, 169, 112578 CrossRef CAS PubMed.
- L. Rubio, L. Galipienso and I. Ferriol, Front. Plant Sci., 2020, 11, 1092 CrossRef PubMed.
- C. J. Menendez-Botet, Clin. Chem., 2003, 49, 1424–1425 CrossRef CAS.
- D. Mabey, R. W. Peeling, A. Ustianowski and M. D. Perkins, Nat. Rev. Microbiol., 2004, 2, 231–240 CrossRef CAS PubMed.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- X. A. Zhang, I.-T. Chen and C.-H. Chang, Nanotechnology, 2019, 30, 352002 CrossRef CAS PubMed.
- P. L. Mage, B. S. Ferguson, D. Maliniak, K. L. Ploense, T. E. Kippin and H. T. Soh, Nat. Biomed. Eng., 2017, 1, 0070 CrossRef.
- E. W. A. Visser, J. Yan, L. J. van Ijzendoorn and M. W. Prins, Nat. Commun., 2018, 9, 2541 CrossRef PubMed.
- J. Heikenfeld, A. YJajack, B. Feldman, S. W. Granger, S. Gaitonde, G. Begtrup and B. A. Katchman, Nat. Biotechnol., 2019, 37, 407–419 CrossRef CAS PubMed.
- M. U. Ahmed, I. Saaem, P. C. Wu and A. S. Brown, Crit. Rev. Biotechnol., 2014, 34, 180–196 CrossRef PubMed.
- D. Ho, S. R. Quake, E. R. McCabe, W. J. Chng, E. K. Chow, X. Ding, B. D. Gelb, G. S. Ginsburg, J. Hassenstab, C.-M. Ho, W. C. Mobley, G. P. Nolan, S. T. Rosen, P. Tan, Y. Yen and A. Zarrinpar, Trends Biotechnol., 2020, 38, 497–518 CrossRef CAS PubMed.
- S. Roh, T. Chung and B. Lee, Sensors, 2011, 11, 1565–1588 CrossRef PubMed.
- J. Wang, A. J. Drelich, C. M. Hopkins, S. Mecozzi, L. Li, G. Kwon and S. Hong, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1754 CrossRef CAS PubMed.
- S. J. Park, S. H. Cha, G. A. Shin and Y. H. Ahn, Biomed. Opt. Express, 2017, 8, 3551–3558 CrossRef CAS PubMed.
- Z. Li, L. Leustean, F. Inci, M. Zheng, U. Demirci and S. Wang, Biotechnol. Adv., 2019, 37, 107440 CrossRef CAS PubMed.
- J. Lee, K. Takemura and E. Y. Park, Sensors, 2017, 17, 2332 CrossRef PubMed.
- D. G. Kotsifaki and S. Nic Chormaic, Nanophotonics, 2019, 8, 1227–1245 CrossRef CAS.
- J.-H. Qu, A. Dillen, W. Saeys, J. Lammertyn and D. Spasic, Anal. Chim. Acta, 2020, 1104, 10–27 CrossRef CAS PubMed.
- C. M. Das, Y. Guo, L. Kang, H.-p. Ho and K.-T. Yong, Adv. Theory Simul., 2020, 3, 2000074 CrossRef CAS PubMed.
- E. Mauriz, Sensors, 2020, 20, 4745 CrossRef CAS PubMed.
- D. G. Kotsifaki and S. Nic Chormaic, Nanophotonics, 2022, 11, 2199–2218 CrossRef CAS.
- L. Krejcova, L. Nejdl, M. A. M. Rodrigo, M. Zurek, M. Matousek, D. Hynek, O. Zitka, P. Kopel and V. Adam, Biosens. Bioelectron., 2014, 54, 421–427 CrossRef CAS PubMed.
- O. Adegoke, T. Kato and E. Y. Park, Biosens. Bioelectron., 2016, 80, 483–490 CrossRef CAS PubMed.
- L. La Spada and L. Vegni, Materials, 2018, 11, 603 CrossRef PubMed.
- Y. Pang, J. Jian, T. Tu, Z. Yang, J. Ling, Y. Li, X. Wang, Y. Qiao, H. Tian, Y. Yang and T.-L. Ren, Biosens. Bioelectron., 2018, 116, 123–129 CrossRef CAS PubMed.
- D. G. Kotsifaki, M. Makropoulou and A. A. Searfetinides, Eur. Phys. J.: Appl. Phys., 2019, 86, 30501 CrossRef CAS.
- R. Abdel-Karim, Y. Reda and A. Abdel-Fattah, J. Electrochem. Soc., 2020, 167, 037554 CrossRef CAS.
- J. Burkhartsmeyer, Y. Wang, K. S. Wong and R. Gordon, Appl. Sci., 2020, 10, 394 CrossRef CAS.
- V. Yesudasu, H. S. Pradhan and R. J. Pandya, Heliyon, 2021, 7, e06321 CrossRef CAS PubMed.
- A. Abbas, M. J. Linman and Q. Cheng, Biosens. Bioelectron., 2011, 26, 1815–1824 CrossRef CAS PubMed.
- Y. Li, K. Zhao, H. Sobhani, K. Bao and P. Nordlander, J. Phys. Chem. Lett., 2013, 4, 1352–1357 CrossRef CAS PubMed.
- M. Couture, S. S. Zhao and J.-F. Masson, Phys. Chem. Chem. Phys., 2013, 15, 11190–11216 RSC.
