Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Diarylheptanoid analogues from the rhizomes of Zingiber officinale and their anti-tumour activity†
Ting Li‡
 a,
Da-bo Pan‡
a,
Da-bo Pan‡ ab,
Qian-qian Pang
ab,
Qian-qian Pang a,
Mi Zhou
a,
Mi Zhou a,
Xiao-jun Yao
a,
Xiao-jun Yao d,
Xin-sheng Yao
d,
Xin-sheng Yao a,
Hai-bo Li
a,
Hai-bo Li *c and
Yang Yu
*c and
Yang Yu *a
*a
aInstitute of Traditional Chinese Medicine & Natural Products, Guangdong Province Key Laboratory of Pharmacodynamic Constituents of TCM and New Drugs Research, College of Pharmacy, Jinan University, Guangzhou 510632, P. R. China. E-mail: 1018yuyang@163.com
bDepartment of Medical Technology, Qiandongnan Vocational & Technical College for Nationalities, Kaili, Guizhou 556000, P. R. China
cKanion Pharamaceutical Co. Ltd, State Key Laboratory of New-tech for Chinese Medicine Pharamaceutical Process, Lianyungang 222001, People's Republic of China. E-mail: lihaibo1985124@sina.com
dState Key Laboratory of Applied Organic Chemistry, Department of Chemistry, Lanzhou University, Lanzhou 730000, P. R. China
First published on 2nd September 2021
Abstract
Eight previously undescribed diarylheptanoids (1–8), together with fifteen known analogues (9–23), were isolated from the rhizomes of Zingiber officinale. Their structures were unambiguously determined by comprehensive spectroscopic analyses and electronic circular dichroism (ECD) calculations. It is worth mentioning that 1–3 are the first reported structures of diaryl ether heptanoids in Z. officinale, whereas 15–17 were isolated from Zingiber for the first time. Furthermore, a gene enrichment analysis of the interacting targets indicated that diarylheptanoids were mainly associated with the anti-tumor activity by affecting DNA damage signaling pathway. The results showed that 6, 16–19 had remarkable inhibitory effects against five tumor cell lines (A549, HepG2, HeLa, MDA-MB-231, and HCT116) with IC50 values ranging from 6.69–33.46 μM, and showed down-regulating the expression of ATR (ataxia telangiectasia mutated and RAD3-related) and CHK1 (checkpoint kinase 1) levels in HCT116 and A549 cell lines. Our studies not only enrich the structural diversity of diarylheptanoids in nature, but also discover several natural compounds for anti-tumor agents.
1. Introduction
Ginger (Zingiber officinale Roscoe; family: Zingiberaceae) has long been used worldwide as an important cooking spice, condiment, and dietary supplement.1,2 It is also valued as a medicinal herb for its prevention and treatment of a wide range of diseases, such as colds, stomachaches, headaches, nausea, diarrhoea, indigestion, rheumatic, cardiopathy and cholera.3–7 Gingerol-related compounds as well as diarylheptanoids are the major classes of biologically active natural products in ginger. In the past decades, the extensive investigations have been made on ginger and gingerols derivatives.8,9 Diarylheptanoids, known as the other significant phenolic components in ginger, have recently attracted widespread attentions for their structural diversity and potential pharmacological effects.Diarylheptanoids are structurally diverse and have the skeletal structure of two aromatic rings conjugated with seven carbon chains, and is categorized into linear, cyclic, and dimeric diarylheptanoids, or diarylheptanoids with special moieties. Numerous pharmaceutical research revealed that diarylheptanoids possess anti-inflammatory,10 anti-oxidant,11 anti-tumor,12 leishmanicidal,13 melanogenesis,14 hepatoprotective,15 and neuroprotective activities.16 A variety of diarylheptanoids have been isolated from seeds, fruits, leaves and roots of plants of different families.17 Ginger is one of the most important producers of diarylheptanoids in nature. In our previous study, gingerol derivatives including six new compounds were reported from ginger rhizomes.18 As a part of a continuing search for new bioactive agents,21 diarylheptanoids including eight undescribed ones (1–8), were identified from the ginger. To rapidly reveal the biological activity, a network pharmacology-based approach was constructed to guide the discovery of targets and biological functions of diarylheptanoid compounds. The results demonstrated that the isolated diarylheptanoids were mainly associated with the antitumor activity by affecting DNA damage signalling pathway. Consequently, cytotoxicity and enzyme activity assays were designed to validate the predictions, suggesting that compounds 6, 17, and 18 may exert anticancer effects by regulating the ATR/CHK1 signalling pathway. Herein, the isolation, structural elucidation, targets prediction, cytotoxic evaluation, enzymatic activity assays, and molecular simulation of these compounds are discussed.
2. Materials and methods
2.1. Instrumentation and reagents
Ultraviolet spectra (UV) were recorded on a JASCO V-550 UV spectrometer (JASCO, Tokyo, Japan). Optical rotations were determined in CHCl3 using a JASCO P-1020 polarimeter (JASCO, Tokyo, Japan). Circular dichroism (CD) spectra were tested by JASCO J-810 circular dichroism spectrometer (Jasco, Tokyo, Japan). Infrared spectra (IR) were measured with a JASCO FT/IR-480 plus spectrometer (JASCO, Tokyo, Japan). 1D and 2D NMR spectra were acquired on a Bruker AV 600 (Bruker Co. Ltd, Bremen, German) using solvent signals (CDCl3: δH 7.26/δC 77.2; CD3OD: δH 3.31/δC 49.0) as internal references. Deuterated solvents were purchased from Cambridge Isotope Laboratories, Inc. (Saint Louis, Missouri, USA). The HR-ESI-MS spectra were obtained on a Micromass Q-TOF mass spectrometer (Waters Corporation, Milford, USA). Analytical high-performance liquid chromatography (HPLC) was carried out on a Shimadzu HPLC system (Shimadzu, Kyoto, Japan) with an LC-20AB solvent delivery system and an SPD-20A UV/vis detector using a Phenomenex Gemini C18 column (5 μm, Φ 4.6 × 250 mm; Phenomenex Inc., Los Angeles, USA). Semi-preparative HPLC was performed on a Shimadzu HPLC system (Shimadzu, Kyoto, Japan) equipped with LC-6AD solvent delivery system and a SPD-20A detector on a Phenomenex Gemini C18 column (5 μm, Φ 10.0 × 250 mm; Phenomenex Inc., Los Angeles, USA).Column chromatography (CC) was carried out on silica gel (300–400 mesh, Haiyang Chemical Co. Ltd, Qingdao, China), ODS (12 nm, S-50 μm, YMC Ltd, Tokyo, Japan) and Sephadex LH-20 (Amersham Pharmacia Biotech Co. Ltd, Atlanta, USA). Thin-layer chromatography (TLC) was performed on pre-coated silica gel plates (GF254, Qingdao Haiyang Co. Ltd, Qingdao, China). HPLC-grade methanol and acetonitrile (CH3CN) were purchased from Oceanpak Alexative Chemicals Co. Ltd. (Gothenburg, Sweden). All analytical grade reagents were from Concord Chemicals Co. Ltd (Tianjin, China).
2.2. Plant materials
Dried rhizomes of Z. officinale were collected from Jinxiang County, Shandong, China, in September 2016. The plant material was identified by Prof. Guang-Xiong Zhou of Jinan University. A voucher specimen (JNU-ZO-201609) was deposited at the Institute of Traditional Chinese Medicine and Natural Products, College of Pharmacy, Jinan University, Guangzhou, China.2.3. Extraction and isolation
The dried rhizomes (30 kg) of Z. officinale were extracted with 70% ethanol (210 L) under reflux for 2 times at 60 °C, 2 h each time. The combined ethanol extract was evaporated under reduced pressure. The ethanol extract (5.5 kg) was suspended in 95% ethanol (16.5 L) and partitioned successively with petroleum ether and ethyl acetate. The ethyl acetate portion (ZO-2, 920 g) was divided into 22 fractions (Fr. A to Fr. V) on a silica gel column chromatograph (CC) (200–300 mesh, Φ 6.5 × 130 cm) by gradient elution with solvent composed of cyclohexane–EtOAc (v/v, 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 95
3, 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 9
5, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7
2, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 6
3, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and 0
4, and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
Fr. O (22 g) was eluted (CH3OH–H2O, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 → 10
5 → 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) by ODS to yield 11 fractions (Fr. O1–O11). Fr. O7 was separated by silica gel CC (CHCl3–acetone 95
0) by ODS to yield 11 fractions (Fr. O1–O11). Fr. O7 was separated by silica gel CC (CHCl3–acetone 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) to give 4 fractions (Fr. O7A–O7D). Compounds 1 (1.4 mg, tR = 35.7 min), and 2 (1.0 mg, tR = 36.5 min) were obtained from Fr. O7A by semi-preparative HPLC (45% CH3OH). Subsequently, Fr. O9 was purified by preparative HPLC to yield compound 4 (14.1 mg, tR = 84 min, 53% CH3OH).
5, v/v) to give 4 fractions (Fr. O7A–O7D). Compounds 1 (1.4 mg, tR = 35.7 min), and 2 (1.0 mg, tR = 36.5 min) were obtained from Fr. O7A by semi-preparative HPLC (45% CH3OH). Subsequently, Fr. O9 was purified by preparative HPLC to yield compound 4 (14.1 mg, tR = 84 min, 53% CH3OH).
Fr. Q (16.0 g) was subjected to ODS eluting with CH3OH–H2O (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 → 10
5 → 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to afford Fr. Q1–Q17. Among them, compound 7 (2.9 mg, tR = 37.5 min, 58% CH3CN) was purified by semi-preparative HPLC from Fr. Q10. Fr. Q7 was separated by semi-preparative HPLC (29% CH3CN) to yield 9 fractions (Fr. Q7A–Q7I), Fr. Q7I and Q7M were further treated by semi-preparative HPLC to obtain compounds 5 (1.3 mg, tR = 18.2 min, 50–60% CH3OH, 40 min) and 6 (3.0 mg, tR = 19.5 min, 50–70% CH3OH, 40 min), respectively.
0) to afford Fr. Q1–Q17. Among them, compound 7 (2.9 mg, tR = 37.5 min, 58% CH3CN) was purified by semi-preparative HPLC from Fr. Q10. Fr. Q7 was separated by semi-preparative HPLC (29% CH3CN) to yield 9 fractions (Fr. Q7A–Q7I), Fr. Q7I and Q7M were further treated by semi-preparative HPLC to obtain compounds 5 (1.3 mg, tR = 18.2 min, 50–60% CH3OH, 40 min) and 6 (3.0 mg, tR = 19.5 min, 50–70% CH3OH, 40 min), respectively.
Fr. R (29.0 g) was applied to a ODS column using CH3OH–H2O (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 → 10
5 → 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) as elute to obtain 11 fractions (R1–R11). Then, Fr. R4 was treated by semi-preparative HPLC eluting with 23% CH3CN to obtain a mixture (R4G), and further purified by semi-preparative HPLC (50% CH3OH) to yield compound 3 (4.8 mg, tR = 11.9 min). Fr. R9 was treated by semi-preparative HPLC eluting with 42% CH3CN, and compound 8 (11.6 mg, tR = 25.6 min) was obtained. In addition, detailed isolation procedures for known compounds (9–23) and spectroscopic data of isolated new compounds are provided in the ESI.†
0) as elute to obtain 11 fractions (R1–R11). Then, Fr. R4 was treated by semi-preparative HPLC eluting with 23% CH3CN to obtain a mixture (R4G), and further purified by semi-preparative HPLC (50% CH3OH) to yield compound 3 (4.8 mg, tR = 11.9 min). Fr. R9 was treated by semi-preparative HPLC eluting with 42% CH3CN, and compound 8 (11.6 mg, tR = 25.6 min) was obtained. In addition, detailed isolation procedures for known compounds (9–23) and spectroscopic data of isolated new compounds are provided in the ESI.†
2.4. ECD calculation
The ECD spectra of (5R)-3 and (5S)-3 were calculated using Gaussian 09 software. Firstly, predominant conformations were obtained using conformation searches. Secondly, all conformations were optimized at B3LYP/6-31G(d) level. Thirdly, the B3LYP/6-311+G(d,p) level was used to calculate the compound's ECD spectra. Finally, The ECD spectra were combined after Boltzmann weighting based on the distribution of structural energy.2.5. Compound-target network construction and analysis
The targets of the isolated compounds were predicted by using SwissTargetPrediction,19 and the cancer-related targets were extracted from GenCLip3.20 Next, these targets were merged to obtain the overlapped targets, and the compounds-antitumor targets network was built by using the overlapped targets and their corresponding compounds. The key anticancer targets were identified based on the intermediate value of degree. Furthermore, the potential biological activity of these isolated-compounds were predicted by using ClueGO program in Cytoscape 3.5 software.212.6. Assays for cell proliferation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C for 15 min. Protein concentration of supernatant was determined. Equal amounts of lysate protein were separated by SDS–polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% nonfat skim milk in TBST [20 mM Tris–HCl (pH 7.6), 135 mM NaCl and 0.1% Tween 20], and then incubated with specific primary antibodies to ATM, ATR, and P53 (Cell Signalling Technology, Danvers, MA) at 4 °C overnight. β-Actin was used as a loading control. The next day, the membrane was incubated with goat anti-mouse IgG–HRP secondary antibody (Cell Signalling Technology, Danvers, MA) at room temperature for 2 h. The signals were detected by chemiluminescence utilizing the enhanced chemiluminescent reagent and recorded on the gel imaging system (Tanon, Shanghai, China) and relative quantified using Image J software program. The experiment was repeated three times, and A549 cells were tested in the same way. The test results of A549 cell line are provided in the ESI (Fig. S72†).
000g and 4 °C for 15 min. Protein concentration of supernatant was determined. Equal amounts of lysate protein were separated by SDS–polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% nonfat skim milk in TBST [20 mM Tris–HCl (pH 7.6), 135 mM NaCl and 0.1% Tween 20], and then incubated with specific primary antibodies to ATM, ATR, and P53 (Cell Signalling Technology, Danvers, MA) at 4 °C overnight. β-Actin was used as a loading control. The next day, the membrane was incubated with goat anti-mouse IgG–HRP secondary antibody (Cell Signalling Technology, Danvers, MA) at room temperature for 2 h. The signals were detected by chemiluminescence utilizing the enhanced chemiluminescent reagent and recorded on the gel imaging system (Tanon, Shanghai, China) and relative quantified using Image J software program. The experiment was repeated three times, and A549 cells were tested in the same way. The test results of A549 cell line are provided in the ESI (Fig. S72†).3. Results and discussion
3.1. Structural elucidation of new compounds
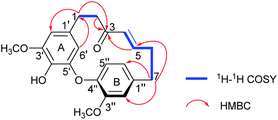
Compound 1 was isolated as a yellow oil. The molecular formula was determined to be C21H22O5 on the basis of the HR-ESI-MS (m/z 355.1539 [M + H]+, calcd 355.1545). The 1H NMR spectrum showed diagnostic signals of one 1,2,4-trisubstituted benzene [δH 6.92 (1H, d, J = 8.0 Hz, H-5′′), 6.80 (1H, d, J = 1.8 Hz, H-2′′), and 6.73 (1H, dd, J = 8.0, 1.8 Hz, H-6′′)] and one 1,2,3,5-tetrasubstituted benzene [δH 6.38 (1H, d, J = 1.8 Hz, H-2′), and 4.99 (1H, d, J = 1.8 Hz, H-6′)], a trans-substituted double bond [δH 6.58 (1H, ddd, J = 15.2, 8.6, 7.5 Hz, H-5), 5.86 (1H, d, J = 15.2 Hz, H-4)], and two methoxyls [δH 3.82 (3H, s, 3′-OCH3), 3.68 (3H, s, 3′′-OCH3)]. The 13C NMR and DEPT data revealed the presence of 21 carbon signals (Table 1), including eight quaternary carbons (δC 201.1, 154.0, 149.9, 149.6, 144.1, 140.8, 134.1, 132.6), seven methines (δC 147.9, 131.8, 125.1, 122.5, 115.8, 106.5, 106.3), four methylenes, and two methoxy carbon signals. The above NMR data supported 1 to be a cyclic diarylheptanoid.22 The structural elucidation of 1 was accomplished by analysis of COSY, HSQC and HMBC data (Fig. 2). In particular, the 1H–1H COSY correlations of H2-1/H2-2 and H-4/H-5/H2-6/H2-7, combined with the HMBC correlations of H2-1, H2-2/C-3, H-4, H-5/C-3, and H2-7/C-5 revealed the presence of a heptane chain moiety. Further, two meta-coupling doublets, associated with chemical shift characteristics, and key HMBC peaks allowed the assignments of aromatic ring A. In aromatic B, HMBC correlations from a broad ortho doublet H-5′′ to C-1′′, 3′′, 4′′, from a narrow meta doublet H-2′′ to C-3′′, 4′′, 1′′, 6′′, and from a doublet of doublets to C-1′′, 2′′, 4′′, completed its assignments of the signals. The linkage of alkyl and aromatic groups was built by the HMBC correlations of H2-7 to C-1′′, 2′′, 6′′, H2-2′′, 6′′ to C-7, as well as H-2′, 6′/C-1, and H2-1/C-1′, 2′, 6′, indicating the connection of C-1/C-1′and C-7/C-1′′. The HMBC correlations between δH 3.82 (3′-OCH3) with δC 149.6(C-3′) and δH 3.68 (3′′-OCH3) with δC 154.0 (C-3′′), demonstrated that the methoxy groups were located at C-3′ and C-3′′, respectively. Moreover, the NMR data the H-6′ (δH 4.99) signal appeared abnormally up-field from other aromatic proton signals, which is the distinguished characteristic of diphenyl ether type cyclic diarylheptanoid. Combined with HR-ESI-MS data, it is confirmed that 1 is diphenyl ether type cyclic diarylheptanoid.23 Furthermore, 1 showed positive optical rotation ([α]25D +22.7), Since 1 has a chiral plane in the molecule.24–26 Thus, 1 was elucidated as shown in Fig. 1 and named (+)-cyclogingerenone A. Unfortunately, the absolute stereochemistry remained undetermined.| Pos. | 1a | 2b | 3b | |||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| a Measured in CD3OD.b Measured in CDCl3. | ||||||
| 1 | 29.2 | 2.85, ddd (16.4, 8.7, 2.4) | 33.6 | 3.01 | 32.9 | 2.96, ddd (12.9, 5.8, 2.9) |
| 2.95 | 3.04, dt (12.9, 5.8) | |||||
| 2 | 42.0 | 2.49, ddd (15.0, 8.7, 2.4) | 43.9 | 2.64 | 44.6 | 2.51 |
| 2.79, dt (12.2, 5.8) | ||||||
| 3 | 201.1 | 200.8 | 215.6 | |||
| 4 | 131.8 | 5.86, d (15.2) | 131.4 | 5.73, dt (15.3, 1.5) | 52.4 | 1.89, d (18.3) |
| 2.22, dd (18.3, 10.5) | ||||||
| 5 | 147.9 | 6.58, ddd (15.2, 8.6, 7.5) | 148.2 | 6.56, dt (15.3, 7.0) | 66.1 | 3.34, br t (8.4) |
| 6 | 35.1 | 2.44 | 32.8 | 2.34 | 34.8 | 1.65 |
| 2.59, dq (12.6, 6.3) | 1.33 | |||||
| 7 | 34.6 | 2.95 | 32.0 | 2.70 | 29.0 | 2.49 |
| 2.72, ddd (16.8, 11.4, 2.1) | ||||||
| 1′ | 132.6 | 132.7 | 131.4 | |||
| 2′ | 106.5 | 6.38, d (1.8) | 105.7 | 6.28, d (1.7) | 105.9 | 6.34, br s |
| 3′ | 149.6 | 147.3 | 147.9 | |||
| 4′ | 134.1 | 132.9 | 133.1 | |||
| 5′ | 149.9 | 147.9 | 147.6 | |||
| 6′ | 106.3 | 4.99, d (1.8) | 106.8 | 5.52, d (1.7) | 107.4 | 5.33, br s |
| 1′′ | 140.8 | 138.5 | 138.0 | |||
| 2′′ | 115.8 | 6.80, d (1.8) | 114.9 | 6.77, d (1.8) | 116.2 | 6.59, d (1.8) |
| 3′′ | 154.0 | 152.7 | 151.9 | |||
| 4′′ | 144.1 | 143.5 | 145.2 | |||
| 5′′ | 125.1 | 6.92, d (8.0) | 124.6 | 7.01, d (8.0) | 124.7 | 7.21, d (8.0) |
| 6′′ | 122.5 | 6.73, dd (8.0, 1.8) | 121.8 | 6.83, dd (8.0, 1.8) | 120.2 | 6.93, dd (8.0, 1.8) |
| 3′-OCH3 | 56.7 | 3.82, s | 56.4 | 3.87, s | 56.4 | 3.87, s |
| 3′′-OCH3 | 56.6 | 3.68, s | 56.5 | 3.72, s | 56.3 | 3.59, s |
Compound 2 was separated as a yellow oil. The molecular formula of 2 was deduced as C21H22O5 according to HR-ESI-MS ion peak at m/z 355.1553 [M + H]+ (calcd for C21H23O5, 355.1545). Comprehensive analysis of the NMR data (see Table 1) demonstrated that the structure of 2 was quite similar to 1, and the difference between them was the connection sites of the benzenes and heptane moiety. The HMBC correlations from H2-1 to C-3, 2′′, 6′′, H2-7 to C-2′, 6′, 5, and H-4, H-5/C-3, 7, inferred that C-1 and C-7 were connected to C-1′′ and C-1′, respectively. In addition, 2 showed negative optical rotation ([α]25D −17.5). Therefore, 2 was elucidated as a new diaryl ether heptanoid and named (−)-cyclogingerenone B.
The molecular formula of compound 3 was determined as C21H24O6 by HR-ESI-MS (m/z 373.1654 [M + H]+; calcd 373.1651). It was disclosed that the only structural difference between 3 and 2 was concerned the change of the heptane chain moiety. The NMR data of 3 revealed the presence of one more oxymethine [δH 3.34 (1H, br t, J = 8.4 Hz, H-5); δC 66.1 (C-5)] and one more methylene [δH 1.89 (1H, d, J = 18.3 Hz, H-4a), 2.22 (1H, dd, J = 18.3, 10.5 Hz, H-4b); δC 52.4 (C-4)], but one less trans-double bond by comparing with 2. Consequently, the detailed analysis of the spectroscopic data allowed the planar structure of 3.
The absolute configuration of C-5 in 3 was established by comparison of the calculated ECD spectra [(5R)-3 and (5S)-3] with the experimental counterpart (see ESI†). The calculated ECD spectrum (Fig. S70 and 71†) (5R)-3 showed good consistency with the experimental one. In addition, 3 showed positive optical rotation ([α]25D +2.7). Thus, the structure of 3 was assigned as (+)-cyclogingerenone C.
Compound 4 was obtained as a yellow gum. The HR-ESI-MS of 4 showed a quasimolecular peak at m/z 403.2110 [M + H]+ (calcd for C23H30O6, 403.2121), corresponding to the molecular formula of C23H30O6. Careful analysis of its 1H and 13C NMR data (Table 2) indicated that 4 had a similar structure to 13. The main difference found between them concerned the substituent group at C-5. A hydroxyl group at C-5 in 13 was changed to an ethoxy group in 4. This deduction was supported by 1H–1H COSY correlations of H2-8/H3-9 and HMBC cross peaks from H-8 [δH 3.42 (2H, qd, J = 7.1, 2.0 Hz)] to C-5 (δC 76.3). The absolute configuration of C-5 of 4 was determined by comparing the optical rotation with analogues.27–30 The negative optical rotation ([α]25D −10.7) of 4 indicated 5R configuration. On the basis of the above evidence, the structure of 4 was determined (Fig. 1) and named (5R)-5-ethoxyhexahydrocurcumin.
| Pos. | 4a,c | 5b,d | 6b,d | |||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| a Measured in CD3OD.b Measured in CDCl3.c 400 MHz for 1H, 100 MHz for 13C.d 600 MHz for 1H, 150 MHz for 13C. | ||||||
| 1 | 30.3 | 2.74 | 29.9 | 2.82, t (7.4) | 30.8 | 2.86, t (7.0) |
| 2 | 46.4 | 2.74 | 46.0 | 2.73 | 41.7 | 2.81, t (7.0) |
| 3 | 211.7 | 208.9 | 200.1 | |||
| 4 | 48.7 | 2.67, dd (15.8, 7.1) | 47.6 | 2.45, dd (15.9, 5.3) | 131.1 | 6.07, d (15.8) |
| 2.55, dd (15.8, 7.1) | 2.71, dd (15.9, 7.1) | |||||
| 5 | 76.3 | 3.72, dt (12.1, 6.1) | 76.8 | 3.69 | 147.0 | 6.77, dd (15.8, 6.9) |
| 6 | 37.5 | 1.71 | 36.2 | 1.76 | 34.3 | 2.47 |
| 7 | 32.1 | 2.54 | 31.2 | 2.55, ddd (15.6, 9.8, 6.6) | 33.8 | 2.64, t (7.1) |
| 2.63, ddd (15.6, 9.8, 5.8) | ||||||
| 8 | 65.6 | 3.42, qd (7.1, 2.0) | 57.2 | 3.31, s | — | — |
| 9 | 15.8 | 1.10, t (7.1) | — | — | — | — |
| 1′ | 134.0 | 132.3 | 137.3 | |||
| 2′ | 113.1 | 6.75, d (1.5) | 105.1 | 6.39, s | 105.6 | 6.39, s |
| 3′ | 148.9 | 147.1 | 153.3 | |||
| 4′ | 145.6 | 133.1 | 136.3 | |||
| 5′ | 116.2 | 6.68, d (8.0) | 147.1 | 153.3 | ||
| 6′ | 121.8 | 6.58, dd (8.0, 1.5) | 105.1 | 6.39, s | 105.6 | 6.39, s |
| 1′′ | 134.8 | 133.9 | 133.6 | |||
| 2′′ | 113.2 | 6.73, d (1.6) | 111.0 | 6.67, br s | 115.5 | 6.63, br s |
| 3′′ | 148.9 | 146.5 | 143.9 | |||
| 4′′ | 145.8 | 143.9 | 142.1 | |||
| 5′′ | 116.2 | 6.70, d (8.0) | 114.4 | 6.82, d (7.9) | 115.5 | 6.75, d (6.9) |
| 6′′ | 121.8 | 6.61, dd (8.0, 1.6) | 121.0 | 6.65, br d (7.9) | 120.7 | 6.55, br d (6.9) |
| 3′-OCH3 | 56.4 | 3.80, s | 56.4 | 3.86, s | 56.3 | 3.83, s |
| 4′-OCH3 | — | — | — | — | 61.1 | 3.82, s |
| 5′-OCH3 | — | — | 56.4 | 3.86, s | 56.3 | 3.83, s |
| 3′′-OCH3 | 56.4 | 3.82, s | 56.0 | 3.87, s | — | — |
Compound 5 showed a molecular formula of C23H30O7 by HR-ESI-MS (m/z 419.2060 [M + H]+, calcd 419.2070). A comparison of the spectral data (Table 2) of 5 and 4 revealed that an ethoxy group in 4 was changed to a methoxyl group in 5, which was confirmed by HMBC correlations from H3-8 (δH 3.31, s) to C-5 (δC 76.8). Moreover, an additional methoxyl group was substituted at C-5′ due to the equivalent aromatic protons [δH 6.39 (2H, s, H-2′, 6′)] and a symmetrical substituted aromatic ring (δC 132.3, 147.1 × 2, 105.1 × 2, 133.1). Similarly to 4, according to the specific optical rotation of 5 as [α]25D −8.3, the absolute configuration of C-5 is assigned to R. To sum up, 5 was identified as (5R)-5-methoxy-1-(4-hydroxy-3,5-dimethoxy-phenyl)-7-(4-hydroxy-3-methoxyphenyl) heptan-3-one.
Compound 6 possessed a molecular formula of C22H26O6 based on HR-ESI-MS (m/z 387.1794 [M + H]+, calcd 387.1808). The 1H and 13C NMR data (Table 2) of 6 was high similarity to those of 20. The only difference between them was that a hydroxyl group in 20 was changed to a methoxyl group at C-4′ in 6, which was further deduced by HMBC correlations between 3.82 (3H, s, 4′-OCH3) and C-4′ (δC 136.3). Therefore, the structure of 6 was defined as (E)-7-(3,4-dihydroxyphenyl)-1-(3,4,5-trimethoxyphenyl) hept-4-en-3-one.
Compound 7 had the molecular formula of C22H28O6 inferred from the HR-ESI-MS at m/z 411.1767 ([M + Na]+, calcd 411.1784), corresponding to an index of hydrogen deficiency of 9. Its NMR data (Table 3) possessed similar signals to those of 10 except for the loss of one acetyl group located at C-5. The acetyl group was substituted at C-3, which was evidenced by HMBC correlations from H-9 (δH 2.03) to C-8 (δC 171.4), and H-3 (δH 4.90) to C-8 (δC 171.4). According to Brewster rule of secondary carbinol,31–33 7 showed a negative optical rotation value of 10.5, which indicated that the acetyl moiety at C-3 is predicted to be R configuration. Therefore, 7 was determined and named as (3R)-3-acetoxy-7-(3,4-dihydroxy-phenyl)-1-(4-hydroxy-3-methoxyphenyl) heptane.
| Pos. | 7 | 8 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 31.6 | 2.58 | 32.1 | 2.59 |
| 2.51, td (7.0, 2.7) | 2.51, td (7.2, 2.0) | |||
| 2 | 36.2 | 1.84 | 36.0 | 1.86 |
| 1.76 | 1.76 | |||
| 3 | 73.8 | 4.90 | 73.5 | 4.92 |
| 4 | 34.0 | 1.56 | 33.8 | 1.56 |
| 5 | 24.7 | 1.30 | 24.7 | 1.29 |
| 6 | 31.1 | 1.51 | 30.9 | 1.49 |
| 7 | 35.0 | 2.46, td (7.5, 1.7) | 34.9 | 2.4, t (7.6) |
| 1′ | 133.7 | 132.9 | ||
| 2′ | 111.2 | 6.66, br s | 105.3 | 6.40, s |
| 3′ | 146.5 | 147.0 | ||
| 4′ | 143.8 | 132.9 | ||
| 5′ | 114.5 | 6.83, d (7.9) | 147.0 | |
| 6′ | 120.9 | 6.65 | 105.3 | 6.40, s |
| 1′′ | 135.4 | 135.4 | ||
| 2′′ | 115.5 | 6.64, br s | 115.4 | 6.59, d (1.5) |
| 3′′ | 143.5 | 143.4 | ||
| 4′′ | 141.8 | 141.8 | ||
| 5′′ | 115.2 | 6.76, d (7.6) | 115.2 | 6.75, d (8.0) |
| 6′′ | 121.0 | 6.56, br d (7.6) | 120.9 | 6.56, dd (8.0, 1.5) |
| 3-OAc | 171.4 | 171.3 | ||
| 21.4 | 2.03, s | 21.4 | 2.04, s | |
| 3′-OCH3 | 56.1 | 3.87, s | 56.5 | 3.87, s |
| 5′-OCH3 | — | — | 56.5 | 3.87, s |
The molecular formula of compound 8 was determined as C23H30O7 by HR-ESI-MS (m/z 441.1895 [M + Na]+, calcd 441.1889). Extensive comparison of the NMR data of 8 and 7 (Table 3) suggested that an additional methoxyl group was located at C-5′ due to the equivalent aromatic protons [δH 6.40 (2H, s, H-2′, 6′)] and a symmetrical substituted aromatic ring (δC 147.0 × 2, 105.3 × 2, 132.9 × 2). Using the same method as above, it was determined that the absolute configuration of C-3 is R based on an optical rotation of [α]25D −16.9. Thus, the structure of 8 was assigned, and named (3R)-3-acetoxy-7-(3,4-dihydroxyphenyl)-1-(4-hydroxy-3,5-dimethoxyphenyl)heptane.
The 15 known compounds (9–23) were identified as (3R*,5S*)-3,5-diacetoxy-1,7-bis(4-hydroxy-3-methoxyphenyl) heptane (9),34 (3S*,5S*)-3,5-diacetoxy-7-(3,4-dihydroxyphenyl)-1-(4-hydroxy-3-methoxyphenyl) heptane (10),35 (3R*,5S*)-3,5-diacetoxy-1-(4-hydroxy-3,5-dimethoxyphenyl)-7-(4-hydroxy-3-methoxyphenyl) heptane (11),35 (3S*,5S*)-3,5-diacetoxy-1-(4-hydroxy-3,5-dimethoxyphenyl)-7-(4-hydroxy-3-methoxyphenyl) heptane (12),28 hexahydrocurcumin (13),36 5-acetyl hexahydro-curcumin D (14),29 7-(3,4-dihydroxyphenyl)-1-(4-hydroxy-3-methoxyphenyl) heptan-3-one (15),37 platyphyllenone (16),38 isogingerenone C (17),39 gingerenone A (18),29 (E)-7-(3,4-dihydroxyphenyl)-1-(4-hydroxy-3-methoxyphenyl) hept-4-en-3-one (19),40 isogingerenone B (20),41 gingerenone B (21),41 (1S*,3R*,5S*) 3-acetoxy-1,5-epoxy-1-(3,4-dihydroxy-5-methoxy-phenyl)-7-(4-hydroxy-3-methoxyphenyl) heptane (22),42 and (1S*,3S*,5S*) 3-acetoxy-1,5-epoxy-1-(3,4-dihydroxy-5-methoxy-phenyl)-7-(4-hydroxy-3-methoxyphenyl) heptane (23).42
3.2. Predicted targets of isolated compounds
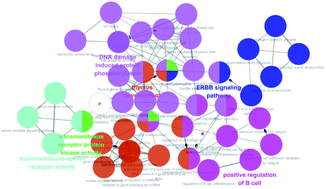
357 targets were discovered as potential targets based on the probability in SwissTargetPrediction (3 and 13 did not have the potential targets). Then, 1381 cancer-related targets were extracted from GenCLiP3 with “cancer” as a keyword. Among them, there are 159 targets were duplicates. A compound-cancer target network consisting of 180 nodes (159 nodes of targets and 21 nodes of compounds) and 598 edges was constructed by using Cytoscape 3.5 software (Fig. 3). Thirty key targets were identified based on the intermediate value of degree, and their biological functions were associated with four pathways: DNA damage induced protein phosphorylation, ERGG pathway, positive regulation of B cell and transmembrane receptor protein kinases activity (Fig. 4). CHEK1 (CHK1), CHEK2 (CHK2) and ABL1 played an important role in DNA damage. Next, we focused on the protein phosphorylation induced by DNA damage pathway and conducted the activity verification.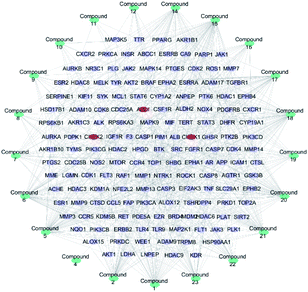 | ||
| Fig. 3 Potential anti-tumour target-compound network (blue stands for the compounds, purple represents the cancer related targets, and red means key targets). | ||
3.3. Diarylheptanoids suppressed cell proliferation
All isolated diarylheptanoids were tested for their inhibitory activities against A549, HepG2, HeLa, MDA-MB-231, and HCT116 lines by using MTT method, and the results are shown in Table 4. Compounds 6, 16–19 showed cytotoxic effects against five human tumour cell lines with IC50 values ranging from 6.69–33.46 μM. Among them, compound 17 displayed more superior cytotoxicity against HCT116 cell lines with IC50 value of 6.69 ± 0.09 μM, compared to the positive control (curcumin, IC50: 14.88 ± 0.14 μM).| Compound | Cytotoxic activity IC50/μmol L−1 | ||||
|---|---|---|---|---|---|
| A549 | HepG2 | HeLa | MDA-MB-231 | HCT116 | |
| a Paclitaxel was used as a control. | |||||
| 6 | 17.49 ± 0.09 | 33.46 ± 0.50 | 30.46 ± 1.20 | 17.83 ± 0.11 | 14.70 ± 0.63 |
| 16 | 13.74 ± 0.48 | 19.67 ± 0.09 | 11.30 ± 0.36 | 10.78 ± 0.12 | 13.88 ± 0.18 |
| 17 | 11.49 ± 0.11 | 15.98 ± 0.14 | 10.41 ± 0.43 | 9.18 ± 0.18 | 10.61 ± 0.21 |
| 18 | 8.63 ± 0.42 | 13.06 ± 0.13 | 11.56 ± 0.21 | 10.51 ± 0.14 | 6.69 ± 0.09 |
| 19 | 17.27 ± 0.21 | 20.93 ± 0.41 | 12.20 ± 0.16 | 13.60 ± 0.05 | 11.64 ± 0.18 |
| Curcumina | 21.86 ± 0.25 | 27.37 ± 0.30 | 19.71 ± 0.22 | 19.03 ± 0.11 | 14.88 ± 0.14 |
The cytotoxic activity of compound 13 was nearly identical to compounds 4, 14 and 15, which suggested the cytotoxic activity of diarylheptanoids might be less dependent on hydroxy groups of heptane chain. The results of MTT assay for compounds 6, 16–19 indicated that the substituted phenyl ring might not be a critical structural determinant for the cytotoxic activities.43 The superior cytotoxic effect of 18 compared with 13 might be attributed to the presence of an α, β-unsaturated carbonyl moiety in 18, as reported earlier.44,45 Previously, it had been suggested that the α, β-unsaturated carbonyl compounds-mediated toxicity was attributable to formation of Michael-type adducts with the nucleophilic sulfhydryl groups of protein thiols.46,47 The above results were not sufficient to further clarify the structure–activity relationship between the diarylheptanoid derivatives and/or other components. More research may be required to clarify their potential selective cytotoxic activity.
3.4. Effects of diarylheptanoids on the ATR/CHK1 signalling pathway
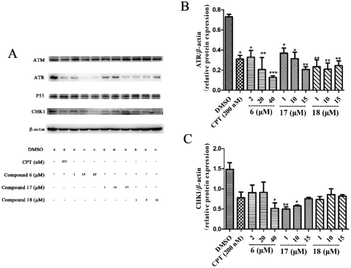
Eukaryotic cells have such a cumbersome DNA damage response (DDR) to protect the integrity of the genome, which contains two main signal cascades, namely the ATM/CHK2 and ATR/CHK1 cascades. As the core protein and kinase of DDR, ATR can activate downstream CHK1 to form ATR/CHK1 signalling pathway, so that bodies have enough time to repair the wrong DNA before turning to the next stage or going to apoptosis. If the damage of the cell cannot be repaired, the DDR will lead the damaged cell to apoptosis.48If the damage of the cell cannot be repaired, the DDR will lead the damaged cell to apoptosis.48 To understand the anti-tumour mechanism of diarylheptanoids in ginger, the effect of compounds 6, 17, and 18 (which showed superior suppressed cell proliferation activity) on the ATR/CHK1 signalling pathway was investigated. As shown in Fig. 5, compounds 6, 17, and 18 significantly suppressed the total protein levels of ATR and CHK1 in HCT116 cells. Our results indicated that diarylheptanoids had potential anti-tumour effects, and they might act through the regulation of the ATR/CHK1 signalling pathway.
4. Conclusions
In summary, 23 diarylheptanoids including eight undescribed ones (1–8), were isolated and identified from an important natural dietary ingredient ginger. It is worth mentioning that 1–3 are the first reported structures of diarylether heptanoids in Z. officinale, whereas 15–17 were isolated from Zingiber for the first time. Network pharmacology demonstrated that diarylheptanoids were mainly associated with the antitumor activity by affecting DNA damage signalling pathway. Cytotoxic and enzymatic activity assays discovered that compounds 6, 17, and 18 may exert anticancer effects by regulating the ATR/CHK1 signalling pathway.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are thankful to financial grants from the National Natural Science Foundation of China for financial support to this research (No. 81803347) and Natural Science Foundation of Guangdong Province 2018A030313707. We are grateful for the high-performance computing platform at Jinan University, which was used to carry out this study.Notes and references
- M. Liu, X. Xia, G. Chou, D. Liu, A. Zuberi, J. Ye and Z. Liu, Variations in the contents of gingerols and chromatographic fingerprints of ginger root extracts prepared by different preparation methods, J. AOAC Int., 2014, 97, 50–57 CrossRef CAS PubMed.
- H. A. Schwertner and D. C. Rios, High-performance liquid chromatographic analysis of 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol in ginger-containing dietary supplements, spices, teas, and beverages, J. Chromatogr. B, 2007, 856, 41–47 CrossRef CAS PubMed.
- S. Das, R. Bordoloi and N. Newar, A review on immune modulatory effect of some traditional medicinal herbs, J. Pharm., Chem. Biol. Sci., 2014, 2, 33–42 CAS.
- I. R. Kubra and L. J. M. Rao, An impression on current developments in the technology, chemistry, and biological activities of ginger (Zingiber officinale Roscoe), Crit. Rev. Food Sci. Nutr., 2012, 52, 651–688 CrossRef CAS PubMed.
- I. Lete and J. Alluέ, The effectiveness of ginger in the prevention of nausea and vomiting during pregnancy and chemotherapy, Integr. Med. Insights, 2016, 11, S36273 CrossRef PubMed.
- R. B. Semwal, D. K. Semwal, S. Combrinck and A. M. Viljoen, Gingerols and shogaols: Important nutraceutical principles from ginger, Phytochemistry, 2015, 117, 554–568 CrossRef CAS PubMed.
- M. S. Baliga, R. Haniadka, M. M. Pereira, K. R. Thilakchand, S. Rao and R. Arora, Radioprotective effects of Zingiber officinale Roscoe (ginger): past, present and future, Food Funct., 2012, 3, 714–723 RSC.
- R.-H. Ma, Z.-J. Ni, Y.-Y. Zhu, K. Thakur, F. Zhang, Y.-Y. Zhang, F. Hu, J.-G. Zhang and Z.-J. Wei, A recent update on the multifaceted health benefits associated with ginger and its bioactive components, Food Funct., 2021, 12, 519–542 RSC.
- R. Kiyama, Nutritional implications of ginger: chemistry, biological activities and signaling pathways, J. Nutr. Biochem., 2020, 108486 CrossRef CAS PubMed.
- V. S. Honmore, A. D. Kandhare, P. P. Kadam, V. M. Khedkar, A. D. Natu, S. R. Rojatkar and S. L. Bodhankar, Diarylheptanoid, a constituent isolated from methanol extract of Alpinia officinarum attenuates TNF-α level in Freund's complete adjuvant-induced arthritis in rats, J. Ethnopharmacol., 2019, 229, 233–245 CrossRef CAS PubMed.
- A. Cerulli, G. Lauro, M. Masullo, V. Cantone, B. Olas, B. Kontek, F. Nazzaro, G. Bifulco and S. Piacente, Cyclic diarylheptanoids from Corylus avellana green leafy covers: determination of their absolute configurations and evaluation of their antioxidant and antimicrobial activities, J. Nat. Prod., 2017, 80, 1703–1713 CrossRef CAS PubMed.
- J. Fan, M. Wu, J. Wang, D. Ren, J. Zhao and G. Yang, 1, 7-Bis (4-hydroxyphenyl)-1, 4-heptadien-3-one induces lung cancer cell apoptosis via the PI3K/Akt and ERK1/2 pathways, J. Cell. Physiol., 2019, 234, 6336–6349 CrossRef CAS PubMed.
- M. Takahashi, H. Fuchino, S. Sekita and M. Satake, In vitro leishmanicidal activity of some scarce natural products, Phytother Res., 2004, 18, 573–578 CrossRef CAS PubMed.
- T. Matsumoto, S. Nakamura, S. Nakashima, M. Yoshikawa, K. Fujimoto, T. Ohta, A. Morita, R. Yasui, E. Kashiwazaki and H. Matsuda, Diarylheptanoids with inhibitory effects on melanogenesis from the rhizomes of Curcuma comosa in B16 melanoma cells, Bioorg. Med. Chem. Lett., 2013, 23, 5178–5181 CrossRef CAS PubMed.
- P. Chuaicharoen, T. Charaslertrangsi, A. Chuncharunee, A. Suksamrarn and P. Piyachaturawat, Non-Phenolic Diarylheptanoid from Curcuma comosa Protects Against Thioacetamide-Induced Acute Hepatotoxicity in Mice, Pharm. Sci. Asia, 2020, 47, 74–85 CrossRef CAS.
- Y. Yang, Q. Gong, W. Wang, Y.-L. Mao, X.-R. Wang, S. Yao, H.-Y. Zhang, C. Tang and Y. Ye, Neuroprotective and Anti-inflammatory Ditetrahydrofuran-Containing Diarylheptanoids from Tacca chantrieri, J. Nat. Prod., 2020, 83, 3681–3688 CrossRef CAS PubMed.
- Y. Jahng and J. G. Park, Recent studies on cyclic 1, 7-diarylheptanoids: Their isolation, structures, biological activities, and chemical synthesis, Molecules, 2018, 23, 3107 CrossRef PubMed.
- D. Pan, C. Zeng, W. Zhang, T. Li, Z. Qin, X. Yao, Y. Dai, Z. Yao, Y. Yu and X. Yao, Non-volatile pungent compounds isolated from Zingiber officinale and their mechanisms of action, Food Funct., 2019, 10, 1203–1211 RSC.
- D. Gfeller, A. Grosdidier, M. Wirth, A. Daina, O. Michielin and V. Zoete, SwissTargetPrediction: a web server for target prediction of bioactive small molecules, Nucleic Acids Res., 2014, 42, W32–W38 CrossRef CAS PubMed.
- J.-H. Wang, L.-F. Zhao, H.-F. Wang, Y.-T. Wen, K.-K. Jiang, X.-M. Mao, Z.-Y. Zhou, K.-T. Yao, Q.-S. Geng and D. Guo, GenCLiP 3: mining human genes’ functions and regulatory networks from PubMed based on co-occurrences and natural language processing, Bioinformatics, 2019, 36, 1973–1975 Search PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res., 2003, 13, 2498–2504 CrossRef CAS PubMed.
- M. Masullo, A. Cerulli, B. Olas, C. Pizza and S. Piacente, Giffonins A–I, antioxidant cyclized diarylheptanoids from the leaves of the hazelnut tree (Corylus avellana), source of the Italian PGI Product “Nocciola di Giffoni”, J. Nat. Prod., 2015, 78, 17–25 CrossRef CAS PubMed.
- W. Jin, X. F. Cai, M. Na, J. J. Lee and K. Bae, Diarylheptanoids from Alnus hirsuta inhibit the NF-kB activation and NO and TNF-α production, Biol. Pharm. Bull., 2007, 30, 810–813 CrossRef CAS PubMed.
- M. Morihara, N. Sakurai, T. Inoue, K.-i. Kawai and M. Nagai, Two novel diarylheptanoid glucosides from Myrica gale var. tomentosa and absolute structure of plane-chiral galeon, Chem. Pharm. Bull., 1997, 45, 820–823 CrossRef CAS.
- J.-X. Liu, D.-L. Di, X.-N. Wei and Y. Han, Cytotoxic diarylheptanoids from the pericarps of walnuts (Juglans regia), Planta Med., 2008, 74, 754–759 CrossRef CAS PubMed.
- M. Q. Salih and C. M. Beaudry, Chirality in diarylether heptanoids: Synthesis of myricatomentogenin, jugcathanin, and congeners, Org. Lett., 2012, 14, 4026–4029 CrossRef CAS PubMed.
- H. Itokawa, H. Morita, I. Midorikawa, R. Aiyama and M. Morita, Diarylheptanoids from the rhizome of Alpinia officinarum Hance, Chem. Pharm. Bull., 1985, 33, 4889–4893 CrossRef CAS.
- J. Ma, X. Jin, L. Yang and Z.-L. Liu, Diarylheptanoids from the rhizomes of Zingiber officinale, Phytochemistry, 2004, 65, 1137–1143 CrossRef CAS PubMed.
- T. Feng, J. Su, Z.-H. Ding, Y.-T. Zheng, Y. Li, Y. Leng and J.-K. Liu, Chemical constituents and their bioactivities of “Tongling White Ginger”(Zingiber officinale), J. Agric. Food Chem., 2011, 59, 11690–11695 CrossRef CAS PubMed.
- Y. Yang, K. Kinoshita, K. Koyama, K. Takahashi, S. Kondo and K. Watanabe, Structure-antiemetic-activity of some diarylheptanoids and their analogues, Phytomedicine, 2002, 9, 146–152 CrossRef CAS PubMed.
- J. H. Brewster, A useful model of optical activity. I. Open chain compounds, J. Am. Chem. Soc., 1959, 81, 5475–5483 CrossRef CAS.
- M. Nagai, N. Kenmochi, M. Fujita, N. Furukawa and T. Inoue, Studies on the constituents of Aceraceae plants. VI.: revised stereochemistry of (−)-centrolobol, and new glycosides from Acer nikoense, Chem. Pharm. Bull., 1986, 34, 1056–1060 CrossRef.
- A. M. El-Halawany, R. S. El Dine, N. S. El Sayed and M. Hattori, Protective effect of Aframomum melegueta phenolics against CCl 4-induced rat hepatocytes damage; role of apoptosis and pro-inflammatory cytokines inhibition, Sci. Rep., 2014, 4, 1–9 Search PubMed.
- G. Sabitha, G. Chandrashekhar, K. Yadagiri and J. S. Yadav, Synthesis of diarylheptanoids,(5S)-5-acetoxy-1, 7-bis (4-hydroxy-3-methoxyphenyl)-3-heptanone and (3S, 5S)-3, 5-diacetoxy-1, 7-bis (4-hydroxy-3-methoxyphenyl) heptane, Tetrahedron: Asymmetry, 2011, 22, 1729–1735 CrossRef CAS.
- H. Kikuzaki, M. Kobayashi and N. Nakatani, Diarylheptanoids from rhizomes of Zingiber officinale, Phytochemistry, 1991, 30, 3647–3651 CrossRef CAS.
- Y. Hori, T. Miura, Y. Hirai, M. Fukumura, Y. Nemoto, K. Toriizuka and Y. Ida, Pharmacognostic studies on ginger and related drugs—part 1: five sulfonated compounds from Zingiberis rhizome (Shokyo), Phytochemistry, 2003, 62, 613–617 CrossRef CAS PubMed.
- A. M. El-Halawany and M. Hattori, Anti-oestrogenic diarylheptanoids from Aframomum melegueta with in silico oestrogen receptor alpha binding conformation similar to enterodiol and enterolactone, Food Chem., 2012, 134, 219–226 CrossRef CAS.
- J. Chen, J. J. Karchesy and R. F. González-Laredo, Phenolic diarylheptenones from Alnus rubra bark, Planta Med., 1998, 64, 74–75 CrossRef CAS PubMed.
- K.-S. Lee, G. Li, S. H. Kim, C.-S. Lee, M.-H. Woo, S.-H. Lee, Y.-D. Jhang and J.-K. Son, Cytotoxic Diarylheptanoids from the Roots of Juglans mandshurica, J. Nat. Prod., 2002, 65, 1707–1708 CrossRef CAS PubMed.
- G. Fu, W. Zhang, D. Du, Y. P. Ng, F. C. Ip, R. Tong and N. Y. Ip, Diarylheptanoids from rhizomes of Alpinia officinarum inhibit aggregation of α-synuclein, J. Agric. Food Chem., 2017, 65, 6608–6614 CrossRef CAS PubMed.
- K. Endo, E. Kanno and Y. Oshima, Structures of antifungal diarylheptenones, gingerenones A, B, C and isogingerenone B, isolated from the rhizomes of Zingiber officinale, Phytochemistry, 1990, 29, 797–799 CrossRef CAS.
- H. Kikuzaki and N. Nakatani, Cyclic diarylheptanoids from rhizomes of Zingiber officinale, Phytochemistry, 1996, 43, 273–277 CrossRef CAS.
- S. Gamre, M. Tyagi, S. Chatterjee, B. S. Patro, S. Chattopadhyay and D. Goswami, Synthesis of Bioactive Diarylheptanoids from Alpinia officinarum and Their Mechanism of Action for Anticancer Properties in Breast Cancer Cells, J. Nat. Prod., 2021, 84, 352–363 CrossRef CAS PubMed.
- Q.-Y. Wei, J.-P. Ma, Y.-J. Cai, L. Yang and Z.-L. Liu, Cytotoxic and apoptotic activities of diarylheptanoids and gingerol-related compounds from the rhizome of Chinese ginger, J. Ethnopharmacol., 2005, 102, 177–184 CrossRef CAS PubMed.
- F. Peng, Q. Tao, X. Wu, H. Dou, S. Spencer, C. Mang, L. Xu, L. Sun, Y. Zhao and H. Li, Cytotoxic, cytoprotective and antioxidant effects of isolated phenolic compounds from fresh ginger, Fitoterapia, 2012, 83, 568–585 CrossRef CAS PubMed.
- R. M. LoPachin, D. S. Barber and T. Gavin, Molecular mechanisms of the conjugated α, β-unsaturated carbonyl derivatives: relevance to neurotoxicity and neurodegenerative diseases, Toxicol. Sci., 2008, 104, 235–249 CrossRef CAS PubMed.
- T. Nakayachi, E. Yasumoto, K. Nakano, S. R. M. Morshed, K. Hashimoto, H. Kikuchi, H. Nishikawa, M. Kawase and H. Sakagami, Structure-activity relationships of α, β-unsaturated ketones as assessed by their cytotoxicity against oral tumor cells, Anticancer Res., 2004, 24, 737–742 CAS.
- K. Ando, Y. Nakamura, H. Nagase, A. Nakagawara, T. Koshinaga, S. Wada and M. Makishima, Co-inhibition of the DNA damage response and CHK1 enhances apoptosis of neuroblastoma cells, Int. J. Mol. Sci., 2019, 20, 3700 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra03592d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |