Open Access Article
Open Access ArticleStructural features localizing the ferroptosis inhibitor GIF-2197-r to lysosomes†
Yoko Hirata *a,
Tomohiro Hashimotob,
Kaori Ando
*a,
Tomohiro Hashimotob,
Kaori Ando b,
Yuji O. Kamatari
b,
Yuji O. Kamatari aef,
Hiroshi Takemori
aef,
Hiroshi Takemori cde and
Kyoji Furuta*c
cde and
Kyoji Furuta*c
aLife Science Research Center, Institute for Advanced Study, Gifu University, Yanagido, Gifu, 501-1193, Japan. E-mail: morita.yoko.m1@f.gifu-u.ac.jp
bFaculty of Regional Studies, Gifu University, Yanagido, Gifu 501-1193, Japan
cDepartment of Chemistry and Biomolecular Science, Faculty of Engineering, Gifu University, Yanagido, Gifu, 501-1193, Japan
dGraduate School of Natural Science and Technology, Gifu University, Yanagido, Gifu, 501-1193, Japan
eUnited Graduate School of Drug Discovery and Medical Information Sciences, Gifu University, Yanagido, Gifu, 501-1193, Japan
fInstitute for Glyco-core Research (iGCORE), Gifu University, 1-1 Yanagido, Gifu 501-1193, Japan
First published on 2nd November 2023
Abstract
We previously reported that N,N-dimethylaniline derivatives are potent ferroptosis inhibitors. Among them, the novel aminoindan derivative GIF-2197-r (the racemate of GIF-2115 (R-form) and GIF-2196 (S-form)) is effective at a concentration of 0.01 μM due to its localization to lysosomes and ferrous ion coordination capacity. The current study demonstrates that the aliphatic tertiary amine moiety of GIF-2197-r is responsible for lysosomal localization. Although N,N-dimethylaniline derivatives cannot form chelate structures with Fe2+, density functional theory computation demonstrates that they can form stable monodentate complexes with a hydrated ferrous ion, likely due to the highly electron-rich nature of the (dialkylamino)phenyl ring. Furthermore, the results suggest that the aliphatic tertiary amine moiety contributes to stabilizing the complexation. These findings could prove useful for developing improved lysosomotropic ferroptosis inhibitors for neurodegenerative diseases.
Introduction
Oxidative stress is a central pathogenic condition underlying numerous neurodegenerative, neoplastic, and metabolic disorders. Neurons are particularly susceptible to oxidative stress, so interventions that can prevent the generation of reactive oxygen species (ROS) or enhance endogenous antioxidant capacity may be effective treatments for neurodegenerative diseases. However, both clinical and preclinical studies have found that direct or indirect scavenging of ROS by exogenous antioxidants alone fails to slow disease progression or prolong the lives of patients.1 Oxidative stress occurs when the production of ROS exceeds removal by antioxidant defense mechanisms, resulting in oxidative damage to vital macromolecules (e.g., proteins, membrane lipids, and DNA) and ensuing activation of cell death pathways, including apoptosis. In addition, recent studies have defined a non-apoptotic form of regulated cell death termed ferroptosis, which is so-named due to its dependence on cellular free ferrous ions.2,3The neuron-derived cell line HT22 is a well-established model to investigate the effects of oxytosis and ferroptosis.4 Treatment of HT22 cells with glutamate or erastin blocks the membrane cystine-glutamate antiporter, thereby depleting the intracellular L-cysteine required for synthesis of the antioxidant glutathione (GSH, γ-L-glutamyl-L-cysteinylglycine). This depletion results in ROS accumulation, lipid peroxidation, and ultimately oxytosis or ferroptosis.5,6 It has been suggested that oxytosis and ferroptosis are actually the same cell death pathway as the underlying molecular pathways appear similar if not identical,7 although both are morphologically, biochemically, and genetically distinct from apoptosis, necrosis, and autophagy, suggesting sensitivity to distinct chemical inhibitors.7 In fact, our previous study showed that various ferroptosis inhibitors, such as the oxindole–curcumin hybrid compound GIF-2165X-G1, N,N-dimethylaniline derivatives GIF-2114 and GIF-2115, and the commercially available ferrostatin-1, did not prevent oxidative stress and apoptosis induced by rotenone, which is widely used to induce Parkinson's disease-like pathology in cells and model animals.8 Given emerging evidence that oxytosis/ferroptosis also contributes to neurodegenerative diseases, interventions targeting ferroptosis may be an effective treatment strategy. Therefore, it is essential to identify chemical structures responsible for ferroptosis inhibition.