- H. Guner, E. Ozgur, G. Kokturk, M. Celik, E. Esen, A. E. Topal, S. Ayas, Y. Uludag, C. Elbuken and A. Dana, Sens. Actuators, B, 2017, 239, 571–577 CrossRef CAS.
- L. Novotny and B. Hecht, Principles of Nano-Optics, Cambridge University Press, 2nd edn, 2012 Search PubMed.
- R. Wood, Philos. Mag., 1902, 4, 396–402 Search PubMed.
- L. Rayleigh, Philos. Mag., 1907, 13, 214–232 Search PubMed.
- R. H. Ritchie, Phys. Rev., 1957, 106, 874–881 CrossRef CAS.
- C. J. Powell and J. B. Swan, Phys. Rev., 1960, 118, 640–643 CrossRef CAS.
- A. Otto, Z. Phys., 1968, 216, 398–410 CrossRef CAS.
- E. Kretschmann and H. Raether, Z. Naturforsch., A: Astrophys., Phys. Phys. Chem., 1968, 23, 2135–2136 CrossRef CAS.
- T. D. Bouloumis and S. Nic Chormaic, Appl. Sci., 2020, 10, 1375 CrossRef.
- M. Fleischmann, P. Hendra and A. McQuillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- D. L. Jeanmaire and R. P. Van Duyne, J. Electroanal. Chem. Interfacial Electrochem., 1977, 84, 1–20 CrossRef CAS.
- B. Liedberg, C. Nylander and I. Lunström, Sens. Actuators, 1983, 4, 299–304 CrossRef CAS.
- S. Scarano, M. Mascini, A. P. Turner and M. Minunni, Biosens. Bioelectron., 2010, 25, 957–966 CrossRef CAS PubMed.
- S. Mohammadzadeh-Asl, A. Keshtkar, J. Ezzati Nazhad Dolatabadi and M. de la Guardia, Biosens. Bioelectron., 2018, 110, 118–131 CrossRef CAS PubMed.
- M. Bocková, J. Slabý, T. Špringer and J. Homola, Annu. Rev. Anal. Chem., 2019, 12, 151–176 CrossRef PubMed.
- K. A. Willets and R. P. Van Duyne, Annu. Rev. Phys. Chem., 2007, 58, 267–297 CrossRef CAS PubMed.
- P. B. Johnson and R. W. Christy, Phys. Rev. B: Solid State, 1972, 6, 4370–4379 CrossRef CAS.
- H. U. Yang, J. D'Archangel, M. L. Sundheimer, E. Tucker, G. D. Boreman and M. B. Raschke, Phys. Rev. B: Condens. Matter Mater. Phys., 2015, 91, 235137 CrossRef.
- N. J. Halas, S. Lal, W.-S. Chang, S. Link and P. Nordlander, Chem. Rev., 2011, 111, 3913–3961 CrossRef CAS PubMed.
- H. Raether, in Surface plasmons on smooth surfaces, Springer Berlin Heidelberg, Berlin, Heidelberg, 1988, pp. 4–39 Search PubMed.
- M. Li, S. K. Cushing and N. Wu, Analyst, 2015, 140, 386–406 RSC.
- C. M. Miyazaki, F. M. Shimizu and M. Ferreira, Nanocharacterization Techniques, William Andrew Publishing, 2017, pp. 183–200 Search PubMed.
- V. G. Kravets, A. V. Kabashin, W. L. Barnes and A. N. Grigorenko, Chem. Rev., 2018, 118, 5912–5951 CrossRef CAS PubMed.
- F. Ian, M. Nigel, S. Margaret and H. Catherine, Gold Bull., 2008, 40, 270–277 Search PubMed.
- P. Chen, M. T. Chung, W. McHugh, R. Nidetz, Y. Li, J. Fu, T. T. Cornell, T. P. Shanley and K. Kurabayashi, ACS Nano, 2015, 9, 4173–4181 CrossRef CAS PubMed.
- A. G. Brolo, Nat. Photonics, 2012, 6, 709–713 CrossRef CAS.
- S. S. Aćimović, M. A. Ortega, V. Sanz, J. Berthelot, J. L. Garcia-Cordero, J. Renger, S. J. Maerkl, M. P. Kreuzer and R. Quidant, Nano Lett., 2014, 14, 2636–2641 CrossRef PubMed.
- D. G. Kotsifaki, M. D. Mackenzie, G. Polydefki, A. K. Kar, M. Makropoulou and A. A. Serafetinides, Opt. Eng., 2017, 56, 124111 Search PubMed.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- J. Kundu, F. Le, P. Nordlander and N. J. Halas, Chem. Phys. Lett., 2008, 452, 115–119 CrossRef CAS.
- S. Fayyaz, M. Tabatabaei, R. Hou and F. Lagugné-Labarthet, J. Phys. Chem. C, 2012, 116, 11665–11670 CrossRef CAS.
- A. B. Taylor and P. Zijlstra, ACS Sens., 2017, 2, 1103–1122 CrossRef CAS PubMed.
- B. Luk'yanchuk, N. I. Zheludev, S. A. Maier, N. J. Halas, P. Nordlander, H. Giessen and T. C. Chong, Nat. Mater., 2010, 9, 707–715 CrossRef PubMed.
- A. Ahmadivand, B. Gerislioglu, R. Ahuja and Y. Kumar Mishra, Mater. Today, 2020, 32, 108–130 CrossRef CAS.
- D. G. Kotsifaki, V. G. Truong and S. Nic Chormaic, Nano Lett., 2020, 20, 3388–3395 CrossRef CAS PubMed.
- D. G. Kotsifaki, V. G. Truong and S. Nic Chormaic, Appl. Phys. Lett., 2021, 118, 021107 CrossRef CAS.
- V. G. Veselago, Sov. Phys. Usp., 1968, 10, 509–514 CrossRef.
- J. Pendry, A. Holden, D. Robbins and W. Stewart, IEEE Trans. Microwave Theory Tech., 1999, 47, 2075–2084 CrossRef.
- D. R. Smith, W. J. Padilla, D. C. Vier, S. C. Nemat-Nasser and S. Schultz, Phys. Rev. Lett., 2000, 84, 4184–4187 CrossRef CAS PubMed.
- J. B. Pendry, Phys. Rev. Lett., 2000, 85, 3966–3969 CrossRef CAS PubMed.
- Y. Fang, Y. Ge, C. Wang and H. Zhang, Laser Photonics Rev., 2020, 14, 1900098 CrossRef CAS.
- P. Yu, L. V. Besteiro, Y. Huang, J. Wu, L. Fu, H. H. Tan, C. Jagadish, G. P. Wiederrecht, A. O. Govorov and Z. Wang, Adv. Opt. Mater., 2019, 7, 1800995 CrossRef.
- M. Beruete and I. Jáuregui-López, Adv. Opt. Mater., 2020, 8, 1900721 CrossRef CAS.
- Y. Roh, S.-H. Lee, J. Kwak, H. S. Song, S. Shin, Y. K. Kim, J. W. Wu, B.-K. Ju, B. Kang and M. Seo, Sens. Actuators, B, 2022, 352, 130993 CrossRef CAS.
- R. Zhou, C. Wang, W. Xu and L. Xie, Nanoscale, 2019, 11, 3445–3457 RSC.
- W. Wang, M. Ramezani, A. I. Väkeväinen, P. Törmä, J. G. Rivas and T. W. Odom, Mater. Today, 2018, 21, 303–314 CrossRef.
- M. Fan, G. F. Andrade and A. G. Brolo, Anal. Chim. Acta, 2020, 1097, 1–29 CrossRef CAS PubMed.
- H. Shi, X. Zhu, S. Zhang, G. Wen, M. Zheng and H. Duan, Nanoscale Adv., 2021, 3, 4349–4369 RSC.
- K. C. Bantz, A. F. Meyer, N. J. Wittenberg, H. Im, Ö. Kurtuluş, S. H. Lee, N. C. Lindquist, S.-H. Oh and C. L. Haynes, Phys. Chem. Chem. Phys., 2011, 13, 11551–11567 RSC.
- E. C. Le Ru, E. Blackie, M. Meyer and P. G. Etchegoin, J. Phys. Chem. C, 2007, 111, 13794–13803 CrossRef CAS.
- C. E. Anderson, C. A. Holstein, E.-M. Strauch, S. Bennett, A. Chevalier, J. Nelson, E. Fu, D. Baker and P. Yager, Anal. Chem., 2017, 89, 6608–6615 CrossRef CAS PubMed.
- T. J. Moore, A. S. Moody, T. D. Payne, G. M. Sarabia, A. R. Daniel and B. Sharma, Biosensors, 2018, 8, 46 CrossRef PubMed.
- G. Spoto and M. Minunni, J. Phys. Chem. Lett., 2012, 3, 2682–2691 CrossRef CAS PubMed.
- Y. Zeng, R. Hu, L. Wang, D. Gu, J. He, S.-Y. Wu, H.-P. Ho, X. Li, J. Qu, B. Z. Gao and Y. Shao, Nanophotonics, 2017, 6, 1017–1030 CrossRef.
- Z. Huo, Y. Li, B. Chen, W. Zhang, X. Yang and X. Yang, Talanta, 2023, 255, 124213 CrossRef CAS PubMed.
- J.-S. Lin, X.-D. Tian, G. Li, F.-L. Zhang, Y. Wang and J.-F. Li, Chem. Soc. Rev., 2022, 51, 9445–9468 RSC.
- E. A. Smith and R. M. Corn, Appl. Spectrosc., 2003, 57, 320A–332A CrossRef CAS PubMed.
- D. Kotsifaki and A. Serafetinides, Opt. Laser Technol., 2011, 43, 1448–1452 CrossRef CAS.
- D. Kotsifaki and A. Serafetinides, Opt. Laser Technol., 2009, 41, 365–373 CrossRef CAS.
- D. G. Kotsifaki, M. Makropoulou and A. A. Serafetinides, 16th International School on Quantum Electronics: Laser Physics and Applications, 2011, p. 77470Z Search PubMed.
- D. G. Kotsifaki, S. Aggelopoulos, M. Makropoulou and A. Serafetinides, J. Nanotechnol. Diagn. Treat., 2016, 4, 25–30 CrossRef.
- C. Christophe, G. Tuan and A. Jacques, Anal. Bioanal. Chem., 2015, 407, 3883–3897 CrossRef PubMed.
- Q. Wang and L. Wang, Nanoscale, 2020, 12, 7485–7499 RSC.
- Y. Esfahani Monfared, Biosensors, 2020, 10, 77 CrossRef PubMed.
- Z. Wang, W. Zhang, X. Liu, M. Li, X. Lang, R. Singh, C. Marques, B. Zhang and S. Kumar, Biosensors, 2022, 12, 1016 CrossRef CAS PubMed.
- P. Estrela, N. Bhalla, P. Jolly, N. Formisano and P. Estrela, Essays Biochem., 2016, 60, 1–8 CrossRef PubMed.
