Open Access Article
Open Access ArticleA novel G·G·T non-conventional intramolecular triplex formed by the double repeat sequence of Chlamydomonas telomeric DNA†
Aparna Bansalab,
Priyanka Phogata and
Shrikant Kukreti *a
*a
aNucleic Acids Research Lab, Department of Chemistry, University of Delhi (North Campus), Delhi, 110007, India. E-mail: skukreti@chemistry.du.ac.in; shrikant.kukreti6@gmail.com
bDepartment of Chemistry, Hansraj College, University of Delhi, (North Campus), Delhi, 110007, India
First published on 26th May 2022
Abstract
Competition among DNA non-canonical structures has been widely studied in repetitive DNA sequences. The factors affecting DNA structural polymorphism have always been an important area of research. The Chlamydomonas reinhardtii telomere (TTTTAGGG)n is found to be an exception to the general idea of forming a folded G-quadruplex by a few repeats in any telomeric sequence. Herein, using gel electrophoresis, UV thermal melting, UV thermal difference spectra (TDS), circular dichroism, and fluorescence and NMR spectroscopy. We demonstrated that the double repeat of the C. reinhardtii telomere (TTTTAGGG)2 (Chlm2) adopts an intramolecular non-conventional triplex structure in Na+ solution. We report that the co-existence of reverse Hoogsteen (G·G) and Wobble base pairing (G·T) stabilizes the triplex structure. To the best of our knowledge, such a non-conventional triplex structure formed by any telomeric sequence has not been reported to date.
Introduction
Telomeres, the protective ends of eukaryotic chromosomes, are ubiquitous and conserved DNA structures. The basic structural unit of most telomeres, a repeated minisatellite motif with the consensus sequence (TnAmGo), may vary between different groups, for example, TTAGGG in vertebrates and fungi, TTTAGGG in plants (Arabidopsis-type) or TTAGG in insects. Algae are a heterogeneous group of organisms broadly defined as photosynthetic autotrophic eukaryotes. Studies on several species of green algae have revealed that this group exhibits at least two types of telomeric sequences, a presumably ancestral type shared with land plants (Arabidopsis-type, TTTAGGG) and conserved in, for example, Ostreococcus and Chlorella, and a novel Chlamydomonas-type (TTTTAGGG), which was identified in Chlamydomonas reinhardtii.1,2 Algal telomeres have not been subjects of any systematic study despite their potential to reveal new features of the telomere maintenance system. It has been found that Chlamydomonas reinhardtii telomere repeats form an interstrand G quadruplex under special conditions such as high ionic strength, high G-strand concentration, and at least 49 hours of incubation, which are unlikely to exist in vivo. Thus, the C. reinhardtii telomere is an exception to the idea that all telomeres composed of 5–8 bp tandem repeats share a common G-quadruplex structure in vivo.3,4Chlamydomonas reinhardtii is a unicellular algae that has proved to be a role model for identifying uncharacterized gene-encoding proteins, which might be associated with the function and biogenesis of chloroplasts or eukaryotic flagella.5 Interestingly, a protein identified as the G-strand binding protein (GBP) in the C. reinhardtii cell specifically binds to single-stranded G-strand telomeric DNA sequences and not to double-stranded telomeric DNA sequences or single-stranded C-strand DNA sequences.6
Our study attempts to fill the considerable gap in the knowledge of C. reinhardtii telomere topology. Using a combination of different techniques such as gel electrophoresis, CD, UV thermal melting, UV thermal difference spectroscopy (TDS), and fluorescence and NMR spectroscopy, the double repeat of the C. reinhardtii telomere d(TTTTAGGG) (Chlm2) was investigated, and it depicted an unusual structure formation that seems to be an exception to the general idea of forming folded G-quadruplexes by a few repeats of any telomeric sequence. Here, we report that Chlm2 generates a non-conventional intramolecular DNA triplex stabilized by G·G and G·T mismatch base pairs.
Materials and methods
The oligonucleotides used were procured from Bio Basic Inc., Canada, in a lyophilized powder form on a 1 μM scale with PAGE purification. They were stored at −20 °C. The concentration of the oligonucleotides was determined spectrophotometrically using the extinction coefficient (ε) calculated by the nearest neighbour method and measuring the absorbance at 260 nm. The ε values used for d-(TTTTAGGG)2 (Chlm2), d-(CCCTAAAA)2 (CChlm2), and d-(CGCGAATTCGCG) (PAL12) were 154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, 159
800, 159![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and 113
000, and 113![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1, respectively. Stock solutions of the oligomers were prepared by directly dissolving the lyophilized powder in MilliQ water. Inorganic salts were purchased from Merck and were used without further purification.
700 M−1 cm−1, respectively. Stock solutions of the oligomers were prepared by directly dissolving the lyophilized powder in MilliQ water. Inorganic salts were purchased from Merck and were used without further purification.
Non-denaturing gel electrophoresis
For the gel experiments, native 20% polyacrylamide gel was used. The samples were heat-treated at 95 °C for 5 min followed by slow cooling to room temperature and incubated overnight at 4 °C before loading into the gel. Both the gel and samples contained 20 mM sodium cacodylate (pH 7.4), 0.1 mM EDTA and NaCl (0.1 M in gel, and 0.1 and 0.5 M in samples), whereas the running buffer contained 1× TBE (pH 7.4) and 0.1 mM EDTA and 0.1 M salt. The gels were run at a constant voltage of 65 V in a cold room. The tracking dye used was orange G mixed with glycerol. After electrophoresis, the gels were stained with Sigma ‘Stains-All’ solution prepared in formamide and finally visualized under white light and photographed using an Alphaimager™ 2200 (Alpha Infotech Corp.)UV-thermal melting
Thermal denaturation experiments (Tm) were carried out on a UV-1650PC Shimadzu UV-VIS spectrophotometer (equipped with a Peltier thermo-programmer TMSPC-8 (E)-200). The samples were prepared and incubated overnight at 4 °C. UV-melting curves were acquired at a wavelength of 295 nm by heating the samples from 0–100 °C for different salt concentrations and oligomer concentrations with a heating rate of 0.5 °C min−1 using stoppered quartz cells with 10 mm and 1 mm path lengths and volume capacities of 110 μL and 35 μL, respectively.Thermal difference spectra
The thermal spectra of Chlm2 were obtained by recording the UV absorbance spectra over a range of temperatures and subsequently taking the difference between each spectrum and that at 20 °C. The spectra were recorded between 220 and 320 nm on a UV-2450PC Shimadzu UV-VIS spectrophotometer using quartz cuvettes with an optical path length of 10 mm.Circular dichroism
For secondary structure analysis, CD spectroscopy was used. The CD spectra were recorded on a JASCO-815 spectrophotometer interfaced with an IBM PC compatible computer and calibrated with D-camphor sulphonic acid. The samples were prepared and incubated overnight at 4 °C. CD spectra were collected as an average of three multiple scans between 210 and 350 nm at a scanning rate of 100 nm min−1 using quartz cells with 10 mm path length and a volume of 1 mL. Data were collected in terms of millidegrees versus wavelength.Fluorescence measurements
Fluorescence measurements were recorded on a CARY eclipse, Varian (USA) spectrofluorometer at an excitation wavelength of 470 nm and emission wavelength of 520 nm. The oligonucleotide under study (Chlm2) was tagged with 6-carboxyfluorescein (FAM) at its 5′ end and a Dabcyl quencher at its 3′ end. FAM-Chlm2-Dabcyl (F-C-D) was dissolved in 20 mM sodium cacodylate (pH 7.4), 0.1 mM EDTA and 0.1 M NaCl.Nuclear magnetic resonance
For the NMR experiment, Chlm2 (TTTTAGGG)2 was commercially procured (synthesized on a 10 μM scale) and supplied as a gel-purified sample. The lyophilized Chlm2 oligonucleotide was dissolved in 90% H2O + 10% D2O solvent containing 20 mM sodium cacodylate (pH 7.4) and 0.1 M NaCl in a total volume of 400 μl. 1H NMR spectra were acquired at 278 K on a Bruker Avance 800 MHz FT NMR spectrometer at the National NMR facility of the Tata Institute of Fundamental Research (TIFR), Mumbai. For information on exchangeable protons, the 1D spectrum was recorded. Typical parameters for the 1D NMR with the pulse program were the number of data points = 16k, spectral width = 20 ppm, number of scans = 512, relaxation delay = 1.0 s, and pulse width = 15 μs.Results and discussion
Non-denaturing gel electrophoresis of (T4AG3)2 (Chlm2)
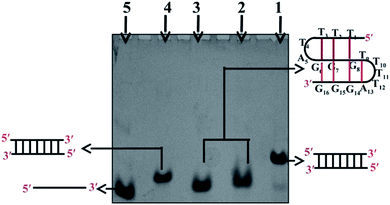
Gel electrophoresis performed at non-denaturing conditions has proven to be a sensitive technique to detect the formation of a DNA structure in terms of its size/molecularity. Fig. 1 shows the electrophoretic mobility pattern of Chlm2 in 20 mM sodium cacodylate (pH 7.4) and 0.1 mM EDTA containing different salt concentrations ranging from 0.1 M to 0.5 M NaCl (lane 2, 3). Gel analysis showed that at each salt concentration used, the oligomer (T4AG3)2 (Chlm2) showed a single band. The other three oligomers namely C3TA4 (CChlm2) (lane 5), (CGCGAATTCGCG) (PAL) (lane 4) and the duplex of T4AG3 (Chlm2) and C3TA4 (CChlm2) (lane 1) were used as size markers to compare the mobility of the structure formed by Chlm2. The bands produced by Chlm2 migrated faster than its duplex and the PAL duplex. The 12 nt long perfect palindromic sequence PAL moves as a 12 mer duplex (24 nt) in the native condition, and the mobility of the duplex of Chlm2 was equivalent to that of a 16 mer duplex (32 nt). The higher mobility of Chlm2, a 16 mer long sequence from PAL and its own duplex, completely ruled out the possibility of its existence as a dimeric structure. Consequently, a unimolecular structure was speculated. Interestingly, the mobility of (T4AG3)2 was found to be retarded when compared with the mobility of its complementary single strand (C3TA4)2. Usually, a structured single strand should migrate faster than an unstructured single strand, but here, the slow mobility of the single-stranded structured form of (T4AG3)2 (Chlm2) might be imitated due to the presence of 8 thymine and 6 guanine nucleotides in the Chlm2 sequence. Our observations agree well with a report that determined that d-T fragments migrate slower than they would be expected to when accounting for their nucleotide number.7 Hence, we considered the presence of an intramolecular structure, as depicted in the gel electrophoretic pattern of (T4AG3)2 (Chlm2). With the double repeat of this telomeric sequence (TTTTAGGGTTTTAGGG), the formation of a unimolecular structure seems difficult to imagine and made us consider whether the Chlm2 double repeat (T4AG3)2 remained as an unstructured single strand under the experimental conditions. It was important to resolve the ambiguity regarding the possibility of a structured or unstructured form of Chlm2. CD has always been a convenient way of following conformational changes in nucleic acids as a function of base sequence and environmental conditions. With this in mind, circular dichroism (CD) was used to determine if any secondary structure was adopted by the Chlm2 sequence.Circular dichroism
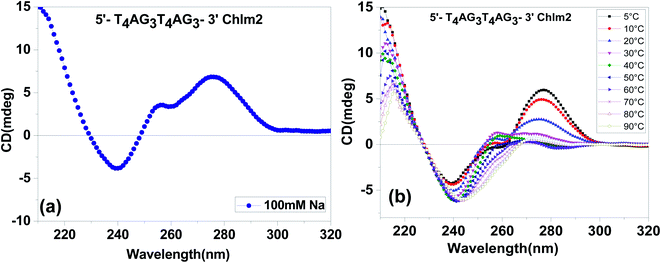
The CD spectra of (T4AG3)2 (Chlm2) in 20 mM sodium cacodylate (pH 7.4), 0.1 mM EDTA and 0.1 M Na+ are shown in Fig. 2(a). The spectra are characterized by two positive peaks at 255 nm and 280 nm, respectively, followed by a negative one at 240 nm. These spectra were found to be almost identical to the CD spectra previously reported for poly [d(G-T)] as a function of salt concentration and temperature by Grey and Ratliff.8 Therefore, it can be determined from the unimolecular species detected in the gel (Fig. 1) and the CD signatures seen in Fig. 2(a) that (T4AG3)2 (Chlm2) adopts a single-stranded structure, possibly stabilized by G·T bonding. The possible existence of G·U(T) base pairs was first introduced by Crick to explain degeneracy in the genetic code.9 Attempts to experimentally demonstrate the formation of wobble base pairs have always been conflicting.10,11 Here, our experimental conditions to demonstrate G·T bonding were supported by the conditions of high salt concentration and low temperature standardized by Grey to show G·T bonding in poly [d(G-T)]. Additionally, it is important to mention that a positive CD band at 255 nm followed by a negative one at 240 nm has been interpreted as CD signatures of the homoduplex formation comprising G·G bonding.12 A careful look at Fig. 2(a) does not rule out the possibility of the presence of G·G bonding in the structure adopted by Chlm2 for the obvious reason that the CD spectra marked the presence of a positive band centered at 255 nm and a negative one at 240 nm. To further substantiate the unimolecular structured form of Chlm2, temperature-dependent CD analysis was carried out (shown in Fig. 2(b)). A decrease in the magnitude of the CD bands with an increase in temperature from 5 °C to 90 °C reflects the disordering of the DNA secondary structure as a result of the destacking of bases. This confirms the existence of a unimolecular structure adopted by (T4AG3)2 (Chlm2).Further, to investigate the presence and stability of G·T and G·G bonding, we performed CD melting experiments at 280 nm and 255 nm, respectively. The CD melting monitored at 280 nm (corresponding to G-T bonding) gave a melting profile with a Tm value of 23 °C (Fig. S1†). Since a positive peak at 255 nm is characteristic of both G·G and G·T bonding and due to the presence of mixed signatures of G·G and G·T bonding at ∼255 nm, a proper melting profile could not be obtained.
However, the possibility of a double repeat sequence of Chlamydomonas forming an intramolecular structure duly formed and stabilized by G·T and G·G non-Watson-Crick hydrogen bonding gives insight towards a better understanding of the stabilization of telomeric ends by alternate DNA structures.
Further, in a separate study, the possibility of the GGT base triplet has been demonstrated by Mishima et al., where they proposed that oligonucleotides consisting of d(GAA) repeats associate intramolecularly, raising the possibility of the formation of a triplex with the third purine strand parallel to the purine strand of the Watson Crick duplex containing d(GGA:TCC) repeats. The triplex between d(GGA:TCC) repeats and d(GGA) repeats involves GGT base triplets as well as GGC and GAT triplets.13
Further, to validate the possibility of the formation of such intramolecular structures by Chlm2, UV thermal difference spectra (TDS) were recorded under the same solution conditions, and were found to exhibit inverted transition at 295 nm (Fig. S2†), displaying the characteristic of non-canonical structures such as Z-DNA, G-quadruplex, i-motif, Hoogsteen duplexes, and pyrimidine triplex.14 Thus, the TDS spectra support our hypothesis of an intramolecular non-canonical structure by a double repeat in the C. reinhardtii telomere.
UV thermal melting
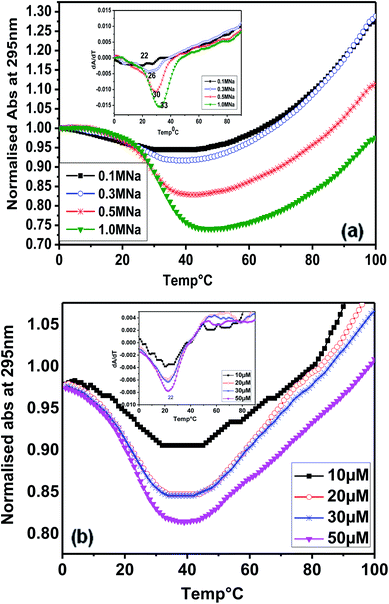
Considering the presence of contiguous stretches of guanines (G) interrupted by intervening bases (T and A), such as in other telomeric sequences, we subsequently studied the thermal denaturation of (T4AG3)2 (Chlm2) monitored at 295 nm. Fig. 3(a) shows the melting profile of Chlm2 monitored at 295 nm, in 20 mM sodium cacodylate buffer (pH 7.4), 0.1 mM EDTA containing 0.1–1 M NaCl. The Tm values of 22–33 °C were calculated from the first derivative (inset) of the melting profile. Even at 1 M salt concentration, the low stability of Chlm2 was credited to G·T mispairing. It has been reported that self-associated forms of the G·T base pair are less thermally stable than the standard base pair.8 Interestingly, the melting of Chlm2 monitored at 295 nm exhibited a nice, inverted sigmoidal curve, evident of non-canonical oligomeric structures. To further confirm the unimolecular structural status of Chlm2, the oligomer concentration-dependent UV-thermal spectra were recorded at 295 nm (Fig. 3(b)). The thermal melting spectra were monitored at 295 nm in 20 mM sodium cacodylate buffer (pH 7.4) and 0.1 mM EDTA containing 0.1 M NaCl. At different oligonucleotide strand concentrations ranging from 10 to 50 μM, the curves consistently revealed monophasic profiles, affording a constant Tm value of 22 °C. The concentration independence of the Tm value thus obtained supported the unimolecular (intramolecular) structure adopted by Chlm2.15Fluorescence spectroscopy
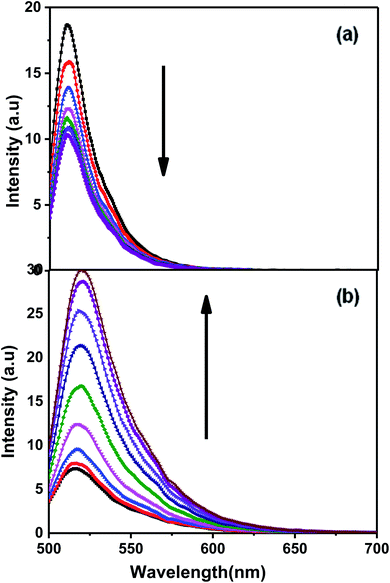
Fluorescence experiments were carried out for Chlm2, tagged with 6-carboxyfluorescein (FAM) at its 5′ end and a Dabcyl quencher at its 3′ end, termed as F-C-D. Fig. 4(a) shows the emission spectra of F-C-D at 50 nM concentration in 20 mM sodium cacodylate buffer (pH 7.4), 0.1 mM EDTA and varying concentrations of NaCl. The emission spectra were recorded from 500 nm to 700 nm, with the excitation wavelength set to 470 nm. It was observed that on increasing the concentration of Na+ (0.1–1 M), the fluorescence intensity decreased at 520 nm (Fig. 4(a)). It is known that within the fluorophore and quencher pairs, the energy absorbed by the fluorophore is transferred to the quencher and dissipated as heat instead of being emitted as light.16 The decrease in the fluorescence intensity at 520 nm indicates the proximity between the donor and the acceptor molecules and thus greater energy transfer from donor to acceptor. This observation supports our finding of a triple helical structure formed by Chlm2. Additionally, the evidence to further support this idea was obtained when the target sequence was hybridized with its complementary C-rich strand (CChlm). Fig. 4(b) demonstrates the fluorescence spectra of F-C-D with successive addition of its C-rich counterpart. The increase in fluorescence intensity with increasing concentration of the complementary strand (0–16 nM) is due to an increase in the distance between donor and acceptor molecule and thus lower energy transfer, resulting in high fluorescence intensity in the donor region. This observation proves the unfolding of an intramolecular structure into a linear one (duplex) on hybridizing with the complementary strand.17,18NMR spectral assignments of Chlm2
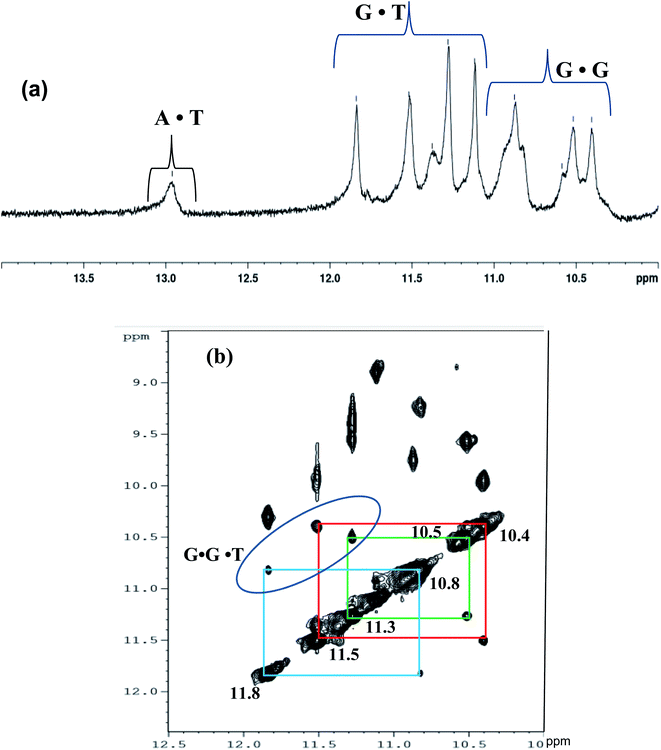
To further authenticate the proposed intramolecular triple helical structure formed by Chlm2, the NMR technique was used. Hydrogen-bonded imino proton resonances are known to scatter over an appreciable spectral region, i.e., from 12.5 to 16 ppm. The imino proton resonances of standard Watson–Crick A·T base pairs are found to be between 13.0 and 14.5 ppm, while those of G·C base pairs occur between 11.5 and 13.5 ppm. Imino protons involved in the G-quadruplex Hoogsteen G·G base pairing are observed between 10 to 12 ppm,19 while a set of exchangeable imino protons from the mismatched sites (GG and TT) are detectable in the range of 10.2 ± 10.7 ppm.20 The sharp signals at 10.85 and 11.85 ppm can be attributed to mismatched G·T base pairs.21 Based on the above-discussed reports, the 1D NMR spectrum of Chlm2 was recorded, as shown in Fig. 5(a). For better resolution, the imino region of the 1D proton spectra was acquired from 10 to 14 ppm. The NMR signals that appeared can be grouped into three regions, one showing peaks between 10.4 to 10.8 ppm, another between 11.3 to 11.8 ppm, and the most downfield region showing peaks at 13 ppm.The downfield signal at 13 ppm is attributed to the imino proton resonance arising from Watson–Crick A·T base pairs,19 while the peaks with chemical shifts between 11.3 to 11.8 ppm correspond to G·T wobble base pairs.21 The resonances of imino protons at 10.4 to 10.8 ppm were assigned to G·G mispairing.20 Thus, our proposal of the formation of a non-conventional intramolecular triple helical structure comprising G·G·T base triplets by Chlm2 predicted from different techniques discussed so far was also validated by the 1D NMR of the exchangeable proton region.
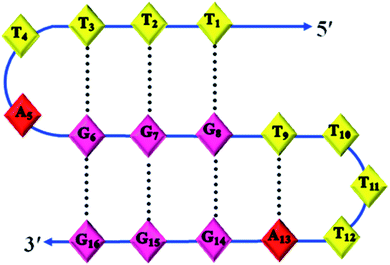
Structural determination by NMR requires identification and quantitative analysis of cross-peaks in the 2D NOESY spectra. It is a more powerful and more efficient method for studying selective NOEs between neighbouring protons in the spatial structure of biological macromolecules. A careful analysis of the cross-peaks in the NOESY spectrum (Fig. 5(b)) revealed the selective NOEs between neighbouring protons in the 10.4/11.5 ppm, 10.5/11.3 ppm and 10.8/11.8 ppm regions. Based on the reports available and the peaks obtained resonating in the imino proton region in the 1D spectrum (Fig. 5(a)), we tried to correlate the cross-peaks of our NOESY spectrum with our hypothesis of the triple helical structure stabilised by G·G and G·T bonding. As shown in Fig. 5(b), the imino proton resonances are at 10.4/11.5 ppm, 10.5/11.3 ppm and 10.8/11.8 ppm in the cross-peak region of the NOESY spectrum and thus present the connectivity between the G·G and G·T base pair regions. Therefore, based on the cross-peak region identified in Fig. 5(b), the resonances at 10.4, 10.5 and 10.8 ppm shown in Fig. 5(a) are assigned to G·G base pairs. Similarly, the peaks with the chemical shifts at 11.5, 11.3 and 11.8 ppm are attributed to G·T base pairs. Hence, the cross-peak regions identified at 10.4/11.5 ppm, 10.5/11.3 ppm and 10.8/11.8 ppm are attributed to three G·G·T triads, which are well in agreement with our proposed structure of the triple helix of Chlm2, as shown in Fig. 6 along with its possible hydrogen bonding pattern.
Conclusions
To summarise, using a combination of gel electrophoresis, circular dichroism, optical melting, fluorescence, and NMR studies, we herein demonstrate that double repeats of the C. reinhardtii telomere (TTTTAGGG)2 (Chlm2) in the presence of Na+ ions, adopt an intramolecular triplex structure stabilized by G·G and G·T mismatch base pairs. We report that the formation of this novel non-conventional intramolecular triple helical structure is an exception to the general idea of forming folded G-quadruplexes by few repeats in any telomeric sequence. To the best of our knowledge, triple helical structures formed by any telomeric repeats have not been characterized till date. We understand that even if we carry sound data and interpretation, we know that there is always scope for more advanced techniques and studies to fully confirm the proposed non-conventional triplex structure of the Chlm2 sequence.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
The authors acknowledge financial support from the DST grant (NO. SP/SO/D-21/2001). AB thanks the University Grant Commission for the JRF and SRF awards.References
- E. J. Richards and F. M. Ausubel, Isolation of a higher eukaryotic telomere from Arabidopsis thaliana, Cell, 1988, 53, 127–136 CrossRef CAS PubMed.
- J. Fulneckova, T. Hasikova, J. Fajkus, A. Lukesova, M. Elias and E. Sykorova, Dynamic evolution of telomeric sequences in the green algal order chlamydomonadales, Genome Biol. Evol., 2012, 4(3), 248–264 CrossRef PubMed.
- M. E. Petracek and J. Berman, Chlamydomonas reinhardtii telomere repeats form unstable structures involving guanine-guanine base pairs, Nucleic Acids Res., 1992, 20, 89–95 CrossRef CAS PubMed.
- T. Hails, O. Huttner and A. Day, Isolation of a chlamydomonas reinhardtii telomere by functional complementation in yeast, Curr. Genet., 1995, 28, 437–440 CrossRef CAS PubMed.
- S. S. Merchant, S. E. Prochnik, O. Vallon and E. H. Harris, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions, Science, 2007, 318, 245–250 CrossRef CAS PubMed.
- M. E. Petracek, L. M. C. Konkel, M. L. Kable and J. A. Berman, A Chlamydomonas protein that binds single-stranded G-strand telomere DNA, EMBO J., 1994, 13(15), 3648–3658 CrossRef CAS PubMed.
- I. Kejnovska, J. Kypr and M. Vorlickova, Oligo (dT) is not a correct native PAGE marker for single stranded DNA, Biochem. Biophys. Res. Commun., 2007, 353, 776–779 CrossRef CAS PubMed.
- D. M. Gray and R. L. Ratliff, Circular dichroism evidence for G.U and G.T base pairing in Poly [r(G-U)] and Poly[d(G-T)], Biopolymers, 1977, 16, 1331–1342 CrossRef CAS PubMed.
- F. H. C. Crick, Codon – anticodon pairing: The Wobble hypothesis, J. Mol. Biol., 1966, 19, 548–555 CrossRef CAS PubMed.
- S. I. Chan, G. C. Y. Lee, C. F. Schmidt and G. P. Kreishman, Guanine-uracil base pairing, Biochem. Biophys. Res. Commun., 1972, 46, 1536–1543 CrossRef CAS PubMed.
- D. M. Gray, I. Tinoco Jr and M. J. Chamberlin, The circular dichroism of synthetic ribonucleic acids and the influence of uracil on conformation, Biopolymers, 1972, 11, 1235–1258 CrossRef CAS PubMed.
- H. Porumb, M. Mannot and S. Fermandjian, Circular dichroism signatures of features simulataneously present in structured guanine-rich oligonucleotide: a combined spectroscopic and electrophoretic approach, Electrophoresis, 2002, 23, 1013–1020 CrossRef CAS PubMed.
- Y. Mishima, T. Suda and R. Kominami, Formation of a Triple- Stranded DNA between d(GGA: TCC) Repeats and d(GGA) repeat oligonucleotides, J. Biochem, 1996, 119, 805–810 CrossRef CAS PubMed.
- J. L. Mergny, J. Li, L. Lacroix, S. Amrane and J. B. Chaires, Thermal difference spectra: a specific signature for nucleic acid structures, Nucleic Acids Res., 2005, 33, e138 CrossRef PubMed.
- L. A. Marky, K. S. Blumenfeld, S. Kozlowski and K. J. Breslauer, Salt-dependent conformational transitions in the self complimentary deoxydodecanucleotide d(CGAATTCGCG): evidence for hairpin formation, Biopolymers, 1983, 22, 1247–1257 CrossRef CAS PubMed.
- S. Tyagi and F. R. Kramer, Molecular Beacons: probes that fluoresce upon hybridisation, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS PubMed.
- N. Kumar and S. Maiti, The effect of osmolytes and small molecule on Quadruplex-WC duplex equilibrium: a fluorescence resonance energy transfer study, Nucleic Acids Res., 2005, 33, 6723–6732 CrossRef CAS PubMed.
- N. Ota, K. I. Hirano, M. Warashina, A. Andrus, B. Mullah, K. Hatanaka and K. Taira, Determination of interactions between structured nucleic acids by fluorescence resonance energy transfer (FRET): selection of target sites for functional nucleic acids, Nucleic Acids Res., 1998, 26, 735–743 CrossRef CAS PubMed.
- M. Hsin-Jui Kuo, Z. F. Wang, T. Y. Tseng, M. H. Li, S. T. D. Hsu, J. J. Lin and T. C. Chang, Conformational transition of a hairpin structure to G-quadruplex within the WNT1 gene promoter, J. Am. Chem. Soc., 2015, 137, 210–218 CrossRef PubMed.
- K. P. Bhattacharya, J. Cha and J. K. Barton, 1H NMR determination of base- pair lifetimes in oligonucleotides containing single base mismatches, Nucleic Acids Res., 2002, 30, 4740–4750 CrossRef PubMed.
- N. Escaja, J. Viladoms, M. Garavis, A. Villasante, E. Pedroso and C. Gonzalez, A minimal i- motif stabilised by minor groove G:T:G:T tetrads, Nucleic Acids Res., 2012, 40, 11737–11747 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra00861k |
| This journal is © The Royal Society of Chemistry 2022 |

![[pentagon open]](https://www.rsc.org/images/entities/char_e1c1.gif) —) 90 °C.
—) 90 °C.