We previously reported that GIF-2197-r, a novel ferroptosis inhibitor, prevented ferroptosis in HT22 cells at a concentration of 0.01 μM, and this potency was associated with superior localization to lysosomes and a decrease in intracellular ferrous ions.9 GIF-2197-r is composed of N,N-dimethyl-p-toluidine, obtained from the oxindole compound GIF-0726-r, and a 1-aminoindane structure obtained from rasagiline.10,11 GIF-2197-r is the racemate; GIF-2115 is in the R form which is a chiral compound with one asymmetric carbon center, and GIF-2196 is in the S form which is its antipode (Fig. 1). Despite the differing stereochemistry around the 1-aminoindane moiety, all three forms demonstrate similar anti-ferroptotic efficacy, suggesting that the stereochemistry does not significantly influence neuroprotection.9 In this paper, we elucidate the structural features responsible for the localization to lysosomes and anti-ferroptotic efficacy. The results indicate that the aliphatic tertiary amine group is essential for localizing GIF-2197-r to lysosomes, while the N,N-dimethylaniline structure is essential for ferrous ion coordination and anti-ferroptotic ability. Moreover, conducted density functional theory (DFT) computations verified the interaction of N,N-dimethylaniline derivatives with ferrous ions.
Experimental methods
Materials
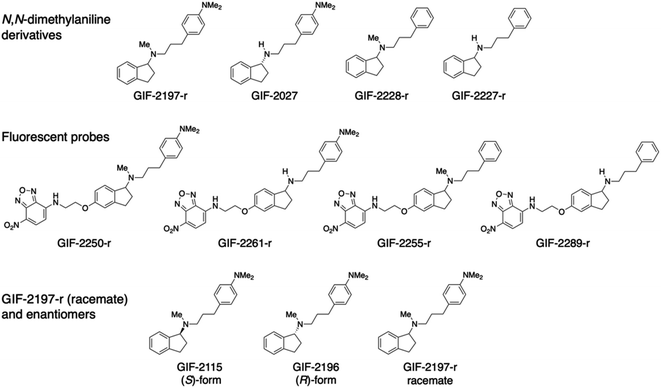
The aminoindan derivatives GIF-2197-r [N-methyl-N-{3-[4-(dimethylamino)phenylpropyl]indan-1-amine}], GIF-2027 [(R)-N-{3-[4-(dimethylamino)phenyl]propyl}indan-1-amine], GIF-2228-r [N-methyl-N-(3-phenylpropyl)indan-1-amine], and GIF-2227-r [N-(3-phenylpropyl)indan-1-amine] were synthesized by reductive amination of aromatic ketones or aldehydes with 3-[4-(dimethylamino)phenyl]propylamine and subsequent reductive methylation of the resultant secondary amines with formaldehyde. GIF-2250-r, GIF-2261-r, GIF-2255-r, and GIF-2289-r were synthesized by nucleophilic substitution reactions of each aminoethoxy derivative of GIF-2197-r, GIF-2027, GIF-2228-r, and GIF-2227-r, respectively, with 4-fluoro-7-nitro-2,1,3-benzoxadiazole (NBD-F; Tokyo Chemical Industry, Tokyo, Japan). All GIF compounds were dissolved in cell culture-grade dimethyl sulfoxide or methanol and stored in the dark at −20 °C.Cell culture
The immortalized mouse hippocampal cell line HT22 (RRID: CVCL_0321), a generous gift from the late Dr David Schubert (The Salk Institute, La Jolla, CA, USA), was cultured in Dulbecco's modified Eagle's medium (DMEM) (Wako Pure Chemicals, Osaka, Japan, Cat#041-29775) supplemented with 5% fetal bovine serum (FBS; Nichirei Biosciences, Tokyo, Japan) at 37 °C in a 5% CO2 incubator. The cells were replaced either at passage 30–40 or every 3 months from the original frozen stocks.Cell death assays
Cell death was determined by measuring the activity of lactate dehydrogenase (LDH) released into the medium using a Cytotoxicity Detection Kit according to the manufacturer's protocol (Dojindo Laboratories, Kumamoto, Japan, Cat# CK12). Briefly, HT22 cells were seeded in 48-well plates at 2–3 × 104 cells per well and cultured for 1 day. After the indicated treatment, LDH activity was measured in the culture medium and cell lysates, and cell death rate (%) was calculated as 100 × (experimental release − blank)/(total release − blank), where the total release represented the LDH activity from cells lysed with 1% Triton X-100.Cell imaging
Cells were seeded at 2 × 105 on a 35 mm glass-bottom dishes and incubated in DMEM supplemented with 5% FBS for 24 h. To label lysosomes, cells were stained with LysoBrite™ Red (AAT Bioquest, Sunnyvale, CA, USA; 1 μM) and fluorescent (NBD-F-conjugated) GIF compounds (1 μM) at 37 °C for 30 min. The subcellular distribution of fluorescence emission was analyzed using a Keyence BZ-X810 microscope equipped with an optical sectioning algorithm system.In vitro (cell-free) ferrous ion assay
Ferrous ions were measured in a cell-free solution using FerroOrange. Diluted compounds (50 μM) were mixed with 50 μM Fe(NH4)2(SO4)2, followed by the addition of 1 μM FerroOrange. Relative fluorescence emission at 572 nm from 532 nm excitation was measured every 1 min as previously reported.12DFT computation
DFT computations were performed using Gaussian 16 software (Gaussian, Inc., Wallingford, CT, 2019) and visualized using GaussView 6 (R. Dennington, T. Keith and J. Millam, GaussView, Version 5.0.8.: Semichem Inc., Shawnee Mission, KS, 2009). All calculations were conducted using the B3LYP hybrid function with LANL2DZ pseudopotential for the Fe atom and the 6-31G(d) basis set for other atoms. All energy values (E) included the zero-point energy (ZPE) correction (E in kcal mol−1). Multiple monodentate coordination structures are possible for the complexes of GIF compounds and Fe(OH2)62+. Complexation releases one H2O molecule around Fe2+ in the monodentate structure. The stabilization energies of complex formation were estimated in the presence of six H2O molecules as the solvent, taking into account hydrogen bond stabilization of the released water molecule and the GIF compounds.Statistical analysis
All statistical analyses were conducted using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA, RRID: SCR_002798). Treatment group means were compared by one-way ANOVA followed by Tukey's tests for pair-wise comparisons. The number of independent experiments and p values are provided in the figure legends.Results and discussion
Chemical structures of GIF-2197-r (−2115 and −2196), −2027, −2228-r, and −2227-r, and their fluorescent probes GIF-2250-r, −2261-r, −2255-r, and −2289-r.To identify the structural features of GIF-2197-r responsible for localization to lysosomes and anti-ferroptotic efficacy, we synthesized a series of derivatives with selective modifications. Hydrophobic weak base drugs, such as aliphatic amines, are known to accumulate at high levels in lysosomes (pH range 4.5 to 5.5) via cation trapping.13,14 While amines with high basicity and hydrogen-bonding capacities, such as secondary amines, typically struggle to penetrate into lipid membranes, lipophilic amine drugs possessing weak base properties easily diffuse across cell membranes at a physiological pH through passive diffusion in their uncharged forms.15 Consequently, it is assumed that demethylation of the N-methyl-aminoindane moiety in GIF-2197-r would attenuate lysosomotropism. This decrease is attributed to the heightened basicity and enhanced hydrogen-bonding tendencies of the resulting secondary amine structure, which acts as a hydrogen bond donor.16 To examine this possibility, we synthesized GIF-2027, which lacks the N-methyl group in the aliphatic tertiary amine moiety of GIF-2197-r (abbreviated as alN-Me) (Fig. 1). We also previously demonstrated that GIF-2228-r, in which the N,N-dimethylamino group of GIF-2197-r is substituted with hydrogen, has lower anti-ferroptotic activity than the parent compound,9 so in addition we synthesized GIF-2227-r, which lacks both the alN-Me and N,N-dimethylamino groups. Further, to directly examine the subcellular distributions of GIF-2027, GIF-2228-r, and GIF-2227-r in treated HT22, we synthesized corresponding 4-amino-7-nitro-2,1,3-benzoxadiazole (NBD-NH)-conjugated fluorescent probes, named GIF-2261-r, GIF-2255-r, and GIF-2289-r, respectively (Fig. 1).
Effects of GIF-2027, GIF-2227-r, and GIF-2228-r on glutamate-induced ferroptosis
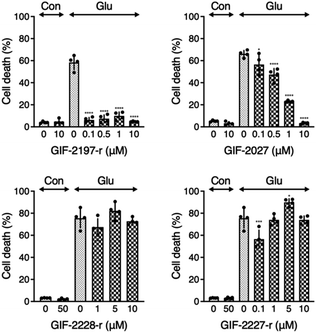
To examine the influence of the alN-Me of GIF-2197-r on anti-ferroptotic activity, we compared LDH release from glutamate-treated HT22 cells in the presence of the described derivative GIF compounds (Fig. 2). GIF-2027 exhibited weaker protection against glutamate-induced ferroptosis than GIF-2197-r but stronger protection than GIF-2227-r and GIF-2228-r. These results strongly suggest that the alN-Me group contributes to anti-ferroptotic ability and further confirms the essential role of the N,N-dimethylaniline structure for anti-ferroptotic activity documented in our previous study.9Localization of GIF-2027, GIF-2228-r, and GIF-2227-r in HT22 cells
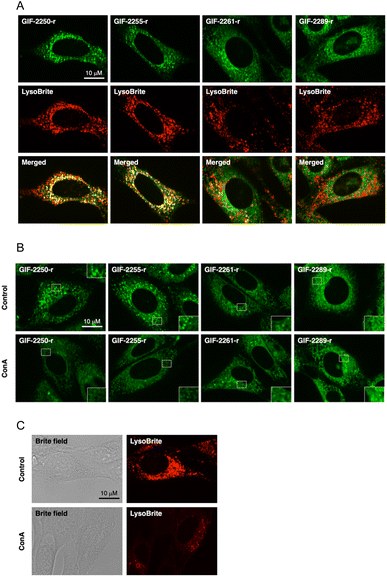
The fluorogenic labeling reagent for amine derivatives NBD-F17 has been used to successfully develop fluorescent derivatives of anti-ferroptotic reagents such as GIF-2114, GIF-2197-r, and haloperidol.9,18 To examine whether the alN-Me group of GIF-2197-r contributes to lysosomal localization, we synthesized GIF-2027-OCH2CH2NH-NBD (GIF-2261-r), GIF-2228-r-OCH2CH2NH-NBD (GIF-2255-r), and GIF-2227-r-OCH2CH2NH-NBD (GIF-2289-r), and examined colocalization of these probes with the lysosome marker LysoBrite™ Red (LysoBrite) in HT22 cells (Fig. 3A). Neither GIF-2261- r nor GIF-2289-r colocalized substantially with LysoBrite. In particular, GIF-2289-r, which lacks both alN-Me and N,N-dimethylamino groups, exhibited fewer intense foci compared to the other compounds. In contrast, GIF-2255-r, which retains the alN-Me group, was observed to colocalize with LysoBrite in a manner similar to the fluorescent GIF-2197-r probe GIF-2250-r. These findings suggest that the aliphatic tertiary amine group alN-Me contributes to lysosomal localization, likely by forming a membrane impermeable cation species under acidic conditions. In addition to the alN-Me group, the N,N-dimethylamino group impacts lysosomal localization as evidenced by the weak colocalization of the fluorescent GIF-2227-r probe GIF-2289-r. To verify these results, we examined the effect of the V-ATPase inhibitor concanamycin A, which reduces lysosomal acidity, on the localization of GIF-OCH2CH2NH-NBD compounds (Fig. 3B). While alkalization of the lysosomal compartment by concanamycin A did not significantly alter the intracellular distribution of GIF-2261-r and GIF-2289-r, it did reduce the punctate fluorescence of GIF-2250-r and GIF-2255-r. These results suggest that GIF-2261-r and GIF-2289-r (lacking alN-Me) do not localize to lysosomes, whereas GIF-2250-r and GIF-2255-r (retaining alN-Me) normally exhibit lysosomal accumulation due to the acidic environment. Further, the lysosome-specific fluorescent probe LysoBrite still selectively accumulated in lysosomes via the lysosome pH gradient (Fig. 3C).19The N,N-dimethylaniline structure is essential for ferrous ion coordination
It has been reported that a major part of cellular redox-active iron is in the acidic vacuolar endosomal/lysosomal compartment, and lysosomal ferric irons, which are reduced to ferrous ions by the metalloreductase Steap3, are required for hydrogen peroxide-induced cell death.20–22 Therefore, the lysosome-targeting ability of GIF-2197-r is important for ferroptosis inhibition. To verify the function of the GIF-2027 N,N-dimethylaniline group in ferrous ion coordination, we measured the reduction in free ferrous ion upon adding the test compound to a cell-free medium fluorometrically using FerroOrange.12 Similar to GIF-2115 (the R form of racemic GIF-2197-r),9 addition of GIF-2027 resulted in a greater reduction in the fluorescence signal than an equivalent concentration of the known iron chelator deferoxamine. Conversely, like GIF-2228-r,9 addition of GIF-2227-r did not cause a detectable reduction in the fluorescence signal (Fig. 4). These results and those previously reported9 suggest that the N,N-dimethylaniline structure confers anti-ferroptotic activity by ferrous ion coordination.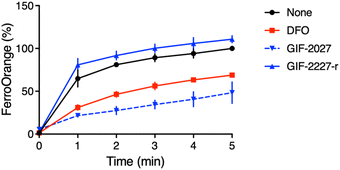 | ||
| Fig. 4 Kinetics of fluorescence response of FerroOrange in the presence of GIF-2027 and GIF-2227-r in vitro (cell-free). Relative fluorescence intensities were measured at 572 nm with excitation at 532 nm every 1 min, as previously described.12 All data were acquired using a 1 μM FerroOrange in the presence of 50 μM of FeSO4 and 50 μM GIF compounds in 50 mM HEPES buffer (pH 7.4). | ||
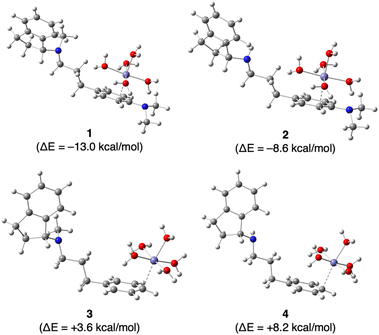
Finally, we investigated the effect of the N,N-dimethylamino group on the complexation of Fe2+ by GIF-2197-r a (containing –N(CH3)2), GIF-2027 b (containing –N(CH3)2), GIF-2228-r c (lacking –N(CH3)2), and GIF-2227-r d (lacking –N(CH3)2) in silico using Gaussian 16 and GaussView 6 programs for DFT computation. Fig. 5 depicts the most stabilized monodentate complexes of GIF-2197-r (1), GIF-2027 (2), GIF-2228-r (3), and GIF-2227-r (4). In these complexes, the Fe2+ coordinates with the benzene ring carbon bound to the N,N-dimethylamino group. The stabilization energies of complexes 1 and 2 were calculated using the formula ΔE = E[1] − (E[a + 6H2O] + E[Fe(OH2)62+] − E[7H2O]) for complex 1, yielding values of −13.0 and −8.6 kcal mol−1, respectively. These values suggest that the complexation of Fe2+ with GIF-2197-r and GIF-2027 results in stabilization. In contrast, the stabilization energies ΔE of complexes 3 and 4, determined using a similar method, yielded values of +3.6 and +8.2 kcal mol−1, respectively, indicating that the complexation of the Fe2+ does not result in stabilization. These calculations indicate that complexation occurs only in GIF compounds including a N,N-dimethylamino group, consistent with the experimental results.
Conclusions
In conclusion, an aliphatic tertiary amine structure is essential for lysosomal localization of the potent ferroptosis inhibitor GIF-2197-r, while an N,N-dimethylamino group is essential for iron complexation and concomitant protection of cells against ferroptosis. These structural features are also observed in another derivative, GIF-2114.9 Collectively, these findings offer valuable clues for the development of improved ferroptosis inhibitors.Author contributions
Y. H. and T. H. performed experiments; K. F. designed and synthesized the compounds; T. H. and K. A. performed DFT computations, and Y. O. K. reviewed and critiqued the manuscript. H. T., K. F., and Y. H. designed experiments; T. H., K. F., and Y. H. wrote the manuscript.Conflicts of interest
The authors declare no conflict of interest in relation to this work.Acknowledgements
The authors would like to express our gratitude to the following individuals: Dr Toshiaki Murai and Dr Masayuki Ninomiya (Gifu University) for their assistance with MS analysis, Dr Makoto Sawada (Nagoya University) for his assistance with fluorescence microscopy study, and Ms Ayame Hasegawa, Ms Mayu Takahashi, and Dr Kentaro Oh-hashi (Gifu University) for their contributions to the initial experiments. The authors are very grateful to Dr Tasuku Hirayama and Dr Hideko Nagasawa (Gifu Pharmaceutical University) for providing the FerroOrange reagent and the late Dr David Schubert for the generous gift of HT22 cells. This work was supported in part by the Japan Society for the Promotion of Science KAKENHI (JP22K11824, Y. H.) and CCI Corporation (Gifu, Japan).References
- E. Niedzielska, I. Smaga, M. Gawlik, A. Moniczewski, P. Stankowicz, J. Pera and M. Filip, Mol. Neurobiol., 2016, 53, 4094–4125 CrossRef CAS.
- E. Radi, P. Formichi, C. Battisti and A. Federico, J. Alzheimer's Dis., 2014, 42(3), S125–S152 Search PubMed.
- B. R. Stockwell, J. P. Friedmann Angeli, H. Bayir, A. I. Bush, M. Conrad, S. J. Dixon, S. Fulda, S. Gascon, S. K. Hatzios, V. E. Kagan, K. Noel, X. Jiang, A. Linkermann, M. E. Murphy, M. Overholtzer, A. Oyagi, G. C. Pagnussat, J. Park, Q. Ran, C. S. Rosenfeld, K. Salnikow, D. Tang, F. M. Torti, S. V. Torti, S. Toyokuni, K. A. Woerpel and D. D. Zhang, Cell, 2017, 171, 273–285 CrossRef CAS PubMed.
- Y. Li, P. Maher and D. Schubert, Neuron, 1997, 19, 453–463 CrossRef CAS PubMed.
- S. Tan, D. Schubert and P. Maher, Curr. Top. Med. Chem., 2001, 1, 497–506 CrossRef CAS PubMed.
- S. J. Dixon, K. M. Lemberg, M. R. Lamprecht, R. Skouta, E. M. Zaitsev, C. E. Gleason, D. N. Patel, A. J. Bauer, A. M. Cantley, W. S. Yang, B. Morrison 3rd and B. R. Stockwell, Cell, 2012, 149, 1060–1072 CrossRef CAS.
- J. Lewerenz, G. Ates, A. Methner, M. Conrad and P. Maher, Front. Neurosci., 2018, 12, 214 CrossRef PubMed.
- Y. Hirata, R. Okazaki, M. Sato, K. Oh-Hashi, H. Takemori and K. Furuta, Eur. J. Pharmacol., 2022, 928, 175119 CrossRef CAS PubMed.
- Y. Hirata, Y. Tsunekawa, M. Takahashi, K. Oh-Hashi, K. Kawaguchi, M. Hayazaki, M. Watanabe, K. I. Koga, Y. Hattori, H. Takemori and K. Furuta, Free Radical Biol. Med., 2021, 174, 225–235 CrossRef CAS PubMed.
- Y. Hirata, C. Yamada, Y. Ito, S. Yamamoto, H. Nagase, K. Oh-Hashi, K. Kiuchi, H. Suzuki, M. Sawada and K. Furuta, Neuropharmacol, 2018, 135, 242–252 CrossRef CAS.
- M. Naoi, W. Maruyama and M. Shamoto-Nagai, J. Neural Transm., 2020, 127, 131–147 CrossRef CAS PubMed.
- T. Hirayama, M. Niwa, S. Hirosawa and H. Nagasawa, ACS Sens., 2020, 5, 2950–2958 CrossRef CAS PubMed.
- A. M. Kaufmann and J. P. Krise, J. Pharm. Sci., 2007, 96, 729–746 CrossRef CAS.
- B. Zhitomirsky and Y. G. Assaraf, Oncotarget, 2017, 8, 45117–45132 CrossRef.
- C. L. Andrew, A. R. Klemm and J. B. Lloyd, Biochim. Biophys. Acta, 1997, 1330, 71–82 CrossRef CAS PubMed.
- C. de Duve, T. de Barsy, B. Poole, A. Trouet, P. Tulkens and F. Van Hoof, Biochem. Pharmacol., 1974, 23, 2495–2510 CrossRef CAS PubMed.
- S. Uchiyama, T. Santa, N. Okiyama, T. Fukushima and K. Imai, Biomed. Chromatogr., 2001, 15, 295–318 CrossRef CAS PubMed.
- Y. Hirata, K. Oka, S. Yamamoto, H. Watanabe, K. Oh-Hashi, T. Hirayama, H. Nagasawa, H. Takemori and K. Furuta, ACS Chem. Neurosci., 2022, 13, 2719–2727 CrossRef CAS PubMed.
- S. D. Goldman, R. S. Funk, R. A. Rajewski and J. P. Krise, Bioanalysis, 2009, 1, 1445–1459 CrossRef CAS PubMed.
- P. E. Starke, J. D. Gilbertson and J. L. Farber, Biochem. Biophys. Res. Commun., 1985, 133, 371–379 CrossRef CAS PubMed.
- T. Kurz, A. Leake, T. Von Zglinicki and U. T. Brunk, Biochem. J., 2004, 378, 1039–1045 CrossRef CAS PubMed.
- K. Iwai, Free Radical Biol. Med., 2019, 133, 64–68 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra06611h |
| This journal is © The Royal Society of Chemistry 2023 |