- Y. Huang, J. Xu, J. Liu, X. Wang and B. Chen, Sensors, 2017, 17, 2375 CrossRef PubMed.
- A. N. Kozitsina, T. S. Svalova, N. N. Malysheva, A. V. Okhokhonin, M. B. Vidrevich and K. Z. Brainina, Biosensors, 2018, 8, 35 CrossRef PubMed.
- B. Špačková, P. Wrobel, M. Bocková and J. Homola, Proc. IEEE, 2016, 104, 2380–2408 Search PubMed.
- M. M. Miller and A. A. Lazarides, J. Phys. Chem. B, 2005, 109, 21556–21565 CrossRef CAS PubMed.
- A. A. Kolomenskii, P. D. Gershon and H. A. Schuessler, Appl. Opt., 1997, 36, 6539–6547 CrossRef CAS PubMed.
- L. S. Live, O. R. Bolduc and J.-F. Masson, Anal. Chem., 2010, 82, 3780–3787 CrossRef CAS PubMed.
- M. Piliarik and J. Homola, Opt. Express, 2009, 17, 16505–16517 CrossRef CAS PubMed.
- D. J. Bergman and M. I. Stockman, Phys. Rev. Lett., 2003, 90, 027402 CrossRef PubMed.
- A. A. Yanik, A. E. Cetin, M. Huang, A. Artar, S. H. Mousavi, A. Khanikaev, J. H. Connor, G. Shvets and H. Altug, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 11784–11789 CrossRef CAS PubMed.
- S. Zhang, K. Bao, N. J. Halas, H. Xu and P. Nordlander, Nano Lett., 2011, 11, 1657–1663 CrossRef CAS PubMed.
- P. Offermans, M. C. Schaafsma, S. R. K. Rodriguez, Y. Zhang, M. Crego-Calama, S. H. Brongersma and J. Gómez Rivas, ACS Nano, 2011, 5, 5151–5157 CrossRef CAS PubMed.
- Y.-F. Chang, W.-H. Wang, Y.-W. Hong, R.-Y. Yuan, K.-H. Chen, Y.-W. Huang, P.-L. Lu, Y.-H. Chen, Y.-M. A. Chen, L.-C. Su and S.-F. Wang, Anal. Chem., 2018, 90, 1861–1869 CrossRef CAS PubMed.
- B. A. Prabowo, R. Y. Wang, M. K. Secario, P.-T. Ou, A. Alom, J.-J. Liu and K.-C. Liu, Biosens. Bioelectron., 2017, 92, 186–191 CrossRef CAS PubMed.
- L. Shi, Q. Sun, J. He, H. Xu, C. Liu, C. Zhao, Y. Xu, C. Wu, J. Xiang, D. Gu, J. Long and H. Lan, Bio-Med. Mater. Eng., 2015, 26, S2207–S2216 Search PubMed.
- S. K. Pushpendra, K. S. Jyoti, S. V. Virendra, B. Utpal, S. S. Shyam, A. I. Syed, D. K. Paban, B. Mannan, G. Kumaran and J. Rajeev, Anal. Bioanal. Chem., 2020, 412, 4101–41120 CrossRef PubMed.
- W. Diao, M. Tang, S. Ding, X. Li, W. Cheng, F. Mo, X. Yan, H. Ma and Y. Yan, Biosens. Bioelectron., 2018, 100, 228–234 CrossRef CAS PubMed.
- M. Rippa, R. Castagna, S. Brandi, G. Fusco, M. Monini, D. Chen, J. Zhou, J. Zyss and L. Petti, ACS Appl. Nano Mater., 2020, 3, 4837–4844 CrossRef CAS.
- A. Ahmadivand, B. Gerislioglu, A. Tomitaka, P. Manickam, A. Kaushik, S. Bhansali, M. Nair and N. Pala, Biomed. Opt. Express, 2018, 9, 373–386 CrossRef CAS PubMed.
- A. Ahmadivand, B. Gerislioglu, Z. Ramezani, A. Kaushik, P. Manickam and S. A. Ghoreishi, Biosens. Bioelectron., 2021, 177, 112971 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- J. Kim, S. Y. Oh, S. Shukla, S. B. Hong, N. S. Heo, V. Bajpai, H. S. Chun, C.-H. Jo, B. G. Choi, Y. S. Huh and Y.-K. Han, Biosens. Bioelectron., 2018, 107, 118–122 CrossRef CAS PubMed.
- T. Lee, G. H. Kim, S. M. Kim, K. Hong, Y. Kim, C. Park, H. Sohn and J. Min, Colloids Surf., B, 2019, 182, 110341 CrossRef CAS PubMed.
- N. S. Heo, S. Y. Oh, M. Y. Ryu, S. H. Baek, T. J. Park, C. Choi, Y. S. Huh and J. P. Park, Biotechnol. Bioprocess Eng., 2019, 24, 318–325 CrossRef CAS.
- A. M. Paul, Z. Fan, S. S. Sinha, Y. Shi, L. Le, F. Bai and P. C. Ray, J. Phys. Chem. C, 2015, 119, 23669–23675 CrossRef CAS PubMed.
- S. A. Camacho, R. G. Sobral-Filho, P. H. B. Aoki, C. J. L. Constantino and A. G. Brolo, ACS Sens., 2018, 3, 587–594 CrossRef CAS PubMed.
- P. P. Austin Suthanthiraraj and A. K. Sen, Biosens. Bioelectron., 2019, 132, 38–46 CrossRef CAS PubMed.
- K. Behrouzi and L. Lin, Biosens. Bioelectron., 2022, 195, 113669 CrossRef CAS PubMed.
- E. Wijaya, C. Lenaerts, S. Maricot, J. Hastanin, S. Habraken, J.-P. Vilcot, R. Boukherroub and S. Szunerits, Curr. Opin. Solid State Mater. Sci., 2011, 15, 208–224 CrossRef CAS.
- J. Liu, M. Jalali, S. Mahshid and S. Wachsmann-Hogiu, Analyst, 2020, 145, 364–384 RSC.
- T. Liyanage, B. Alharbi, L. Quan, A. Esquela-Kerscher and G. Slaughter, ACS Omega, 2022, 7, 2411–2418 CrossRef CAS PubMed.
- G. Nava, G. Zanchetta, F. Giavazzi and M. Buscaglia, Nanophotonics, 2022, 11, 4159–4181 CrossRef CAS.
- D. Kotsifaki, M. Makropoulou and A. A. Serafetinides, Saratov Fall Meeting 2006: Optical Technologies in Biophysics and Medicine VIII, 2007, p. 65351O Search PubMed.
- J. Homola, Chem. Rev., 2008, 108, 462–493 CrossRef CAS PubMed.
- J. Wang, A. J. Drelich, C. M. Hopkins, S. Mecozzi, L. Li, G. Kwon and S. Hong, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1754 CAS.
- D. Schofield and N. Dimmock, J. Virol. Methods, 1996, 62, 33–42 CrossRef CAS PubMed.
- Ravina, A. Dalal, H. Mohan, M. Prasad and C. Pundir, Biosci. Rep., 2020, 40, 1–18 CrossRef PubMed.
- F. Zang, Z. Su, L. Zhou, K. Konduru, G. Kaplan and S. Y. Chou, Adv. Mater., 2019, 31, 1902331 CrossRef PubMed.
- J. A. Jackman, E. Linardy, D. Yoo, J. Seo, W. B. Ng, D. J. Klemme, N. J. Wittenberg, S.-H. Oh and N.-J. Cho, Small, 2016, 12, 1159–1166 CrossRef CAS PubMed.
- Z. Klestova, A. Voronina, A. Yushchenko, O. Vatlitsova, G. Dorozinsky, Y. Ushenin, V. Maslov, T. Doroshenko and S. Kravchenko, Spectrochim. Acta, Part A, 2022, 264, 120236 CrossRef CAS PubMed.
- H.-Y. Hsieh, R. Chang, Y.-Y. Huang, P.-H. Juan, H. Tahara, K.-Y. Lee, D. N. K. Vo, M.-H. Tsai, P.-K. Wei, H.-J. Sheen and Y.-J. Fan, Biosens. Bioelectron., 2022, 195, 113672 CrossRef CAS PubMed.
- H. A. Bethe, Phys. Rev., 1944, 66, 163–182 CrossRef.
- A. A. Yanik, M. Huang, O. Kamohara, A. Artar, T. W. Geisbert, J. H. Connor and H. Altug, Nano Lett., 2010, 10, 4962–4969 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, L. Saleh, J. Tang, T. Gui, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2021, 15, 7536–7546 CrossRef CAS PubMed.
- N. Cennamo, L. Pasquardini, F. Arcadio, L. Lunelli, L. Vanzetti, V. Carafa, L. Altucci and L. Zeni, Talanta, 2021, 233, 122532 CrossRef CAS PubMed.
- N. Cennamo, G. D'Agostino, C. Perri, F. Arcadio, G. Chiaretti, E. M. Parisio, G. Camarlinghi, C. Vettori, F. Di Marzo, R. Cennamo, G. Porto and L. Zeni, Sensors, 2021, 21, 1681 CrossRef CAS PubMed.
- L. Yu, L. Hao, T. Meiqiong, H. Jiaoqi, L. Wei, D. Jinying, C. Xueping, F. Weiling and Z. Yang, RSC Adv., 2019, 9, 9354–9363 RSC.
- L. Huang, L. Ding, J. Zhou, S. Chen, F. Chen, C. Zhao, J. Xu, W. Hu, J. Ji, H. Xu and G. L. Liu, Biosens. Bioelectron., 2021, 171, 112685 CrossRef CAS PubMed.
- M. Forinová, A. Pilipenco, I. Víšová, N. S. J. Lynn, J. Dostálek, H. Mašková, V. Hönig, M. Palus, M. Selinger, P. Kočová, F. Dyčka, J. Štěrba, M. Houska, M. Vrabcová, P. Horák, J. Anthi, C.-P. Tung, C.-M. Yu, C.-Y. Chen, Y.-C. Huang, P.-H. Tsai, S.-Y. Lin, H.-J. Hsu, A.-S. Yang, A. Dejneka and H. Vaisocherová-Lísalová, ACS Appl. Mater. Interfaces, 2021, 13, 60612–60624 CrossRef PubMed.
- B. Yin, W. K. H. Ho, Q. Zhang, C. Li, Y. Huang, J. Yan, H. Yang, J. Hao, S. H. D. Wong and M. Yang, ACS Appl. Mater. Interfaces, 2022, 14, 4714–4724 CrossRef CAS PubMed.
- O. Calvo-Lozano, M. Sierra, M. Soler, M. C. Estévez, L. Chiscano-Camón, A. Ruiz-Sanmartin, J. C. Ruiz-Rodriguez, R. Ferrer, J. J. González-López, J. Esperalba, C. Fernández-Naval, L. Bueno, R. López-Aladid, A. Torres, L. Fernández-Barat, S. Attoumani, R. Charrel, B. Coutard and L. M. Lechuga, Anal. Chem., 2022, 94, 975–984 CrossRef CAS PubMed.
- I. B. Ansah, S. H. Lee, J.-Y. Yang, C. Mun, S. Jung, H. S. Jung, M.-Y. Lee, T. Kang, S. Lee, D.-H. Kim and S.-G. Park, Biosens. Bioelectron., 2023, 220, 114930 CrossRef CAS PubMed.
- G. Palermo, M. Rippa, Y. Conti, A. Vestri, R. Castagna, G. Fusco, E. Suffredini, J. Zhou, J. Zyss, A. De Luca and L. Petti, ACS Appl. Mater. Interfaces, 2021, 13, 43715–43725 CrossRef CAS PubMed.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed.
- C. Ménard-Moyon, A. Bianco and K. Kalantar-Zadeh, ACS Sens., 2020, 5, 3739–3769 CrossRef PubMed.
- N. Rohaizad, C. C. Mayorga-Martinez, M. Fojtů, N. M. Latiff and M. Pumera, Chem. Soc. Rev., 2021, 50, 619–657 RSC.
- M. M. Y. A. Alsaif, K. Latham, M. R. Field, D. D. Yao, N. V. Medehkar, G. A. Beane, R. B. Kaner, S. P. Russo, J. Z. Ou and K. Kalantar-zadeh, Adv. Mater., 2014, 26, 3931–3937 CrossRef CAS PubMed.
- B. Y. Zhang, A. Zavabeti, A. F. Chrimes, F. Haque, L. A. O'Dell, H. Khan, N. Syed, R. Datta, Y. Wang, A. S. R. Chesman, T. Daeneke, K. Kalantar-zadeh and J. Z. Ou, Adv. Funct. Mater., 2018, 28, 1706006 CrossRef.
- T. Low, A. Chaves, J. D. Caldwell, A. Kumar, N. X. Fang, P. Avouris, T. F. Heinz, F. Guinea, L. Martin-Moreno and F. Koppens, Nat. Mater., 2017, 16, 182–194 CrossRef CAS PubMed.
- A. Bolotsky, D. Butler, C. Dong, K. Gerace, N. R. Glavin, C. Muratore, J. A. Robinson and A. Ebrahimi, ACS Nano, 2019, 13, 9781–9810 CrossRef CAS PubMed.
- S.-H. Oh, H. Altug, X. Jin, S. J. K. Tony Low, A. P. Ivanov, J. B. Edel, P. Avouris and M. S. Strano, Nat. Commun., 2021, 12, 3824 CrossRef CAS PubMed.
- M.-Q. Fu, X.-C. Wang, W.-T. Dou, G.-R. Chen, T. D. James, D.-M. Zhou and X.-P. He, Chem. Commun., 2020, 56, 5735–5738 RSC.
- N. A. S. Omar, Y. W. Fen, S. Saleviter, W. M. E. M. M. Daniyal, N. A. A. Anas, N. S. M. Ramdzan and M. D. A. Roshidi, Materials, 2019, 12, 1928 CrossRef CAS PubMed.
- X. Peng, Y. Zhang, D. Lu, Y. Guo and S. Guo, Sens. Actuators, B, 2019, 286, 222–229 CrossRef CAS.
- J. Z. Ou, A. F. Chrimes, Y. Wang, S.-y. Tang, M. S. Strano and K. Kalantar-zadeh, Nano Lett., 2014, 14, 857–863 CrossRef CAS PubMed.
- Y. Liu, Y. Nie, M. Wang, Q. Zhang and Q. Ma, Biosens. Bioelectron., 2020, 148, 111823 CrossRef CAS PubMed.
- S. Barua, H. S. Dutta, S. Gogoi, R. Devi and R. Khan, ACS Appl. Nano Mater., 2018, 1, 2–25 CrossRef CAS.
- V. Yadav, S. Roy, P. Singh, Z. Khan and A. Jaiswal, Small, 2019, 15, 1803706 CrossRef PubMed.
- T. B. A. Akib, S. F. Mou, M. M. Rahman, M. M. Rana, M. R. Islam, I. M. Mehedi, M. A. P. Mahmud and A. Z. Kouzani, Sensors, 2021, 21, 3491 CrossRef PubMed.
- S. Mostufa, T. B. A. Akib, M. M. Rana, I. M. Mehedi, U. M. Al-Saggaf, A. U. Alsaggaf, M. U. Alsaggaf and M. S. Alam, Opt. Continuum, 2022, 1, 494–515 CrossRef CAS.
- N. H. L. Nguyen, S. Kim, G. Lindemann and V. Berry, ACS Nano, 2021, 15, 11743–11752 CrossRef CAS PubMed.
- S. Addanki, I. Amiri and P. Yupapin, Results Phys., 2018, 10, 743–750 CrossRef.
- A. G. Podoleanu, J. Lightwave Technol., 2010, 28, 624–640 Search PubMed.
- Y. Esfahani Monfared, Biosensors, 2020, 10, 77 CrossRef PubMed.
- G. Gauglitz, Anal. Bioanal. Chem., 2020, 412, 3317–3349 CrossRef CAS PubMed.
- X. Zhao, Y.-C. Tsao, F.-J. Lee, W.-H. Tsai, C.-H. Wang, T.-L. Chuang, M.-S. Wu and C.-W. Lin, J. Virol. Methods, 2016, 233, 15–22 CrossRef CAS PubMed.
- H.-Y. Lin, C.-H. Huang, S.-H. Lu, I.-T. Kuo and L.-K. Chau, Biosens. Bioelectron., 2014, 51, 371–378 CrossRef CAS PubMed.
- B. Luo, Y. Xu, S. Wu, M. Zhao, P. Jiang, S. Shi, Z. Zhang, Y. Wang, L. Wang and Y. Liu, Biosens. Bioelectron., 2018, 100, 169–175 CrossRef CAS PubMed.
- D. Murugan, H. Bhatia, V. V. R. Sai and S. Jitendra, Transactions of the Indian National Academy of Engineering, 2020, 5, 211–215 CrossRef.
- M. Aliee and H. M. Mozaffari, Plasmonics, 2022, 17, 1655–1660 CrossRef CAS PubMed.
- S. Yosra, H. G. Mohamed, M. Karine, S. Marwa and B. Hafedh, Plasmonics, 2022, 17, 1489–1500 CrossRef PubMed.
- L. Wu, H. S. Chu, W. S. Koh and E. P. Li, Opt. Express, 2010, 18, 14395–14400 CrossRef CAS PubMed.
- A. Wei, M. Thomas, J. Mehtala and J. Wang, Biomaterials for Cancer Therapeutics, Woodhead Publishing, 2013, pp. 349–389 Search PubMed.
- N. Ibrahim, N. D. Jamaluddin, L. L. Tan and N. Y. Mohd Yusof, Sensors, 2021, 21, 5114 CrossRef CAS PubMed.
- H. Geng, S. Vilms Pedersen, Y. Ma, T. Haghighi, H. Dai, P. D. Howes and M. M. Stevens, Acc. Chem. Res., 2022, 55, 593–604 CrossRef CAS PubMed.
- K. Takemura, O. Adegoke, T. Suzuki and E. Y. Park, PLoS One, 2019, 14, e0211517 CrossRef CAS PubMed.
- A. D. Chowdhury, K. Takemura, I. M. Khorish, F. Nasrin, M. M. N. Tun, K. Morita and E. Y. Park, Nanoscale Adv., 2020, 2, 699–709 RSC.
- J. Luan, A. Seth, R. Gupta, Z. Wang, P. Rathi, S. Cao, H. G. Derami, R. Tang, B. Xu, S. Achilefu, J. J. Morrissey and S. Singamanen, Nat. Biomed. Eng., 2020, 4, 518–530 CrossRef CAS PubMed.
- S. Das, D. K. Agarwal, B. Mandal, V. R. Rao and T. Kundu, ACS Omega, 2021, 6, 17413–17423 CrossRef CAS PubMed.
- H. Liang, H. Wei, D. Pan and H. Xu, Nanotechnol. Rev., 2015, 4, 289–302 Search PubMed.
- T.-H. Kim, M. Kim, H.-S. Park, U. S. Shin, M.-S. Gong and H.-W. Kim, J. Biomed. Mater. Res., Part A, 2012, 100, 1033–1043 CrossRef PubMed.
- Q. Zhang, J. Hong and X. Y. Biofuels, Methods Mol. Biol., 2012, 581, 1–330 Search PubMed.
- S. S. Jeremiah, K. Miyakawa, T. Morita, Y. Yamaoka and A. Ryo, Biochem. Biophys. Res. Commun., 2020, 533, 195–200 CrossRef CAS PubMed.
- A. Salleh, R. Naomi, N. D. Utami, A. W. Mohammad, E. Mahmoudi, N. Mustafa and M. B. Fauzi, Nanomaterials, 2020, 10, 1566 CrossRef CAS PubMed.
- Y. Wang, Y. Kang, W. Y. Wang, Q. Ding, J. Zhou and S. Yang, Nanotechnology, 2018, 29, 414001 CrossRef PubMed.
- M. Tejamaya, I. Römer, R. C. Merrifield and J. R. Lead, Environ. Sci. Technol., 2012, 46, 7011–7017 CrossRef CAS PubMed.
- J. Hong, S. Jun, S. Cha, J. Park, S. Lee, G. Shin and Y. Ahn, Sci. Rep., 2018, 8, 1–8 Search PubMed.
- Y. Lu, Y. Lin, Z. Zheng, X. Tang, J. Lin, X. Liu, M. Liu, G. Chen, S. Qiu, T. Zhou, Y. Lin and S. Feng, Biomed. Opt. Express, 2018, 9, 4755–4766 CrossRef CAS PubMed.
- J. Nan, W. Sun, X. Liu, Y. Che, H. Shan, Y. Yue, J. Liu, L. Wang, K. Liu, W. Xu, W. Zhang, S. Zhang, B. Liu, K. S. Hettie, S. Zhu, J. Zhang and B. Yang, Nano Lett., 2022, 22, 9596–9605 CrossRef CAS PubMed.
- N. R. Blumenfeld, M. A. E. Bolene, M. Jaspan, A. G. Ayers, S. Zarrandikoetxea, J. Freudman, N. Shah, A. M. Tolwani, Y. Hu, T. L. Chern, J. Rogot, V. Behnam, A. Sekhar, X. Liu, B. Onalir, R. Kasumi, A. Sanogo, K. Human, K. Murakami, G. S. Totapally, M. Fasciano and S. K. Sia, Nat. Nanotechnol., 2022, 17, 984–992 CrossRef CAS PubMed.
- T. T. S. Lew, K. M. M. Aung, S. Y. Ow, S. N. Amrun, L. Sutarlie, L. F. P. Ng and X. Su, ACS Nano, 2021, 15, 12286–12297 CrossRef CAS PubMed.
- N. Bhalla, A. F. Payam, A. Morelli, P. K. Sharma, R. Johnson, A. Thomson, P. Jolly and F. Canfarotta, Sens. Actuators, B, 2022, 365, 131906 CrossRef CAS PubMed.
- Y.-J. Yeh, T.-N. Le, W. W.-W. Hsiao, K.-L. Tung, K. K. Ostrikov and W.-H. Chiang, Anal. Chim. Acta, 2023, 1239, 340651 CrossRef CAS PubMed.
- Y. Park, B. Ryu, S. Ki, M. Chen, X. Liang and K. Kurabayashi, medRxiv, 2022, preprint, pp. 1–30, DOI:10.1101/2022.08.01.22278286.
- C. M. Das, Y. Guo, G. Yang, L. Kang, G. Xu, H.-P. Ho and K.-T. Yong, Adv. Theory Simul., 2020, 3, 2000185 CrossRef CAS PubMed.
- E. Zavyalova, O. Ambartsumyan, G. Zhdanov, D. Gribanyov, V. Gushchin, A. Tkachuk, E. Rudakova, M. Nikiforova, N. Kuznetsova and L. Popova, et al. , Nanomaterials, 2021, 11, 1394 CrossRef CAS PubMed.
- M. N. Tripathi, K. Singh, U. Yadav, R. R. Srivastava, M. Gangwar, G. Nath, P. S. Saxena and A. Srivastava, Antiviral Res., 2022, 205, 105382 CrossRef CAS PubMed.
- P. Moitra, M. Alafeef, K. Dighe, M. B. Frieman and D. Pan, ACS Nano, 2020, 14, 7617–7627 CrossRef CAS PubMed.
- Y.-S. Borghei, H. R. Samadikhah and S. Hosseinkhani, Anal. Chem., 2022, 94, 13616–13622 CrossRef CAS PubMed.
- E. Sobhanie, F. Salehnia, G. Xu, Y. Hamidipanah, S. Arshian, A. Firoozbakhtian, M. Hosseini, M. R. Ganjali and S. Hanif, TrAC, Trends Anal. Chem., 2022, 157, 116727 CrossRef CAS PubMed.
- Z. Fan, B. Yao, Y. Ding, D. Xu, J. Zhao and K. Zhang, Chem. Eng. J., 2022, 427, 131686 CrossRef CAS PubMed.
- Z. Zhang, H. Wang, Z. Chen, X. Wang, J. Choo and L. Chen, Biosens. Bioelectron., 2018, 114, 52–65 CrossRef CAS PubMed.
- E. M. Materón, F. R. Gómez, M. B. Almeida, F. M. Shimizu, A. Wong, K. B. R. Teodoro, F. S. R. Silva, M. J. A. Lima, M. K. S. C. Angelim, M. E. Melendez, N. Porras, P. M. Vieira, D. S. Correa, E. Carrilho, O. N. J. Oliveira, R. B. Azevedo and D. Goncalves, ACS Appl. Mater. Interfaces, 2022, 14, 54527–54538 CrossRef PubMed.
- L. In-Ho, Y. Daehan, A. Phaedon, L. Tony and O. Sang-Hyun, Nat. Nanotechnol., 2019, 14, 313–319 CrossRef PubMed.
- X. Lin, Z. Liu, T. Stauber, G. Gómez-Santos, F. Gao, H. Chen, B. Zhang and T. Low, Phys. Rev. Lett., 2020, 125, 077401 CrossRef CAS PubMed.
- W. Yunhao, Z. Yue, B. Audrey, W. Yuru and F. Kin, Nat. Biotechnol., 2021, 39, 1348–1365 CrossRef PubMed.
- X. Wei, D. Ma, Z. Zhang, L. Y. Wang, J. L. Gray, L. Zhang, T. Zhu, X. Wang, B. J. Lenhart, Y. Yin, Q. Wang and C. Liu, ACS Sens., 2020, 5, 1707–1716 CrossRef CAS PubMed.
- R. A. Bull, T. N. Adikari, J. M. Ferguson, J. M. Hammond, I. Stevanovski, A. Beukers, Z. Naing, M. Yeang, A. Verich, H. Gamaarachchi, K. W. Kim, F. Luciani, S. Stelzer-Braid, J. S. Eden, W. D. Rawlinson, S. J. van Hal and I. W. Deveson, Nat. Commun., 2020, 11, 1–8 CrossRef PubMed.
- A. C. Olivieri, N. M. Faber, J. Ferré, R. Boqué, J. H. Kalivas and H. Mark, Pure Appl. Chem., 2006, 78, 633–661 CrossRef CAS.
- F. Cui, M. Rhee, A. Singh and A. Tripathi, Annu. Rev. Biomed. Eng., 2015, 17, 267–286 CrossRef CAS PubMed.
- D. G. Kotsifaki, M. Makropoulou and A. Serafetinides, J. Nanotechnol. Diagn. Treat., 2018, 6, 1–7 CrossRef.
- G. A. Lopez, M.-C. Estevez, M. Soler and L. M. Lechuga, Nanophotonics, 2017, 6, 123–136 CrossRef CAS.
- Q. Liu, H. Yuan, Y. Liu, J. Wang, Z. Jing and W. Peng, J. Biomed. Opt., 2018, 23, 047003 Search PubMed.
- K. Salimiyan rizi, Curr. Opin. Electrochem., 2022, 32, 100925 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2023 |