Open Access Article
Open Access ArticleSERS based Y-shaped aptasensor for early diagnosis of acute kidney injury†
Dan Li‡
ab,
Linlu Zhao‡bc,
Jin Qianbc,
Heng Liu bc,
Jinmao You
bc,
Jinmao You *a,
Ziyi Cheng
*a,
Ziyi Cheng *bc and
Fabiao Yu
*bc and
Fabiao Yu *bc
*bc
aKey Laboratory of Life-Organic Analysis of Shandong Province, School of Chemistry and Chemical Engineering, Qufu Normal University, Qufu, 273165, PR China. E-mail: jmyou6304@163.com
bLaboratory of Neurology, The First Affiliated Hospital of Hainan Medical University, Hainan Medical University, Haikou 571199, China. E-mail: chengziyi@hainmc.edu.cn; yufabiao@hainmc.edu.cn
cKey Laboratory of Hainan Trauma and Disaster Rescue, Key Laboratory of Emergency and Trauma, Ministry of Education, College of Emergency and Trauma, Hainan Medical University, Haikou 571199, China
First published on 26th May 2022
Abstract
Considering the pivotal role of biomarkers in plasma, the development of biomarker specific sensing platforms is of great significance to achieve accurate diagnosis and monitor the occurrence and progress in acute kidney injury (AKI). In this paper, we develop a promising surface-enhanced Raman scattering-based aptasensor for duplex detection of two protein biomarkers in AKI. Exploiting the base-pairing specificity of nucleic acids to form a Y-shaped self-assembled aptasensor, the MGITC labelled gold nanoparticles will be attached to the surface of magnetic beads. In the presence of specific AKI-related biomarkers, the gold nanoparticles will detach from magnetic beads into the supernatant, thus leading to a SERS signal increase, which can be used for the highly sensitive analysis of target biomarkers. In addition, the limit of detection calculated for each biomarker indicates that the SERS-based aptasensor can well meet the detection requirements in clinical applications. Finally, the generality of this sensor in the early diagnosis of AKI is confirmed by using a rat model and spiked plasma samples. This sensing platform provides a facile and general route for sensitive SERS detection of AKI-related biomarkers, which offers great promising utility for in vitro and accurate practical bioassay in AKI early diagnosis.
Introduction
Protein biomarkers, representing the structural, physiological, genetic, or biochemical parameter change, can indicate the presence, severity, or progress of a disease.1–4 Generally, abnormal expression of biomarkers may often be discovered earlier than clinical imaging findings,5,6 showing promising potential in the field of precise diagnosis. Over the past decades, biomarker detection techniques have attracted significant attention and have been widely applied in diagnosing different diseases owing to the unique properties of the biomarkers. For example, various detection methods,7–11 including fluorescence immunoassay, enzyme-linked immunosorbent assay (ELISA), western blot, and surface plasmon resonance, are exploited to detect specific biomarkers directly. However, these traditional techniques still exist some problems in terms of sensitivity, stability, and operational complexity. Moreover, the biomarkers of diseases are usually present in tiny amounts in blood or tissues, especially at the early stage of diseases, which increases the difficulty of accurate detection.12,13 Given this, developing a specific, rapid, and ultrasensitive biomarker detection technique is urgently needed for the early diagnosis of diseases.As an ultrasensitive analytical detection method, the surface-enhanced Raman scattering (SERS) technique can provide fingerprint information of biological substances at the molecular level, quantify the protein concentration and analyse the protein structure, thus realising the accurate results detection of protein biomarkers.14–17 The SERS signal can be highly responsive to the fractal distribution of “hot spots” where the electromagnetic field is powerful due to the unique plasmonic coupling between adjacent nanoparticles.14 Furthermore, SERS-based immunoassay technology utilising functional metal nanoparticles has aroused tremendous interest in the early diagnosis of diseases owing to its excellent sensitivity and multiplex detection ability.18 In our previous work,19,20 we have successfully fabricated SERS probes based on antibody recognition to conduct sensitive detection of several representative protein biomarkers for the accurate diagnosis of prostate cancer and acute myocardial infarction. These findings demonstrate that SERS detection technology for rapid and sensitive disease diagnosis tends to be a research focus via rapid, in situ, and non-destructive testing of biomarkers. Compared to antibodies, aptamers are advantageous in less immunogenicity, repeatable synthesis, low cost, easy modification, structure-switching capability, and long-term stability. Therefore, recent studies are dedicated to constructing SERS-based aptasensors via the strong binding of DNA aptamer and target analytes to realise selective detection of biomarkers from complex mixed samples.
A series of biomarkers such as the neutrophil gelatinase-associated lipocalin (NGAL), interleukin-18, cystatin C (Cys C), N-acetyl-β-D-glycosaminidase, and netrin-1 have been found to be associated with functional kidney diseases.21–23 Regarding acute kidney injury (AKI), it begins by triggering biological and molecular changes and then progresses to cellular damage. This can be early detected by specific biomarkers long before the increase of serum creatinine for diagnosing AKI, especially in elderly and malnourished patients. For example, NGAL, as a type of tubular protein biomarker that is immediately upregulated in response to injury, has been validated by a large number of studies to exhibit prominent early diagnostic performance in diverse clinical situations.24–27 Besides, Cys C, an essential cysteine protease inhibitor in human bodies, has been proposed as a fascinating biomarker of glomerular filtration rate for AKI detection.28,29 Since there are many other physical interference components in the blood or tissues, it is worth noting that a single biomarker always fails to realise the required clinical specificity. Moreover, considering the pivotal role of NGAL and Cys C in the occurrence and progress of AKI, developing SERS-based detection techniques to achieve dual detection of NGAL and Cys C for accurate diagnosis of AKI is of great significance.30
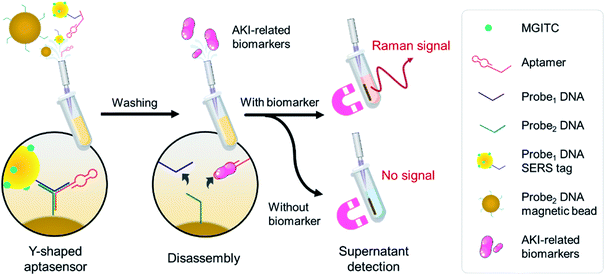
In this work, with the purpose of developing a sensitive and accurate AKI diagnostic technique, we report a unique Y-shaped aptamer-assisted SERS sensing platform based on magnetic separation using a recognition-release mechanism to detect dual biomarkers of AKI. For this, we functionalised Raman reporter-labelled gold nanoparticles with probe DNA.31,32 The SERS tags were then mixed with probe DNA conjugated magnetic beads and selective aptamer for the biomarkers to form a Y-shaped DNA structure. AKI-related biomarkers were detected via SERS-tag released from pre-build Y-shaped structure on magnetic beads to the supernatant.33–35 The SERS scan for supernatant using a Raman spectrometer enabled selective and quantitative detection for dual biomarkers. The proposed SERS-based aptasensor was applied to quantify dual biomarkers in the rat model with high sensitivity and accuracy. From the HE staining results, this novel biosensor could serve as an effective indicator for the early diagnosis of AKI. Besides, the spiked plasma samples were also evaluated to demonstrate the feasibility of a potential clinical application.
Experimental section
Materials
Sodium citrate, gold(III) chloride trihydrate (HAuCl4·3H2O), phosphate-buffered saline (PBS) and human serum albumin (HSA) were purchased from Sigma-Aldrich (USA). Tris(hydroxymethyl)aminomethane (Tris), tris(2-carboxyethyl)phosphine (TCEP) and EDTA-Na were purchased from Shanghai Reagent Company (Shanghai, China). Streptavidin magnetic beads (MBs) were purchased from Zhongke Leiming Technology Company (Beijing, China). Safe Green was provided by Biosharp (Anhui, China). Malachite green isothiocyanate (MGITC) was obtained from Thermo Fisher Scientific (USA). Human recombinant proteins NGAL and Cys C were provided by Cloud-Clone Corp (USA). The aptamer sequence of NGAL or Cys C and other DNA fragments were synthesised by Sangon Biotech (Shanghai, China), among which the DNA sequences were listed in Table S1.† The above purchased analytical reagents were used as received without any treatments. Milli-Q water (18.2 MΩ cm) was used for all the experiments.Synthesis of gold nanoparticles (AuNPs)
As previously reported,36,37 the AuNPs were synthesised using citrate reduction method. 75 mL of 2.2 mM sodium citrate was added to a three-necked flask and heated to boiling. It utilised a condenser to prevent evaporation of the solvent. After this step, 0.5 mL of 25 mM HAuCl4 was added and boiled with stirring. It was found that the colour of the solution first changed from pale yellow to greyish blue and finally to pink within 15 min. The boiling solution was immediately cooled in the same vessel until the temperature reached 90 °C. Next, 0.5 mL of 60 mM of sodium citrate and 0.5 mL of 25 mM of HAuCl4 solution were added sequentially for approximately 2 min. By repeating this process, 12 generations of gold particles of progressively larger size could be obtained. After that, the solution was stirred at 90 °C for another 30 min and cooled to room temperature.Synthesis of DNA functionalised SERS tags
Raman reporters MGITC (1 μL, 10−4 M) were added to 5 mL AuNPs and stirred vigorously at room temperature for 1 h until the MGITC was anchored onto the surface of the AuNPs via Au–S bonds. The centrifugation process (8000 rpm, 25 min) was conducted to remove unbound MGITC. Eventually, prepared MGITC-labelled AuNPs were stored at 4 °C for further use. All DNA probes were pretreated as follows before their conjugation to AuNPs. In short, probe1/probe1′, probe2/probe2′ and aptamer were dispersed in TE buffer (1 M Tris–HCl 5 mL pH 8.0, 0.5 M EDTA 1 mL) and heated to 95 °C for 5 min in water baths. It was cooled to room temperature and stored at 4 °C for further use. Firstly, 100 μL of 10 μM thiolated probe1/probe1′ was incubated with 10 μL of 20 mM TCEP for 60 min to be active at room temperature. Then, 0.5 mL AuNPs were incubated overnight in the mixture at room temperature. 120 μL of PBS (saline) was gradually added to the mix over 24 h. After that, the MGITC labelled AuNPs were washed with Milli-Q water to remove the uncombined DNA. Finally, DNA functionalised SERS tags were obtained and stored in PBS at 4 °C for further use.Construction of Y-shaped aptasensor
The biotinylated probe2/probe2′ was bound to the MBs by the biotin-streptavidin reaction at the 3′ end of probe2/probe2′. First, 100 μL of 10 μM probe2/probe2′ was incubated with 200 μL MBs (300 nm), shaking for 1 h at room temperature. Following this, the functionalised MBs were washed three times with Milli-Q water and dispersed in PBS. Then, probe1/probe1′-functionalised AuNPs were dispersed in the mixture. 100 μL of 10 μM aptamer was added and incubated with the mixture overnight at room temperature to fabricate the aptasensor. The final solution was washed 3 times with Milli-Q water and stored at 4 °C before use.The coupling reactions of the probe2 DNA onto magnetic beads were confirmed using a fluorescent spectrometer. Briefly, the streptavidin-conjugated magnetic beads were divided into two groups. The first group did not undergo any treatment, and the second group was heated to 100 °C for 5 min to inactivate the streptavidin on the surface of the magnetic beads. Next, 100 μL (0.5 mg mL−1) of the above two sets of magnetic beads were incubated with 0, 10, 20, 30, 40, and 50 μM biotinylated probe2 DNA on a rotary shaker at RT. After 1 h, the magnetic beads were collected using a magnet, and the unbound probe2 DNA in the supernatants were discarded by three washing steps using 500 μL TBE buffer. Detection of conjugated probe2 DNA was performed by a microplate reader using one-step safe green nucleic acid dye. The excitation wavelength was selected at 254 nm to obtain the optimal specific fluorescent signals, and the fluorescence intensity at 520 nm was collected for qualitative analysis.
Gel electrophoresis
Briefly, 1 μL of 10 μM NGAL aptamer, 3 μL of 10 μM probe1/probe1′ and 3 μL of 10 μM probe2/probe2′ were added to the hybridisation buffer. Then, their final concentrations were controlled to reach 0.3 μM, 1 μM and 1 μM. Next, the mixture was heated to 95 °C for 10 min, then slowly reduced to 37 °C overnight.Polyacrylamide gel electrophoresis (PAGE) requires the preparation of 20% polyacrylamide. The main components of the reagent were 0.62 mL Milli-Q water, 30% acrylamide 5.3 mL, 4× Tris-SDS 2.0 mL, 10% ammonium persulfate (APS) 80 μL, TEMED 4 μL. Electrophoresis was performed at 80 V for 2 h.
Testing and characterisation
The UV-vis absorption spectra of nanoparticles were measured with a microplate reader (BioTek, USA). The structure and morphology of AuNPs were characterised by an HT7800 transmission electron microscopy (TEM) (HITACHI, Tokyo, Japan) at an accelerating voltage of 120 kV. Dynamic light scattering (DLS) was performed using a Zetasizer Nano (DMNI, Nano Brook).SERS measurement for AKI biomarkers
The Y-shaped aptamer MBs (10 μL) and different concentrations of NGAL or Cys C solutions (5 μL) were mixed for SERS measurements. The Raman scattering signals of the supernatants were measured in the range 600–1700 cm−1 using a Renishaw InVia™ Raman microscope (Renishaw, UK) and a Leica 20× objective (N.A. = 0.4) under the excitation of a 633 nm laser (power 5 mW). SERS signals were generated using single-peak intensities in steps of 10 points, with an integration time of 10 s per point. Raman shift band of the silicon wafer (520 cm−1) was used to calibrate and normalise the obtained spectra. The software package WIRE 5.0 (Renishaw) was used for spectral acquisition and analysis.Collection of animal's blood samples
According to reported,38,39 rats were randomly selected and treated with cisplatin (20 mg kg−1 body weight, i.p. injection) and gentamicin (100 mg kg−1 body weight, i.p. injection). The control group were given saline (0.2 mL) or NAC (400 mg kg−1 body mass, i.p. injection) half an hour before cisplatin treatment. During the treatment, the body weight of rats and the signs of discomfort were monitored daily throughout the experiments. Blood sampling was conducted at different time points after drug treatment. After anaesthesia, 3–5 mL of blood was collected from the medial canthus of the rat. Centrifugation was carried out at 3000 rpm for 15 min, and the supernatant was removed. Blood samples were stored at 4 °C for SERS testing.All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Hainan Medical University and experiments were approved by the Animal Ethics Committee of Hainan Medical University. Male SD rats of 4–6 weeks were housed in a sterile environment and lived in standard rat cages. Rats were provided with rat food and water and were acclimated to the animal facility for seven days prior to experimentation.
Spiked sample detection
First, 10 mL of human plasma was heated into boiling for 5 min to inactivate proteins and then added to the centrifuge tubes. The centrifugation process was carried out at 3000 rpm for 15 min to remove coagulation protein and impurities from the blood sample. NGAL and Cys C were added to total of five blood samples and with final concentrations of 0.1 and 200 ng mL−1. A standard solution of protein NGAL (200 ng mL−1) and Cys C (1 μg mL−1) was added to the blood samples and the concentration was respectively diluted to 35, 45, 25, 40, 45 ng mL−1 and 800, 900, 700, 800, 700 ng mL−1. The above five blood samples (5 μL) and Y-shaped aptasensor (10 μL) were mixed and incubated for one hour at room temperature for SERS measurements.Results and discussion
Mechanism of proposed aptasensor
The MGITC labelled AuNPs were used as the common SERS tags for aptasensor assay of protein biomarkers to improve the detection sensitivity. As illustrated in Fig. 1, MGITC was first introduced to the surface of the constructed AuNPs via Au–S bond. The MGITC-modified AuNPs was then functionalized with a thiolated single-strand DNA (ssDNA) named probe1 to obtain probe1 DNA SERS tag as sensing motif. For the capture part, a biotinylated ssDNA named probe2 was immobilized to the surface of streptavidin magnetic bead via biotin-streptavidin recognition to fabricate probe2 DNA magnetic bead. Part of these two ssDNA (probe1 and probe2) could be complementary via base pairing. Then, thiolated DNA modified AuNPs (probe1), biotinylated DNA functionalised magnetic beads (probe2) and aptamer were then mixed in a centrifuge tube to form a Y-shaped aptamer sensor due to the base complementary pairing of aptamer with both probe1 and probe2. In the presence of the AKI-related biomarkers, the Y-shaped structure would disassemble due to the specific recognition of the aptamer to the targeted protein with the unstable seven bp hybrid chain of probe1 and probe2 released at room temperature. As the concentration of NGAL or Cys C increased, the number of AuNPs on the magnetic beads decreased and more dispersed AuNPs appeared in the supernatant, which resulted in a significant increase in the SERS signal of the supernatant. Quantitative analysis of the AKI biomarkers was then achieved by measuring the calibration curves of different concentrations of biomarkers and corresponding SERS signals.Characterisation of the SERS tags
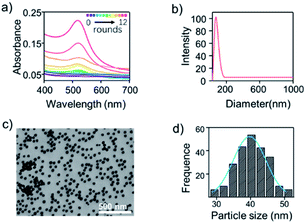
The reproducibility of SERS tags was critical to the performance of the SERS-based analysis assay. AuNPs were easy to synthesise, and the overlapping between the plasmon absorption of AuNPs and the incident laser (633 nm) significantly increased the signal intensity. The synthesis of the Raman-active SERS tag was presented in Fig. 1. Briefly, the AuNPs were prepared by reducing HAuCl4 with sodium citrate under boiling conditions. With 12 rounds of seed growth, the AuNPs had an average diameter of 50 nm determined from the corresponding hydrodynamic diameters (Fig. 2b) and a maximum UV-vis absorption at 520 nm (Fig. 2a). Fig. 2c and S1a† showed the TEM image of AuNPs particles, exhibiting a good dispersibility and morphology at around 40 nm in diameter (Fig. 2d). The green area shown in the EDS diagram (Fig. S1b†) was the distribution of gold elements, and the EDS spectrum (Fig. S1c†) indicated that the nanoparticle mainly contains Au.MGITC was chosen as a Raman reporter molecule because its absorption through the Au–S covalent bonds of AuNPs under the π-conjugated form was stable under various matrix conditions. In addition, SERS quantification was achieved using the characteristic Raman peak (aromatic ring stretching) of MGITC at 1618 cm−1 (I1618). The salt-ageing method was used to attach negatively charged probe1 DNA to negatively charged AuNPs. Where salt was gradually added into the mixture of thiol (SH)-probe1 DNA and AuNPs to reduce charge repulsion. The maximal DLS intensity (Fig. S2†) of AuNPs was about 10 nm increasing after adding MGITC and probe1 DNA with the washing step. These results indicated the successful conjugation of Raman reporter and probe DNA onto AuNPs.
As demonstrated from Fig. S3,† with the increase of biotinylated probe DNA concentration, the fluorescent signals from group 2 are significantly increased. This is because the DNAs are bound to the magnetic sphere by a biotin-streptavidin linkage so that the safe green DNA dye can be attached to the DNA of the streptavidin-coated magnetic beads. On the contrary, due to the inactive streptavidin on the surface of the magnetic beads, the biotinylated probe cannot be conjugated with magnetic beads, resulting in the elimination of fluorescent strength (Fig. S3,† group 1).
Gel electrophoresis characterisation of the Y-shaped DNA assembly
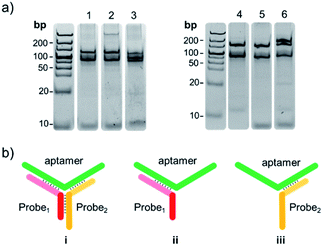
To determine the formation of the Y-shaped aptamer probe, the designed ssDNA sequences in forming DNA assembly was first studied by gel electrophoresis. The designed ssDNA probe1 and probe2 were 14 bp, and the NGAL aptamer and Cys C aptamer were 75 bp and 82 bp, respectively. As shown in lane 1 and lane 2 in Fig. 3a, the probe1/probe2 respectively paired with NGAL aptamer to form a characteristic band of 89 bp (ii and iii) apart from the aptamer band of 75 bp. Notably, when probe1, probe2 and the aptamer existed simultaneously, co-assembly between the two ssDNAs and the Y-shaped DNA unit finally resulted in larger DNA nanostructures. As expected, it presented a new band at the position of higher molecular weight at 103 bp shown in lane 3, demonstrating the formation of the ternary DNA complex. This result indicated that the complementary sequences of the concatemer units (the motif probe1 and probe2) could further hybridise with NGAL aptamer, forming the Y-shaped DNA nano-assembly (Fig. 3b(i)). Similarly, the designed probe1, probe2, and Cys C aptamer exhibited the same binding mode. As shown in lanes 4–6, probe1 or probe2 separately coupled to the Cys C aptamer could only achieve partial hybridisation (Fig. 3b(ii and iii)), while the integration of these three DNA sequences ultimately induced the Y-shaped structure with high stability.Feasibility of the SERS-based aptamer sensor
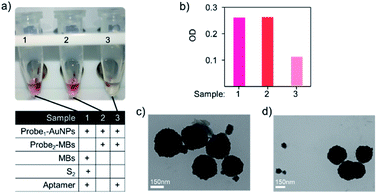
To investigate the feasibility of the aptasensor, we designed three groups of hybridised DNA patterns displayed in Fig. 4a. After activation of the sulfhydryl modified DNA strand probe1 with TCEP, probe1 and probe2 were respectively coupled to the surface of AuNPs@MGITC and streptavidin MBs. Then, specific aptamers were added and co-incubated with AuNPs@MGITC and streptavidin MBs to form the model Y-shaped aptasensor as sample 3. For comparison, we introduced a DNA strand S2 with no complementarily pairing ability with both probe1 and aptamer bases to replace probe2 as sample 1. Besides, we also attempted the DNA pattern of probe1 and probe2 complementary hybridisation without aptamer as sample 2. To evaluate their binding ability, the samples were placed on a magnetic centrifuge for a few seconds, and the streptavidin MBs were aggregated under the magnetic force. The red colour in sample 1 revealed that the free AuNPs@MGITC were dispersed in the supernatant, illustrating that probe2 on the surface of streptavidin MBs was one of the requirements to form a stable nanostructure (Fig. 4a). The same result had also been observed in sample 2, which was mainly attributed to the fragile conjugation of probe1 and probe2 in the absence of aptamer. However, it could be found that the supernatant became colourless in sample 3, revealing that all the AuNPs@MGITC were bound tightly with the MBs (Fig. 4a). This could be explained as the high affinity between the aptamer and its complementary sequences (probe1 and probe2) that were immobilised on the surfaces of AuNPs and MBs, respectively, which induced the assembly to form a Y-shaped structure, thus keeping AuNPs@MGITC and MBs in close proximity to each other.We further characterised the supernatants of these three samples by UV-vis spectroscopy and collected characteristic peak of AuNPs@MGITC at 520 nm. As shown in Fig. 4b, the first two samples exhibited intense absorbance that could reasonably support the invalid binding mode in these two states. As expected, the weak absorption intensity corresponding to negligibly unbound AuNPs@MGITC could further validate the successful generation of stable Y-shaped nanostructure (Fig. 4b). In order to study the biomarker detection behaviour of the constructed Y-shaped aptasensor, we utilised TEM to characterise the binding mode before and after the addition of the targeted biomarker. An apparent dissociation phenomenon could be observed with the existence of the targeted biomarker (Fig. 4d), which showed a sharp contrast of the tightly binding pattern in the aptasensor alone (Fig. 4c), providing direct evidence of the availability of the designed aptasensor for specific biomarker recognition. Therefore, the feasibility of this aptamer-based sensing strategy was verified.
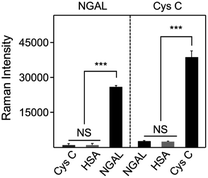
In this aptasensor, determination of AKI biomarker was achieved based on the specific recognition of the corresponding aptamer. There might be other common proteins that coexisted in plasma in actual detection. Thus, the detection specificity of the constructed aptasensor was further evaluated by investigating the cross-reactivity of potential interfering proteins. For the NGAL aptasensor, almost no Raman signals were observed at the characteristic peak of 1618 cm−1 for the introduced interfering components, including HSA and Cys C. In contrast, NGAL led to a dramatic change in Raman signal intensity, revealing satisfactory detecting sensitivity to NGAL in the developed assay (Fig. 5). This could be ascribed to the advantageous structure of the Y-shaped design that endowed the aptasensor with the selective binding ability and high affinity to NGAL biomarkers. Similarly, for Cys C aptasensor, also exhibited outstanding selectivity to Cys C detection with nearly no response to HSA and NGAL.
Quantitative SERS analysis of AKI biomarkers
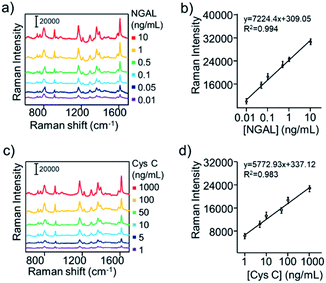
To assess the detection performance of the aptasensor, quantitative analysis of the AKI biomarker was carried out. The featured Raman peak of MGITC (1618 cm−1) was selected for monitoring NGAL and Cys C levels. Fig. 6a showed the typical recording output for testing various concentrations of NGAL utilising the proposed assay. It could be found that the Raman signal intensities gradually rise with the increase of the NGAL concentration in the system (Fig. S5†), representing a sensitive and activable signal response to NGAL. The standard curve based on the increased Raman signal intensity plotted against the NGAL concentration over the range from 0.01 ng mL−1 to 10 ng mL−1 showed a good linear relationship between changed Raman intensity and the NGAL concentration with a correlation coefficient (R2) of 0.994 (Fig. 6b). Furthermore, similar increasing tendencies (Fig. S6†) were also obtained from SERS spectra for the detection of Cys C (Fig. 6c). The SERS intensity as the function of Cys C with the concentration from 1 to 1000 ng mL−1 was plotted in Fig. 6d and portrayed a good linear response (R2 = 0.983). We then analysed the UV spectra of the supernatant with different concentrations of biomarkers and compared them with the SERS results. In Fig. S7,† the absorbance values of UV-vis spectra at 520 nm also increased from different concentrations. It was found that UV-vis spectroscopy could not effectively response at lower biomarker concentrations. In addition, the limit of detection (LOD) of the developed aptasensing platform for NGAL and Cys C were calculated to be 0.052 ng mL−1 and 0.34 ng mL−1, respectively, which could well meet the requirements demanded in clinical applications.Duplex detection and rat plasma matrix application
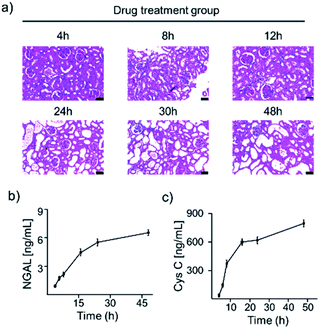
To verify the performance of the duplex assay and its application in rat plasma matrices. We first constructed a rat model of AKI and studied it by observing H&E staining of kidney tissue sections and SERS assays at various time points after drug treatment (Fig. 7a). Above all, H&E staining (Fig. S8†) showed that the brush border and hyaline casting disappear in kidney sections at 24 h after cisplatin treatment. In contrast, the control and blank groups showed normal kidney tissue structure. This demonstrates the success of the AKI rat modelling. As shown in Fig. 7b and c, AKI rats (4, 6, 8, 16, 24 and 48 h) plasma was tested by in vitro SERS assay, incubated with the probe at room temperature, and the supernatant was taken out for UV-vis, SERS testing. In order to quantify the concentration of AuNPs in the supernatant for quantitative analysis of AKI-related biomarkers, the supernatant of rat plasma was taken for SERS testing after incubation with the probe, and the I1618 values were substituted into the equation, the NGAL, Cys C concentrations were then obtained as shown in Fig. 7b and c. The graph showed that the target protein concentration levels in the blood of the rats increased significantly between 4 and 6 h AKI, which allowed SERS to detect changes in target protein concentration levels at an earlier stage than UV-vis and H&E results. This also suggested that Y-shaped aptamer probes could be used for in vitro SERS detection in rats with AKI and that there was a significant response in the early stages of AKI.Spiked samples detection
Due to the well performance of proposed method, we evaluate the performance of the developed SERS-based platform for the duplex detection of NGAL and Cys C in a clinical scenario. We applied the platform to the duplex quantification of different concentrations of each biomarker spiked into the plasma. The concentrations selected within the linear dynamic range of the calibration curve were 0.01–10 ng mL−1 and 1–1000 ng mL−1. Different concentrations of the biomarkers were mixed and added to the plasma. The SERS signal intensity increased as expected with increasing NGAL and Cys C concentrations following duplex detection of spiked samples using the Y-shaped aptasensor. Quantitative analysis of the spiked NGAL and Cys C samples was performed by monitoring the SERS signal intensity at 1618 cm−1 for the corresponding test lines. The linear regression equations derived from the NGAL and Cys C calibration curves were used to calculate the recovered concentrations of biomarkers in the spiked samples based on their SERS signal intensities.Then approximate concentrations of NGAL and Cys C for each sample could be determined by the proposed biosensor based on the established calibration curve. These results were summarised in Table 1. It could be found that the concentration of NGAL and Cys C for AKI clinical scenario samples ranged from 28.93 to 44.01 ng mL−1 and 904 to 1201 ng mL−1. By analysing the concordance between concentrations measured by the two different assays, we found that the concentration of NGAL and Cys determined by the SERS assay was highly correlated with that measured by the ELISA assay. Furthermore, the detection deviation calculated by (CSERS − CELISA)/CELISA × 100% was shown in Table 1. These results demonstrated that the SERS assay was capable of achieving a comparable detection accuracy obtained by the ELISA assay for NGAL and Cys C. Besides, the SERS assay has a more extensive dynamic range, as well as less blood sample requirement (only 5 μL) in comparison with the commonly used ELISA method (100 μL). These exciting performances of aptasensors based on SERS suggested that our approach held the promising potential to be an alternative tool for NGAL and Cys C detection in clinical practice.
| Sample number | Background (ng mL−1) | Spiked (ng mL−1) | SERS assay (ng mL−1) | ELISA (ng mL−1) | Recovery (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| NGAL | Cys C | NGAL | Cys C | NGAL | Cys C | NGAL | Cys C | NGAL | Cys C | |
| 1 | 0.1 | 200 | 35 | 800 | 36.38 | 991 | 36.97 | 1013 | 103.66 | 98.88 |
| 2 | 0.1 | 200 | 45 | 900 | 41.12 | 1201 | 40.58 | 1156 | 91.16 | 111.22 |
| 3 | 0.1 | 200 | 25 | 700 | 28.93 | 904 | 29.53 | 889 | 115.32 | 100.57 |
| 4 | 0.1 | 200 | 40 | 800 | 40.12 | 973 | 40.59 | 1004 | 100.05 | 96.63 |
| 5 | 0.1 | 200 | 45 | 700 | 44.01 | 898 | 43.47 | 916 | 97.58 | 99.71 |
Conclusion
In summary, in this study, we successfully developed a duplex SERS-based aptasensing platform for the specific and sensitive detection of NGAL and Cys C biomarkers for the early and accurate AKI diagnosis. Highly selective SERS nanotags for NGAL and Cys C have been fabricated by functionalising Raman reporter-labelled AuNPs, integrated with MBs and specific aptamers by complementary base pairing to achieve activable biomarker detection using a recognition-release mechanism. The detailed sensing mechanism and feasibility of the designed nanoprobe in biomarker detection have been well validated by gel electrophoresis, DLS, and TEM. By testing the characteristic SERS signal change in 1618 cm−1, the designed aptasensors exhibited excellent selectivity to NGAL and Cys C to detect the supernatant collected upon magnetic separation and showed good quantitative analytical ability with LOD of 0.052 ng mL−1 and 0.34 ng mL−1, respectively. Moreover, this aptasensor also showed good reproducibility and selectivity towards its targets in both rat models and spiked plasma samples. Compared with the traditional ELISA method, the SERS-based aptasensor could realise a comparable detection accuracy with a more extensive dynamic range and less blood sample requirement of only 5 μL. It is believed that the constructed aptasensing platform could be further modified with other specific aptamers and thus easily extended for detecting a wide range of biomarkers to broaden its clinical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Hainan Province Science and Technology Special Fund (ZDYF2021SHFZ245, ZDYF2021SHFZ219 and ZDYF2020133), National Natural Science Foundation of China (22064009 and 22164009), Hainan Provincial Natural Science Foundation of China (820RC641, 820RC655, and 322QN309), the Talent Program of Hainan Medical University (HYPY2020017), Natural Science Research Talent Project of Hainan Medical University (Grant JBGS202101), and Hainan Province Clinical Medical Center (2021).References
- T. Burki, Lancet Oncol., 2017, 18, 511 CrossRef.
- A. Krückel, A. Moreira and W. Fröhlich, BMC Cancer, 2019, 19, 207–213 CrossRef PubMed.
- D. Margel, M. Pesvner-Fischer and J. Baniel, Eur. Urol., 2011, 59, 113–119 CrossRef CAS PubMed.
- T. Notter, J. M. Coughlin and A. Sawa, Mol. Psychiatry, 2018, 23, 36–47 CrossRef CAS PubMed.
- J. K. Schaefer, D. E. Angelini and A. Hawley, Blood, 2016, 128, 3804 CrossRef.
- K. A. Schindlbeck and E. David, Lancet Neurol., 2018, 17, 629–640 CrossRef PubMed.
- Y. Lv, Y. Qin and F. Svec, Biosens. Bioelectron., 2016, 80, 433–441 CrossRef CAS PubMed.
- R. Yu, W. Ma and X. Liu, Theranostics, 2016, 6, 1732–1739 CrossRef CAS PubMed.
- L.-J. Zhao, R.-J. Yu and W. Ma, Theranostics, 2017, 7, 876–883 CrossRef CAS PubMed.
- X. Liu, D. Song and Q. Zhang, Sens. Actuators, B, 2006, 117, 188–195 CrossRef CAS.
- Q. Wu, S. Li and S. Ying, Microchim. Acta, 2017, 184, 1–8 CrossRef.
- V. Jill, V. Raymond and N. Evi, Nephrol., Dial., Transplant., 2013, 28, 254–273 CrossRef PubMed.
- J. Vanmassenhove, G. Glorieux and E. Hoste, Crit. Care, 2013, 17, 1–10 CrossRef PubMed.
- S. Bell, G. Charron and E. Cortés, Angew. Chem., Int. Ed., 2020, 59, 5454–5462 CrossRef CAS PubMed.
- S. Y. Ding, E. M. You and Z. Q. Tian, Chem. Soc. Rev., 2017, 46, 4042–4076 RSC.
- S. Schluecker, Cheminform, 2014, 45, 4756–4795 CrossRef.
- S. Liu, Y. Ying and Y. Long, Chin. Chem. Lett., 2020, 31, 473–475 CrossRef CAS.
- C. Qiu, Z. Cheng, C. Lv, R. Wang and F. Yu, Chin. Chem. Lett., 2021, 32, 2369–2379 CrossRef CAS.
- Z. Cheng, R. Wang and Y. Xing, Analyst, 2019, 144, 6533–6540 RSC.
- R. Gao, Z. Cheng and A. J. Demello, Lab Chip, 2016, 16, 1–26 RSC.
- J. L. Koyner, A. X. Garg and S. G. Coca, J. Am. Soc. Nephrol., 2012, 23, 905–914 CrossRef CAS PubMed.
- E. Kwiatkowska, L. Domański and J. Bober, Acta Biochim. Pol., 2014, 61, 275–280 CrossRef PubMed.
- W. Woyniec, W. Ratkowski and J. Renke, Int. J. Mol. Sci., 2020, 21, 5673 CrossRef PubMed.
- P. Devarajan, Nephrology, 2010, 15, 419–428 CrossRef PubMed.
- J. Mishra, C. Dent and R. Tarabishi, Lancet, 2005, 365, 31–38 CrossRef.
- K. Mori, H. T. Lee and D. Rapoport, J. Clin. Invest., 2005, 115, 610–621 CrossRef CAS PubMed.
- E. Singer, A. Elger and S. Elitok, Kidney Int., 2011, 80, 405–414 CrossRef CAS PubMed.
- S. M. Bagshaw and R. Bellomo, Curr. Opin. Crit. Care, 2010, 16, 533–539 CrossRef PubMed.
- S. Song, M. Marko and T. Türk, Nephrol., Dial., Transplant., 2009, 24, 1157–1161 CrossRef CAS PubMed.
- L. Lei, L. P. Li and Z. Zeng, Sci. Rep., 2018, 8, 7962 CrossRef PubMed.
- H. Kooshki, R. Abbaszadeh and R. Heidari, Anal. Biochem., 2019, 584, 113386–113408 CrossRef CAS PubMed.
- X. Hong, H. Yan and F. Xie, J. Transl. Med., 2019, 17, 204–217 CrossRef PubMed.
- K. Zhang, J. Cao and Y. Wu, Microchim. Acta, 2019, 186, 120 CrossRef PubMed.
- F. Wang, S. Liu and M. Lin, Biosens. Bioelectron., 2015, 68, 475–480 CrossRef CAS PubMed.
- Y. Zhao, X. Yang and H. Li, Chem. Commun., 2015, 51, 16908–16911 RSC.
- Y. Jin and S. Dong, J. Phys. Chem. B, 2003, 107, 12902–12905 CrossRef CAS.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef PubMed.
- B. Florova, D. Rajdl and J. Racek, Physiol. Res., 2020, 69, 307–317 CAS.
- J. Huang, J. Li and L. Yan, Nat. Mater., 2019, 18, 1133–1143 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02813a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |