Open Access Article
Open Access ArticleReactions of proteins with a few organopalladium compounds of medicinal interest†
Lara Massai a,
Thomas Scattolinb,
Matteo Tarchia,
Fabiano Visentin
a,
Thomas Scattolinb,
Matteo Tarchia,
Fabiano Visentin *b and
Luigi Messori
*b and
Luigi Messori *a
*a
aDepartment of Chemistry, University of Florence, Via della Lastruccia 3-13, 50019 Sesto Fiorentino, Italy. E-mail: luigi.messori@unifi.it
bDipartimento di Scienze Molecolari e Nanosistemi, Università Ca’ Foscari, Campus Scientifico Via Torino 155, 30174 Venezia-Mestre, Italy. E-mail: fvise@unive.it
First published on 21st September 2022
Abstract
Pd compounds form a promising class of experimental anticancer drug candidates whose mechanism of action is still largely unknown; in particular, a few organopalladium compounds seem very attractive. To gain mechanistic insight into medicinal palladium compounds, we have explored here – through ESI MS analysis – the interactions of four organopalladium agents (1–4) – showing remarkable in vitro antiproliferative properties – with a few representative model proteins, i.e., lysozyme (HEWL), ribonuclease A (RNase), and carbonic anhydrase (hCAI). The tested panel included three Pd allyl compounds with one or two carbene ligands and a palladacyclopentadienyl complex. Notably, the Pd allyl compounds turned out to manifest, on the whole, a modest tendency to react with the above proteins. Only complex 3 produced small amounts of characteristic adducts with hCAI bearing either one or two Pd allyl groups. In contrast, the palladacyclopentadienyl complex 4 manifested a greater and peculiar reactivity with all the above proteins generating invariably protein adducts with a mass increase of +256 Da where a butadienyl group – with no associated Pd – is attached to the proteins. Afterwards, we extended our investigations to the C-terminal dodecapeptide of thioredoxin reductase bearing the –Cys–Sec– reactive motif. In this latter case adducts were formed with all tested Pd compounds; however, complex 4 manifested towards this dodecapeptide a type of reactivity deeply different from that observed with HEWL, RNase A and hCAI. The mechanistic implications of these findings are discussed.
Introduction
Nowadays, palladium compounds are being considered with great attention in the field of medicinal inorganic chemistry as they might represent a valuable alternative to clinically established anticancer platinum drugs.1 Indeed, a number of palladium complexes with innovative features and encouraging anticancer properties were recently prepared, characterized and tested by Visentin and co-workers.2 In particular, the most interesting compounds contain the palladium-η3-allyl and palladacyclopentadienyl fragments.Palladium allyl complexes are particularly promising since they generally exhibit good to excellent antiproliferative activities towards different cancer cell lines and, in some cases, a far lower cytotoxicity towards normal cells.2b–e In-depth immunofluorescence studies suggested that primary damage occurs at the level of mitochondria rather than on DNA as typically found in the case of cisplatin and its second- and third-generation derivatives.2b Interestingly, an excellent antitumor activity was observed for almost all the investigated Pd-allyl compounds, regardless of the ancillary ligand used. This evidence suggests that the Pd-allyl fragment, with its well-known reactivity towards nucleophiles,3 might represent the key factor in the biological activity of this fascinating family of compounds. Another possibility is offered by the nucleophilic attack on the metal center and the consequent formation of biotarget-Pd-allyl adducts.
Palladacyclopentadienyl complexes exhibit a remarkable anticancer activity when strong ancillary ligands (e.g. N-heterocyclic carbenes, phosphines and isocyanides) are present in the coordination sphere of palladium.2f,g Their anticancer activity seems to involve a strong non-covalent interaction with DNA, similarly to many organic anticancer drugs (i.e. doxorubicin).2g The palladacyclopentadienyl fragment under ordinary conditions usually exhibits a lower reactivity than the Palladium-allyl moiety. In fact, it can only be attacked by halogens or reactive alkyl/aryl halides leading to the corresponding Pd(II)-butadienyl complexes.4 The butadienyl fragment can then be released from the metal center by a further oxidative addition and consecutive reductive elimination. However, no studies have been performed so far on the reactivity of this particular palladacyclic fragment towards proteins. In fact, it is always worth remembering the ability of many proteins to promote reactions that are difficult to foresee under other operating conditions.
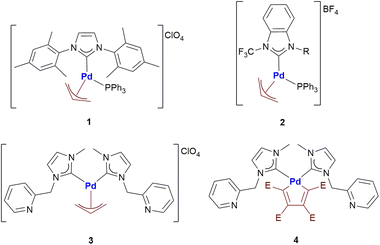
With the aim of investigating even in more detail these two classes of promising anticancer agents, herein we report the first systematic study of the reactivity of four different organopalladium complexes (Scheme 1) with a group of biologically relevant proteins.
In the Metmed laboratory, in Florence, a general ESI MS based protocol has been developed in recent years to characterize the protein interactions of metal-based drugs. The protocol is grounded on the detailed comparative ESI MS analysis of the metallodrug protein adducts that are formed when incubating the various proteins with each metallodrug under well-defined experimental conditions.5,6 In principle, ESI MS experiments allow to establish the nature of the metallic fragments that are present in the adducts, the extent of adduct formation and the binding stoichiometry. Owing to these favourable features the ESI MS based protocol turns out to be a more straightforward and informative method with respect to other bioanalytical protocols based, for instance, on hyphenation of LC with ICP MS detection.
Here, we have applied this type of approach to the analysis of the interactions occurring between the palladium compounds reported in Scheme 1 and three selected model proteins, i.e., Hen Egg White Lysozyme (HEWL), Bovine pancreatic ribonuclease A (RNase A) and human carbonic anhydrase I (hCAI). The obtained results are compared with those emerging from the reactions of the same palladium compounds with the C-terminal dodecapeptide of TrxR1.7
Results and discussion
The reactions of panel Pd compounds with RNase and HEWL
The selected Pd compounds were initially challenged against the small proteins RNase A and Hen Egg White Lysozyme that are often used for this kind of investigations and the respective ESI MS spectra were recorded.8Bovine pancreatic ribonuclease A is a pancreatic ribonuclease enzyme that cleaves single-stranded RNA, specifically after pyrimidine nucleotides. RNase A is one of the classic model systems of protein science since it is extremely stable and can be purified in large quantities. RNase A is a rather small protein (124 residues, ∼13.7 kDa). RNase A has four disulfide bonds in its native state, i.e., Cys26–84, Cys58–110, Cys40–95 and Cys65–72. These disulfide bonds are relatively well exposed to the solvent.
Hen Egg White Lysozyme is an enzyme characterized by the ability to break down the bacterial cell wall to improve protein or nucleic acid extraction efficiency. HEWL is a small (129 amino acids) and stable enzyme, that is ideal for research concerning protein structure and function.
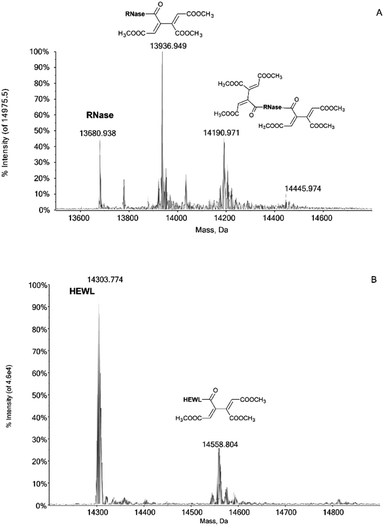
Notably, the Pd compounds 1, 2 and 3 do not show any significant reactivity with RNase A and lysozyme; indeed, no peaks assignable to of protein adducts were detected in the ESI MS spectra after 24 or 72 h of incubation (data not shown). Unlike the previous compounds, 4 manifests a significant reactivity towards both proteins. Indeed, its 24 h and 72 h deconvoluted spectra show a very intense signal at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 936 Da which is shifted of +256 Da from the RNase A peak at 13
936 Da which is shifted of +256 Da from the RNase A peak at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 Da (Fig. 1A). In the same way, a peak with a shift of +256 Da is observed in the 24 and 72 h deconvoluted spectra taken on HEWL samples reacted with 4 (Fig. 1B). This mass shift has been attributed to the protein binding of a butadienyl fragment; remarkably, this behaviour is identical to that observed in the case of hCAI (see later).
680 Da (Fig. 1A). In the same way, a peak with a shift of +256 Da is observed in the 24 and 72 h deconvoluted spectra taken on HEWL samples reacted with 4 (Fig. 1B). This mass shift has been attributed to the protein binding of a butadienyl fragment; remarkably, this behaviour is identical to that observed in the case of hCAI (see later).
Reactions of panel Pd compounds with hCA 1
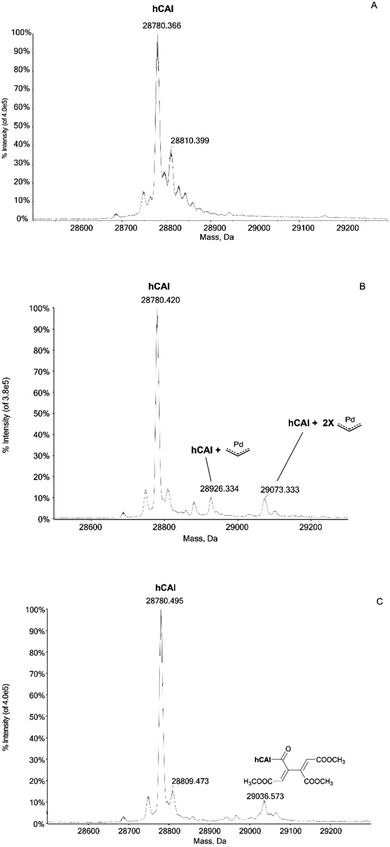
In humans there are 14 different carbonic anhydrase (CA) isoenzymes with different subcellular localization and tissue distribution. High levels of CA I and II expression have been found in blood cells and are necessary for maintaining the physiological blood pH. CA I consists of 261 amino acids with a molecular weight of about 28 kDa and contains a Zn(II) ion essential for the catalysis.9 The Zn(II) ion is coordinated by three histidine residues (His94, 96 and 119) and by a water molecule/hydroxide ion. Moreover, the enzyme presents a free cysteine (Cys213) as a potential anchoring site for metals. Carbonic Anhydrases can play a crucial role in controlling the pH in the tumor microenvironment. In fact, cancer cells can go to a state of hypoxia through the process known as the “Warburg effect”, defined as the increase in glucose consumption through glycolysis with the production of lactic acid. When this happens, there is a decrease in the pH of the extracellular environment due to the efflux of lactic acid and free protons outside the tumor cell. The decrease in external pH promotes the survival and proliferation of cancer cells. Several hypotheses have been proposed in which both the membrane CAs and the cytosolic ones contribute to regulating and promoting this particular pH difference.10 It has been observed that CA I is over-regulated in human pancreatic cancer and that high levels of CA I mRNA have been found in patients with myeloid leukaemia, colorectal and kidney cancers.11The important role played by CA in these cancer-related processes justifies the great attention for this protein. We have analyzed here the reaction of the four Pd compounds with hCA I (the deconvoluted spectrum of hCA I is shown in Fig. 2A). Interestingly, we found that 1 and 2 do not interact with hCA I as the deconvoluted spectra taken after 24 or 48 hours do not exhibit any new signal with a mass greater than the free protein; instead, adducts formation was clearly observed upon reacting hCA I with 3 and 4. In the deconvoluted spectrum of hCAI reacted with 3 (Fig. 2B), two distinct signals are indeed detected at greater mass values; in particular, the signal at 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 926 Da (shift = +146 Da) that well corresponds to the adduct of CA I with the Pd-allyl group and the signal at 29
926 Da (shift = +146 Da) that well corresponds to the adduct of CA I with the Pd-allyl group and the signal at 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 073 Da (shift = +293 Da) due to the formation of a protein adduct bearing two Pd-allyl fragments. These new signals remain visible in the deconvoluted spectrum taken at 72 h.
073 Da (shift = +293 Da) due to the formation of a protein adduct bearing two Pd-allyl fragments. These new signals remain visible in the deconvoluted spectrum taken at 72 h.
In the deconvoluted spectrum of hCAI reacted with 4 a signal at 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 036 Da (shift = +256 Da from the apo hCA I) is evident, which is attributed to the protein binding of a butadienyl fragment; notably, in this case, the interaction does not involve any metal moiety (Fig. 2C). The 72 h deconvoluted spectrum does not differ substantially from that taken at 24 h, indicating that stable adducts are fully formed already in the first 24 hours of incubation.
036 Da (shift = +256 Da from the apo hCA I) is evident, which is attributed to the protein binding of a butadienyl fragment; notably, in this case, the interaction does not involve any metal moiety (Fig. 2C). The 72 h deconvoluted spectrum does not differ substantially from that taken at 24 h, indicating that stable adducts are fully formed already in the first 24 hours of incubation.
The reactions with dTrxR
The enzyme thioredoxin reductase is unanimously considered one of the main targets for cytotoxic gold compounds.12–14So, we selected the C-terminal dodecapeptide that mimics the TrxR1 active site, to study its reactivity with Pd complexes. This peptide possesses the –Cys–Sec– reactive motif as the most probable binding site for a soft Lewis metal.15 The intramolecular –S–Se– bridge between the aforesaid residues needs to be reduced in order to favour the reaction with metal compounds. So, the reducing agent dithiothreitol (DTT) was added, in 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DTT/peptide ratio, 30 minutes before the incubation of the dodecapeptide with each Pd compound. The mass/charge spectrum reported in Fig. 3A shows the signals of the peptide and of its adducts with Na+ and K+ ions from the solution. Upon reaction with the four Pd complexes, two distinct reactivity trends were identified:
1 DTT/peptide ratio, 30 minutes before the incubation of the dodecapeptide with each Pd compound. The mass/charge spectrum reported in Fig. 3A shows the signals of the peptide and of its adducts with Na+ and K+ ions from the solution. Upon reaction with the four Pd complexes, two distinct reactivity trends were identified:
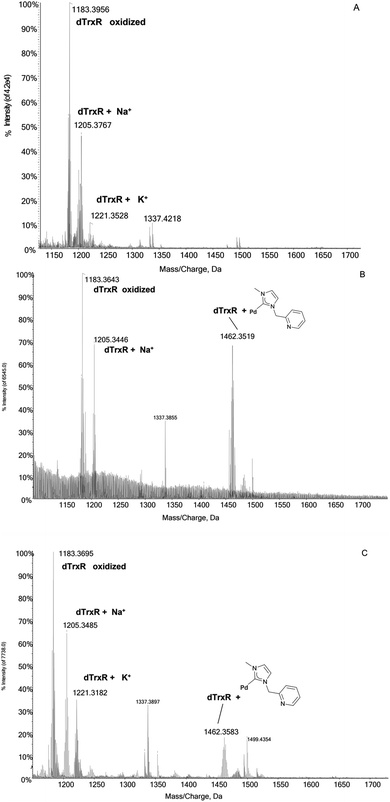 | ||
Fig. 3 (A) ESI mass spectra of dTrxR 5 × 10−7 M incubated at 37 °C for 2 h with (B) 3 and (C) 4 at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3:10 peptide-to-palladium-to-DTT ratio. 0.1% v/v of formic acid was added just before infusion. 3:10 peptide-to-palladium-to-DTT ratio. 0.1% v/v of formic acid was added just before infusion. | ||
(a) The case of complex 2 which binds the dodecapeptide through the [Pd-PPh3] fragment; instead, upon interaction with 1 the dTrxR peptide binds the [allyl-Pd-PPh3] moiety (see ESI†).
(b) The case of complex 3 and 4 where the adducts originate from the interaction between dTrxR and 1-methylpyridine-3-methyl-imidazole-2-ylidene Pd(II) (Fig. 3B and C).
Conclusions
Palladium compounds are attracting a renewed interest as potential cytotoxic drugs for cancer treatment. Indeed, the palladium(II) center manifests a close structural similarity to the medicinally relevant platinum(II) center but is characterized by a profoundly different ligand exchange kinetics. Various series of palladium compounds were recently developed. The mode of action of these metallodrugs to produce their antiproliferative effects is still unclear. To gain mechanistic information we decided to test systematically the interactions of some of them with a few selected model proteins. Accordingly, four representative organopalladium compounds were selected for this investigation. Studies conducted on the smaller proteins lysozyme and RNase revealed that the tested palladium compounds, with the exception of 4, are not able to form adducts with these proteins. Instead, a characteristic reactivity was observed when incubating 3 with hCAI: the latter reaction results into the binding to the protein of one or two Pd-allyl fragments. The presence of cysteine residues on the hCAI suggests the formation of one or two Pd–S bonds. The poor reactivity of complexes 1 and 2 may be ascribed to the greater steric hindrance of their ancillary ligands (one PPh3 combined with one bulky NHC) that prevents the access of the cysteine residue to the metal centre. Conversely, 4 was able to form adducts with the above three proteins: in fact, in all cases, an adduct characterized by a constant increase in molecular mass of +256 was detected. Quite surprisingly, these adducts do not contain Pd. We best interpret this reactivity in terms of protein binding of an organic moiety originating from the butadienyl fragment. A plausible explanation is represented by the nucleophilic attack of an amino/imidazole or hydroxyl group (the former is present in lysine or histidine, the latter in serine or tyrosine side chain of the amino acid sequences of the three proteins) on one of the ester moieties of the butadienyl fragment and the consequent formation of a new amidic or ester bond, respectively. To understand this variegate reactivity pattern in more depth we have extended our studies to the C-terminal dodecapeptide of the enzyme thioredoxin reductase bearing the –Cys–Sec– reactive motif. Interestingly, this dodecapeptide manifested a very different reactivity profile towards the selected Pd compounds in comparison to model proteins; indeed, adducts were formed with all Pd compounds. In addition, in the case of 4, a different type of adduct was formed that does not correspond to those previously observed, all characterised by a mass shift of +256 Da.In conclusion, the present study reveals that the panel palladium complexes are on the whole not particularly reactive with proteins while showing rather complex reactivity patterns. The greater reactivity manifested by 4 towards the three model proteins is traced back to the binding of an organic moiety to the interacting proteins with no transfer of the metal. On the other hand, the reactions taking place between 4 and the C-Terminal dodecapeptide of thioredoxin reductase highlight a very different type of reactivity that is probably determined by the presence on the peptide of the Cys–Sec– reactive motif. We believe that disclosing in depth these patterns of interaction is crucial to understand in more detail the reactions of medicinal palladium complexes with their presumed biomolecular targets.
Experimental
Materials
The palladium complexes 1–4 were synthesized according to previously published procedures.2b,c,4fLyophilized human carbonic anhydrase (hCA I), HEWL and RNase A were purchased from Merck and used without further purification or manipulation. The C-terminal dodecapeptide of thioredoxin reductase (dTrxR), were synthesized in the MetMed Laboratories at the Department of Chemistry, University of Florence, following already established procedures.7,15 LC-MS materials (water, acetonitrile) were purchased from Honeywell.
ESI-Q-TOF mass spectrometer
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The mass range of the instrument is between 50 Da and 40 kDa. The ESI source parameters have been optimized for each protein. The following parameters were used for protein analysis alone and for metallation studies:
000. The mass range of the instrument is between 50 Da and 40 kDa. The ESI source parameters have been optimized for each protein. The following parameters were used for protein analysis alone and for metallation studies:- hCA I: positive polarity, ionspray voltage floating (ISFV) 5500 V, temperature (TEM) 0, gas 1 (GS1) 50 L min; gas 2 (GS2) 0; curtain gas (CUR) 20 L min−1, declustering potential (DP) 50 V, collision energy (CE) 10 V; range 600–1400 m/z.
- RNase A: positive polarity, ion spray voltage floating (ISFV) 5500 V, temperature (TEM) 0, gas 1 (GS1) 40 L min; gas 2 (GS2) 0; curtain gas (CUR) 15 L min−1, declustering potential (DP) 100 V, collision energy (CE) 10 V; range 1000–2600 m/z.
- HEWL: positive polarity, ionspray voltage floating (ISFV) 5500 V, temperature (TEM) 0, gas 1 (GS1) 40 L min; gas 2 (GS2) 0; curtain gas (CUR) 20 L min−1, declustering potential (DP) 100 V, collision energy (CE) 10 V; range 1000–2800 m/z.
- dTrxR: positive polarity, ion spray voltage floating 5500 V, temperature 100 °C, ion source gas 1 (GS1) 25 L min; ion source gas 2 (GS2) 25 L min; CUR 30 L min−1, CE 10 V; DP 50 V, acquisition range 1000–2000.
For the acquisition, the Analyst TF 1.7.1 (Sciex) software is used and deconvolved spectra are obtained using the Bio Tool Kit v.2.2 incorporated in the PeakViewTM v.2.2 (Sciex) software.
Sample preparation
Stock solutions of hCA I 10−4 M, of HEWL, RNase and dTrxR 10−3 M, Mb, were prepared, dissolving the proteins and the peptide in H2O LC-MS grade. Stock solutions 10−2 M of the Pd compounds were prepared dissolving the samples in acetonitrile.For all experiments, solutions of the protein/peptide 10−5 M and Pd complexes at protein to metal ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 were prepared in 2 mM ammonium acetate buffer at pH 6.8. The mixtures were then incubated at 37 °C up to 72 h.
3 were prepared in 2 mM ammonium acetate buffer at pH 6.8. The mixtures were then incubated at 37 °C up to 72 h.
After the incubation time, all solutions were sampled and diluted to a final protein concentration of 7 × 10−7 M for hCA I and dTrxR and 10−7 M for HEWL and RNase using ammonium acetate buffer. In the final solutions were also added with 0.1% v/v of formic acid just before the infusion in the mass spectrometer.
Author contributions
Conceptualization, Luigi Messori, Fabiano Visentin; validation, Lara Massai, Luigi Messori, Thomas Scattolin, Fabiano Visentin investigation, Lara Massai, Matteo Tarchi, resources Luigi Messori; writing—original draft preparation, Luigi Messori, Lara Massai, Fabiano Visentin, Thomas Scattolin writing—review and editing, Luigi Messori, Lara Massai, supervision, Luigi Messori, Fabiano Visentin; project administration, Luigi Messori funding acquisition, Luigi Messori. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
LM and LMa thank Ente Cassa Risparmio Firenze (ECR) and AIRC for funding the project “Advanced mass spectrometry tools for cancer research: novel applications in proteomics, metabolomics and nanomedicine” (Multi-user Equipment Program 2016, Ref. code 19650). The CIRCMSB (Consorzio Interuniversitario di Ricerca in Chimica dei Metalli nei Sistemi Biologici, Italy) is, also, gratefully acknowledged by LM and LMa.Notes and references
- (a) T. Scattolin, V. A. Voloshkin, F. Visentin and S. P. Nolan, Cell Rep. Phys. Sci., 2021, 2, 100446 CrossRef CAS; (b) A. R. Kapdi and I. J. Fairlamb, Chem. Soc. Rev., 2014, 43, 4751 RSC.
- (a) T. Scattolin, E. Bortolamiol, S. Palazzolo, I. Caligiuri, T. Perin, V. Canzonieri, N. Demitri, F. Rizzolio, L. Cavallo, B. Dereli, M. V. Mane, S. P. Nolan and F. Visentin, Chem. Commun., 2020, 56, 12238 RSC; (b) T. Scattolin, E. Bortolamiol, F. Visentin, S. Palazzolo, I. Caligiuri, T. Perin, V. Canzonieri, N. Demitri, F. Rizzolio and A. Togni, Chem.–Eur. J., 2020, 26, 11868 CrossRef CAS PubMed; (c) T. Scattolin, E. Bortolamiol, I. Caligiuri, F. Rizzolio, N. Demitri and F. Visentin, Polyhedron, 2020, 186, 114607 CrossRef CAS; (d) T. Scattolin, E. Bortolamiol, F. Rizzolio, N. Demitri and F. Visentin, Appl. Organomet. Chem., 2020, 34, e5876 CrossRef CAS; (e) T. Scattolin, I. Caligiuri, L. Canovese, N. Demitri, R. Gambari, I. Lampronti, F. Rizzolio, C. Santo and F. Visentin, Dalton Trans., 2018, 47, 13616 RSC; (f) T. Scattolin, S. Giust, P. Bergamini, I. Caligiuri, L. Canovese, N. Demitri, R. Gambari, I. Lampronti, F. Rizzolio and F. Visentin, Appl. Organomet. Chem., 2019, 33, e4902 CrossRef; (g) T. Scattolin, I. Caligiuri, N. Mouawad, M. El Boustani, N. Demitri, F. Rizzolio and F. Visentin, Eur. J. Med. Chem., 2019, 179, 325 CrossRef CAS PubMed; (h) T. Scattolin, N. Pangerc, I. Lampronti, C. Tupini, R. Gambari, L. Marvelli, F. Rizzolio, N. Demitri, L. Canovese and F. Visentin, J. Organomet. Chem., 2019, 899, 120857 CrossRef CAS; (i) T. Scattolin, G. Moro, F. Rizzolio, C. Santo, L. M. Moretto and F. Visentin, ChemistrySelect, 2019, 4, 10911 CrossRef CAS.
- (a) B. M. Trost and M. L. Crawley, Chem. Rev., 2003, 103, 2921 CrossRef CAS PubMed; (b) L. Canovese, F. Visentin, T. Scattolin, C. Santo and V. Bertolasi, Polyhedron, 2016, 119, 377 CrossRef CAS; (c) T. Schlatzer, J. Kriegesmann, H. Schröder, M. Trobe, C. Lembacher-Fadum, S. Santner, A. V. Kravchuk, C. F. W. Becker and R. Breinbauer, J. Am. Chem. Soc., 2019, 141, 14931 CrossRef CAS; (d) A. M. Johns, M. Utsunomiya, C. D. Incarvito and J. F. Hartwig, J. Am. Chem. Soc., 2006, 128, 1828 CrossRef CAS PubMed.
- (a) R. Van Belzen, C. J. Elsevier, A. Didieu, N. Veldman and A. L. Speck, Organometallics, 2003, 22, 722 CrossRef CAS; (b) L. Canovese, F. Visentin, T. Scattolin, C. Santo and V. Bertolasi, J. Organomet. Chem., 2016, 808, 48 CrossRef CAS; (c) L. Canovese, F. Visentin, T. Scattolin, C. Santo and V. Bertolasi, Polyhedron, 2016, 113, 25 CrossRef CAS; (d) T. Scattolin, F. Visentin, C. Santo, V. Bertolasi and L. Canovese, Dalton Trans., 2016, 45, 11560 RSC; (e) L. Canovese, F. Visentin, T. Scattolin, C. Santo and V. Bertolasi, Dalton Trans., 2015, 44, 15049 RSC; (f) L. Canovese, C. Santo, T. Scattolin, F. Visentin and V. Bertolasi, J. Organomet. Chem., 2015, 794, 288 CrossRef CAS; (g) F. Visentin, C. Santo, T. Scattolin, N. Demitri and L. Canovese, Dalton Trans., 2017, 46, 10399 RSC.
- L. Massai, C. Zoppi, D. Cirri, A. Pratesi and L. Messori, Front. Chem., 2020, 8, 581648 CrossRef CAS PubMed.
- F. Sacco, M. Tarchi, G. Ferraro, A. Merlino, G. Facchetti, I. Rimoldi, L. Messori and L. Massai, Int. J. Mol. Sci., 2021, 22(19), 10551 CrossRef CAS PubMed.
- A. Pratesi, C. Gabbiani, E. Michelucci, M. Ginanneschi, A. M. Papini, R. Rubbiani, I. Ott and L. Messori, J. Inorg. Biochem., 2014, 136, 161 CrossRef CAS PubMed.
- (a) G. Ferraro, T. Marzo, M. E. Cucciolito, F. Ruffo, L. Messori and A. Merlino, Int. J. Mol. Sci., 2019, 20(3), 520 CrossRef CAS; (b) G. Ferraro, N. Demitri, L. Vitale, G. Sciortino, D. Sanna, V. Ugone, E. Garribba and A. Merlino, Inorg. Chem., 2021, 60(24), 19098 CrossRef CAS PubMed; (c) C. Pelosi, F. Saitta, C. Zerino, G. Canil, T. Biver, A. Pratesi, C. Duce, D. Fessas, C. GabbianI and M. R. Tiné, Molecules, 2021, 26(8), 2376 CrossRef CAS PubMed.
- V. M. Krishnamurthy, G. K. Kaufman, A. R. Urbach, I. Gitlin, K. L. Gudiksen, D. B. Weibel and G. M. Whitesides, Chem. Rev., 2008, 108(3), 946 CrossRef CAS PubMed.
- C. T. Supuran, A. Scozzafava and A. Casini, Med. Res. Rev., 2003, 3, 146 CrossRef PubMed.
- M. Y. Mboge, B. P. Mahon, R. McKenna and S. C. Frost, Metabolites, 2018, 8, 19 CrossRef PubMed.
- V. Gandin, A. Potamitou Fernandes, M. P. Rigobello, B. Dani, F. Sorrentino, F. Tisato, M. Björnstedt, A. Bindoli, A. Sturaro, R. Rella and C. Marzano, Biochem. Pharmacol., 2010, 79(2), 90 CrossRef CAS.
- Y. Cheng and Y. Qi, Med. Chem., 2017, 17(8), 1046 CAS.
- E. Vergara, A. Casini, F. Sorrentino, O. Zava, E. Cerrada, M. P. Rigobello, A. Bindoli, M. Laguna and P. J. Dyson, ChemMedChem, 2010, 5, 96 CrossRef CAS PubMed.
- A. Pratesi, C. Gabbiani, M. Ginanneschi and L. Messori, Chem. Commun., 2010, 46(37), 7001 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Additional ESI-MS spectra. See https://doi.org/10.1039/d2ra05332b |
| This journal is © The Royal Society of Chemistry 2022 |