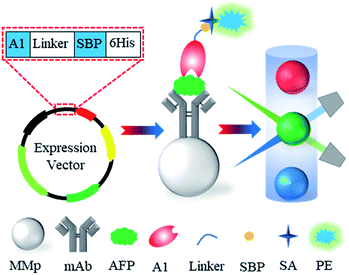
Open Access Article
Open Access ArticleDevelopment of a nanobody tagged with streptavidin-binding peptide and its application in a Luminex fluoroimmunoassay for alpha fetal protein in serum†
Qi Chena,
Danyang Suna,
Hua Peib,
Benchao Sua,
Kunlu Baoa,
Hongmei Caoa,
Chenghui Zhanga,
Bruce D. Hammockc and
Xing Liu *a
*a
aCollege of Food Science and Engineering, Hainan University, Haikou 570228, China. E-mail: qichen@hainanu.edu.cn; sdy@hainanu.edu.cn; benchao312@hainanu.edu.cn; baokunlu@hainanu.edu.cn; hmcao@hainanu.edu.cn; zchlm@163.com; xliu@hainanu.edu.cn
bDepartment of Clinical Laboratory, The Second Affiliated Hospital of Hainan Medical University, Haikou 570311, China. E-mail: phzmh61@aliyun.com
cDepartment of Entomology and Nematology and UCD Comprehensive Cancer Center, University of California, Davis, California 95616, USA. E-mail: bdhammock@ucd.edu
First published on 22nd June 2020
Abstract
Sensitive and accurate detection of disease-related biomarkers can promote the early screening and diagnosis of cancers for improving the prognosis and survival of patients. Herein alpha fetal protein (AFP) was selected as the model macromolecule antigen and we developed AFP-specific alpaca nanobodies (Nbs) from an immunized phage-displayed Nb library. Then Nbs tagged with streptavidin-binding peptide (Nb-SBP) were constructed and used to develop an Nb-SBP-mediated fluoroimmunoassay based on the Luminex-200 system (NS-LFIA). Based on the optimal experimental conditions, the NS-LFIA has a limit of detection of 0.237 ng mL−1 with a linear detection range of 0.49–125 ng mL−1. The average recovery rate and relative standard derivation were in the range of 98.2–110% and 2.8–13.8%, respectively. The NS-LFIA is highly selective for AFP and ignorable cross-reaction was observed with the other biomarkers. The content of AFP in clinical serum samples was determined by both the developed NS-LFIA and the Roche E601 automatic chemiluminescence immunoassay analyzer and a good correlation was obtained between the two methods (R2 = 0.9894). Moreover, the Nb-SBP can significantly improve the homogeneity of the fluorescent signals tested by the Luminex-200 system compared with the biotinylated conventional monoclonal antibodies, which could reduce the magnetic microsphere consumption and test cost by decreasing the repetitions of each sample. Thus the results demonstrated that the Nb-SBP was a very promising immunological diagnostic reagent and indicated the applicability and reliability of the NS-LFIA for sensitive detection of AFP and other disease-related biomarkers.
1. Introduction
According to statistics from the Global Cancer Observatory in 2018, cancer is a major cause of death in most countries around the world, and caused one out of seven global disease deaths.1 From the statistics of the National Cancer Centre of China, cancer has become the leading cause of disease death in China since 2010. Among them, liver cancer is one of the cancers with the highest morbidity and mortality. There are an estimated 841![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases and 782
000 new cases and 782![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths annually worldwide, and half of them occur in China.2 Thus, liver cancer has caused enormous economic losses and posed a huge threat to public health.
000 deaths annually worldwide, and half of them occur in China.2 Thus, liver cancer has caused enormous economic losses and posed a huge threat to public health.
Numerous studies have shown that early screening and diagnosis of liver cancer and other malignant tumors could significantly improve the prognosis and survival of the patient.3–5 Compared with clinical imaging and pathological diagnosis, the detection of biomarkers in serum can determine the existence of tumor earlier. Alpha fetal protein (AFP) is the most widely used clinical biomarker, and has been used in the early diagnosis of liver cancer since the 1970s.6,7 Currently, the common methods for the detection of AFP mainly include enzyme-linked immunosorbent assay (ELISA) and chemiluminescent immunoassay. These traditional immunoassays are sensitive and rapid. However, there are also two challenges, including the limited repetitions of samples and the interference on the assay accuracy and reproducibility caused by impurities and operation.
As an alternative to the traditional immunoassays for biomarker, the suspension arrays integrate advantages of high-throughput and multiple analytes detection.8,9 Due to the rapid development of nanoscience, many nanotechnologies were introduced into suspension arrays to improve detection capability, sensitivity and reliability. Yuan et al. reported a simple method for the fabrication of plasmonic magnetic microbeads and applied it to suspension array with fluorescence enhancement of about 60-fold.10 Lowe et al. developed a signal amplification approach for suspension array where the streptavidin-phycoerythrin (SA-PE) labels binding sites were increased by using the biotinylated dendrimers.11 It is worth noting that most studies were focused on new materials,12,13 signal amplification strategies14 and non-specificity suppression.15 However, there are few reports focusing on the biorecognition probe and conjugation mode. Sensitive and specific recognition of target is vital in suspension array, and the most commonly used biorecognition probe is antibody. Currently, the Luminex system has been widely used for rapid and high-throughput detection of biomarkers,8 where the fluorescent signals were introduced into the system through the binding of biotinylated detection antibodies with SA-PE. However, the traditional chemical conjugation could cause loss of antibody activity and randomness of the amount of biotin bound to each antibody molecule.16 This results in the increased antibody consumption and uneven fluorescent protein bound to each detection antibody via the biotin–streptavidin interaction.
With the advance in gene engineering and antibody engineering, various miniaturized antibodies have been generated as the alternatives to the conventional intact antibodies, such as the antigen-binding fragment,17 single-chain variable fragment,18 and single-domain antibody (sdAb).19,20 Among these miniaturized antibodies, the sdAb that is the smallest known antigen-binding unit has been very attractive for its nanoscale size, low cost, ease of genetic manipulation, good water-solubility and high thermal stability.21 The sdAb, also namely nanobody (Nb), commonly refers to the VHH or VNAR, which originates from the heavy chain variable domain of the heavy-chain antibodies (HCAbs) in Camelidae or of the immunoglobulin new antigen receptor (IgNAR) in Chondrichthyes.19,22 The smaller size of Nb makes it possible to obtain a higher probe density, faster binding rate and facile to site-specific derivatization by comparison with the conventional intact antibodies. Moreover, numerous derivates of Nb that retain good antibody activity have been generated attributing to its ease of genetic manipulation, such as Nbs fused with alkaline phosphatase, horseradish peroxidase, nanoluciferase, and peptide tag.23 These derivates are cloneable and could be reproduced with low cost in the prokaryotic expression system, eliminating the loss of antibody activity and batch variation in the traditional chemical conjugation.
In the present study, a phage-displayed Nb library was constructed from an immunized alpaca, and the anti-AFP Nbs were successfully selected from the library by biopanning. After antibody pair test with a commercial anti-AFP monoclonal antibody, a construct of the selected Nb and streptavidin-binding peptide (SBP) was expressed as a fusion protein (Nb-SBP) probe for AFP. As a concept-of-proof, the performance of Nb-SBP probe in the Luminex system using AFP as the model molecule was evaluated (Fig. 1). We found that the uniformity of the results was improved, which indicates that the repetitions of each sample can be further decreased and demonstrates the potential to reduce magnetic microspheres consumption and test costs.
2. Experimental
2.1. Reagent and materials
Freund's complete/incomplete adjuvant was purchased from Sigma-Chemical Co. (St. Louis, MO). N-Hydroxysulfosuccinimide (NHSS) and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) were from Aladdin Chemistry Co., Ltd. (Shanghai, China). Restriction enzyme Sfi I, RNA extraction kit, RT-PCR Kit and DNA polymerase were purchased from Takara (Dalian, China). Vectors pComc3XSS and SBP-pET25b containing streptavidin-binding peptide (SBP) gene (MDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREP) were obtained from Nb-Biolab (Chengdu, China). Helper phage M13KO7 was obtained from New England Biolabs LTD. (Ipswich, MA). pET25b+ vector, E. coli ER2738 and E. coli Rosetta were saved in our laboratory. Primers were synthesized by Sangon Biotech (Shanghai, China). HRP-conjugated anti-M13 phage antibody was obtained from Abcam (Cambridge, UK). Standards of cancer biomarker AFP, carbohydrate antigen 19-9 (CA19-9), carbohydrate antigen 15-3 (CA15-3), carbohydrate antigen 12-5 (CA12-5), carcinoembryonic antigen (CEA), cytokeratin 19 fragment (CY21-1), human epididymis protein 4 (HE4), prostate-specific antigen (PSA) and squamous cell carcinoma antigen (SCCA) were purchased from Linc-Bio Science (Shanghai, China). Anti-AFP monoclonal antibodies were purchased from HyTest (Turku, Finland). SA-PE and magnetic microspheres were purchased from Luminex (Austin, TX). PageRuler Plus Prestained Protein Ladder was purchased from Thermo Fisher Scientific (Waltham, MA). All organic solvents and inorganic chemicals were reagent grade. HydroFlex magnetic microplate washer was from Tecan (Männedorf, Switzerland). Cobas E601 electrochemiluminescence immunoassay analyzer was from Roche (Basel, Switzerland). Luminex-200 system was from Luminex Co. (Austin, TX).2.2. Alpaca immunization and phage-displayed library construction
The alpaca immunization was conducted by Nb-Biolab (Chengdu, China). Briefly, a two-year-old female alpaca was immunized with 500 μg of human AFP in complete Freund's adjuvant by subcutaneous injection for the first time, and three additional injections mixed with incomplete Freund's adjuvant were performed at 2 weeks intervals. One week after the final booster, 150 mL of blood was drawn and peripheral blood lymphocytes (PBLs) were isolated by the Lymphocyte Separation Medium (Solarbio Life Sciences, Beijing, China). The cells were washed three times with PBS and total RNA was extracted with a RNAprep pure Cell/Bacteria Kit (TIANGEN Biotech CO., LTD., Beijing, China). The phage-displayed Nb library was constructed as described previously with some modifications.24 Briefly, 10 μg of total RNA were reverse transcribed using the oligodT primers to synthesize the first strand of complementary DNA using a PrimeScript II 1st Strand cDNA Synthesis Kit (Takara Bio, Beijing, China). The VH and VHH genes were amplified by a two-step nested PCR as shown in Tables S1 and S2.† For the first step PCR, primers AlpVh-LD (CTT GGT GGT CCT GGC TGC) and CH2-R (GGT ACG TGC TGT TGA ACT GTT CC) were used to amplify the VH gene. Subsequently, primers Alp-Vh-F1 (CAT GCC ATG ACT GTG GCC CAG GCG GCC CAG KTG CAG CTC GTG GAG TC) and AlpVhh-R1 (CAT GCC ATG ACT CGC GGC CGG CCT GGC CAT GGG GGT CTT CGC TGT GGT GCG) or Alp-Vh-F1 and AlpVhh-R2 (CAT GCC ATG ACT CGC GGC CGG CCT GGC CGT CTT GTG GTT TTG GTG TCT TGG G) were used to amplify the VHH gene with short or long hinge region in the second step PCR. The final products of ∼400 bp DNA fragments were extracted by agarose gel purification and digested by restriction enzyme SfiI. Then the digested products were cloned in the phagemid vector pComb3XSS. A total of 10 μg of the constructed phagemid were transfected into E. coli ER2738 competent cells by electroporation (25 μF, 200 Ω, 2.5 kV). The transformed cells were resuspended in SOC medium with gentle shaking (37 °C, 1 h) for resuscitation and spread on the 2YT-ampicillin agar plates. After culturing at 37 °C for 12 h, the transformants were rinsed with 2YT medium and inoculated into the 2YT medium containing 100 μg mL−1 of ampicillin with vigorous shaking (37 °C, 2 h). Then the cells were infected with helper phage M13KO7 and cultured overnight with shaking at 37 °C. The rescued phage was precipitated by PEG/NaCl and resuspended in PBS. The prepared phage-displayed Nb library was stored at 4 °C prior to use.2.3. Biopanning and identification of Nbs against AFP
The phage-displayed Nb library was subjected to three rounds of biopanning on ELISA microplate wells according to our previously reported protocol with some modification.24 Briefly, the wells were coated with successively decreasing contents of AFP (1, 0.5 and 0.25 μg) overnight at 4 °C and then blocked with 3% BSA or 3% OVA alternatively at 37 °C for 2 h. The phage library (1011 colony forming units) was preincubated with blocking buffer at 37 °C for 1 h before adding to the AFP-coated wells. After 1.5 h incubation at 37 °C, unbound and weak affinity phage was removed by washing with PBS and PBST. Then bound phage was eluted by adding 100 μL of elution buffer (Gly-HCl, 0.2 mol L−1, pH 2.2) with gentle shaking for 8 min and immediately neutralized with 10 μL of Tris–HCl (1 mol L−1, pH 9.1). Neutralized phage was used to infect E. coli ER2738 cells for titration and amplification according to standard protocols. After three rounds of biopanning, 48 independent colonies were rescued using helper phage M13KO7. The characterization of rescued phage particles were identified by indirect phage enzyme-linked immunosorbent assay (phage-ELISA). The detailed parameters of biopanning were found in the ESI.†2.4. Construction, expression and characterization of Nb-SBP
The VHH fragments of positive phage clones were amplified with primers AF-Nco (TCG CGG CCC AGC CGG CCA TGG CCC AGK TGC AGC TCG TGG AGT CNG GNG G) and AR-Not (CGA GTG CGG CCG CTT GTG GTT TTG GTG TCT TGG G), which was flanked by restriction enzyme sites Nco I and Not I (5′–3′), respectively. The purified double enzyme digestion products of the VHH fragments were ligated into the similarly digested expression vector SBP-pET25b with the T4 DNA ligase. The ligation products was transformed into the E. coli Rosetta competent cells by heat shock (42 °C, 90 s), followed by spreading the cells on the 2YT-ampicillin agar plates. The transformants were sent to Sangon Biotech (Shanghai, China) for nucleic acid sequencing. The auto-induction expression and affinity purification of Nb-SBP was performed as described previously.25 To characterize the purified Nb-SBP, the binding affinity of Nb-SBP to AFP was tested by a microscale thermophoresis-based Monolith NT.115 system (NanoTemper Technologies, Munich, Germany) as described previously.252.5. Preparation and characterization of immunomagnetic microspheres
To prepare the immunomagnetic microspheres (IMMps), the carboxylated magnetic microspheres (MMps) with the diameter of 5.6 μm were labeled with the anti-AFP monoclonal antibody (mAb) by the EDC/NHSS method according to the manufacturer's manual with some modification. The stock vial of MMps was shaken on a vortexer for 10 s and sonicated for 10 s to disperse the microspheres prior to use. Then 200 μL of MMps (5 × 105) were washed twice with 500 μL of activation buffer using a magnetic separator. After washing, the MMps were suspended in 500 μL activation buffer containing Sulfo-NHS (50 μg mL−1) and EDC (80 μg mL−1) for 20 min at room temperature with gently rotated to activate the carboxyl groups. After washing, the MMps were immediately suspended in 500 μL activation buffer containing mAb (10 μg mL−1) and gently rotated at room temperature for 2 h. The modified MMps were separated and rinsed with wash buffer to remove surplus mAb, followed by blocking with 5% BSA for 45 min and washing three times with wash buffer. Then, the IMMps were resuspended in 1 mL of wash buffer and stored at 4 °C until needed. For characterization of the IMMps, the mixture of 25 μL of IMMps and 75 μL of the biotin/goat anti-mouse IgG monoclonal antibody (bio-Ab2) solution was incubated at 37 °C for 1 h. The IMMps were rinsed with PBST and incubated with 75 μL of SA-PE at 37 °C for 30 min. After rinsing with PBST and resuspending in 100 μL of assay buffer, the median fluorescent intensity (MFI) of the IMMps@bio-Ab2@SA-PE complex was determined by the Luminex-200 system.2.6. Procedures of Nb-SBP-mediated Luminex fluoroimmunoassay
In the Nb-SBP-mediated Luminex fluoroimmunoassay (NS-LFIA), the separation and detection of AFP were based on IMMps and fluorescence protein introduced by Nb-SBP, respectively. Procedures of the system were shown as follows, where the washing steps were performed on a magnetic plate washer. First, 25 μL of IMMps and 25 μL of reaction buffer were added into 25 μL of AFP solution with different concentrations and incubated overnight at 4 °C in the dark with shaking. After thrice washing with 300 μL of PBST, 75 μL of Nb-SBP solution was added and incubated at 37 °C for 1 h to form sandwich immune complexes. The unbound Nb-SBP was rinsed three times with 300 μL of PBST, followed by the addition of 75 μL of SA-PE and incubation at 37 °C for 30 min. The immune complexes were washed another three times with 300 μL of PBST and resuspended with 100 μL of assay buffer. Then the fluorescence intensity of immune complexes was measured on the Luminex 200 instrument referring to the user's manual for proper operation. The standard curve for quantitative analysis of AFP was constructed by plotting the MFI against the AFP concentration, where the MFI refers to the median fluorescent intensity of SA-PE on 50 IMMps.2.7. Selectivity of Nb-SBP-mediated Luminex fluoroimmunoassay
To evaluate the selectivity of the NS-LFIA, the MFI of eight common cancer biomarkers (CA19-9, CA15-3, CA12-5, CEA, CY211, HE4, PSA and SCCA, 1 μg mL−1) was tested by the Luminex assay and compared with that of AFP (50 ng mL−1). A blank control test was conducted by replacing cancer biomarker with PBS under the same experimental conditions.2.8. Sample analysis and assay validation
The AFP-negative serum sample and clinical serum samples were collected from the Second Affiliated Hospital of Hainan Medical University. The negative serum sample was used for the recovery experiment and the AFP-spiked serum samples were subjected to the NS-LFIA without further pretreatment. To evaluate the practicability of the proposed method, the clinical serum samples were directly determined by the NS-LFIA and validated by the Cobas E601 electrochemiluminescence immunoassay analyzer in accordance with the manufacturer's instructions.2.9. Live subject statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Hainan University and approved by the Animal Ethics Committee of Hainan University.3. Results
3.1. Selection of AFP-specific VHH phages
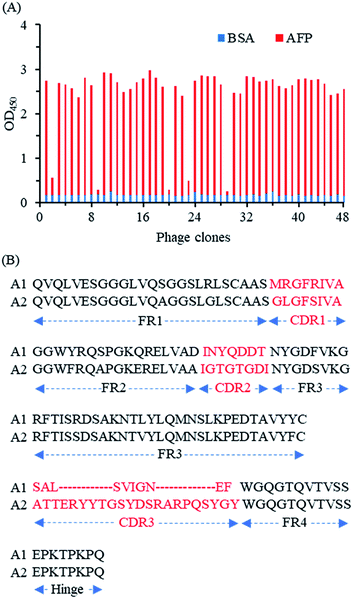
After four immunizations, good immune response was obtained in the alpaca with high antibody titer for AFP (data not shown). Then a phage-displayed VHH library of 1 × 108 individuals was constructed to select AFP-specific VHH phages. In total three rounds of biopanning were performed to isolate the AFP-specific VHH phages, and the biopanning conditions were gradually tightened to ensure high-affinity clones (Table S3†). Compared to the original phage library, significant enrichment of the phages with AFP recognition ability was observed after three rounds of biopanning, where the amount of eluted phage was enriched from an initial 1.8 × 106 cfu to 1.6 × 109 cfu (Table S4†). Then, 48 randomly selected phage clones from the third round were identified by phage-ELISA. Among them, 43 phage clones were identified as positive (Fig. 2A). Thereafter the positive clones were sequenced and analyzed, the DNA sequencing results showed that the 43 positive phage clones contain two different DNA sequences namely as A1 and A2 (Fig. 2B). Both A1 and A2 have a short hinge and an intramolecular disulfide bond between framework 1 (FR1) and FR3. However, they are significantly different in complementary determining regions (CDRs), especially the CDR3, which could lead to distinct properties of both clones.3.2. Preparation and characterization of Nb-SBP
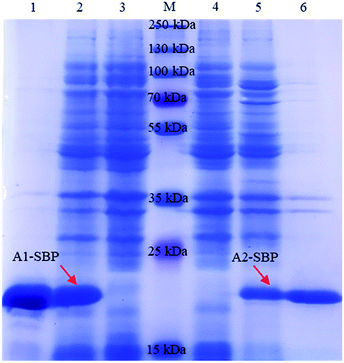
For the expression of Nb-SBP, the VHH fragments of A1 and A2 were subcloned into the expression vector SBP-pET25b, respectively. The positive recombinant vectors confirmed by colony PCR and DNA sequencing were transformed into E. coli Rosetta cells. After auto-induction, soluble fusion protein in the cells was released into the supernatant by ultrasonication and then purified by Ni-NTA affinity chromatography. The expression and purification of the fusion protein were characterized by 12% SDS-PAGE (Fig. 3). A clear target band of approximately 20 kDa was detected in the induced cells, while it was absent in the noninduced cells. The A1-SBP has a higher expression yield (100 mg L−1) in comparison with the A2-SBP (30 mg L−1). In addition, both fusion proteins were purified well with purity exceeding 90% (Fig. 3). Moreover, the A1-SBP shows stronger affinity than the A2-SBP (data not shown), therefore the following experiments were conducted with the A1-SBP.3.3. Screening of paired antibodies based on Luminex-200 system
To obtain paired antibodies that can recognize different epitopes on AFP, the A1-SBP (detection antibody) and 14 commercial mAbs (capture antibody) were tested according to standard Luminex-200 protocols. The mAbs (M3008-1, 100015, LDZL0001, H036001, 4A3, 5H7, 100014, H036002, 5153-01M, XA2, 6483-01M, XFPA, and JFZL0002) were covalently immobilized on 14 MMps encoded as 12, 14, 18, 20, 22, 26, 28, 44, 69, 72, 74, 76, 78 and 80, respectively. The IMMps were confirmed to be well prepared since the MFI of the IMMps@bio-Ab2@SA-PE complex increased as the concentration of bio-Ab2 increased as shown in Fig. S1.† Moreover, dramatically enhanced fluorescent signals were produced in five tested mAbs (M3008-1, 100015, H036001, 5H7 and 6483-01M) with the increase of AFP concentration, indicating that these mAbs and A1-SBP could recognize different epitopes of AFP (Table S5†). Among them, the mAb-5H7 produced the highest fluorescence intensity at both concentrations of AFP (4.88 and 62.7 ng mL−1). Therefore, the mAb-5H7/A1-SBP antibody pair was selected for AFP detection in the subsequent experiments.3.4. Optimization of Nb-SBP-mediated Luminex fluoroimmunoassay
To obtain the highest fluorescence intensity and the best sensitivity, orthogonal experiments of L9 (3)3 were designed to optimize the concentrations of mAb-5H7, A1-SBP and SA-PE when the concentration of target AFP was fixed at 30 ng mL−1. The MFI was recorded to determine the optimal parameters. As shown in Table 1, the optimal combination was selected as follows: 5 μg mL−1 of mAb-5H7, 2 μg mL−1 of A1-SBP, and 3.5 μg mL−1 of SA-PE, under which the MFI was 1178.5. To ensure the reliability of the results, the above optimizations were performed with another concentration of AFP (15 ng mL−1). As shown in Table S6,† the strongest MFI was observed under the same optimal parameters, showing a fairly good consistency.| Treatment name | Factors | Results | ||
|---|---|---|---|---|
| mAb-5H7 (μg mL−1) | A1-SBP (μg mL−1) | SA-PE (μg mL−1) | ||
| 1 | 2.5 | 1 | 1.5 | 786.5 |
| 2 | 2.5 | 2 | 2.5 | 952 |
| 3 | 2.5 | 3 | 3.5 | 1069 |
| 4 | 5 | 1 | 2.5 | 862.5 |
| 5 | 5 | 2 | 3.5 | 1178.5 |
| 6 | 5 | 3 | 1.5 | 818 |
| 7 | 10 | 1 | 3.5 | 1145 |
| 8 | 10 | 2 | 1.5 | 726.5 |
| 9 | 10 | 3 | 2.5 | 796 |
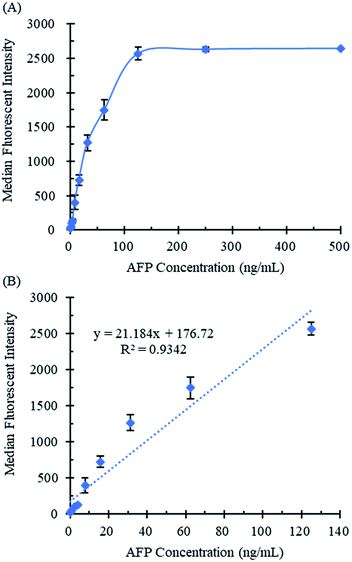
The quantitative curve of the NS-LFIA for AFP was constructed by plotting the MFI against the concentrations of AFP under the optimal experimental conditions, respectively. As shown in Fig. 4A, the MFI increased as the AFP concentration increased from 0 to 125 ng mL−1. Moreover, the fluorescent signal remained stable when the AFP concentration exceeded 125 ng mL−1. It could be attributed to the fluorescence saturation of IMMps since the number of monoclonal antibody molecule immobilized on the magnetic microspheres is restricted. The linear relationships between the median fluorescence intensity and the concentration of AFP ranging from 0 ng mL−1 to 125 ng mL−1 was shown in Fig. 4B and described as y = 21.184x + 176.72, with a reliable correlation coefficient R2 = 0.9342, where y is the MFI and x is the AFP concentration. Moreover, the limit of detection for AFP was 0.237 ng mL−1 which was calculated as the mean value plus threefold standard deviations of 20 blank samples.
3.5. Selectivity of Nb-SBP-mediated Luminex fluoroimmunoassay
The selectivity of the proposed NS-LFIA for AFP was evaluated by comparing the MFI of AFP (50 ng mL−1) with 8 common cancer biomarkers (1 μg mL−1), including CA19-9, CA15-3, CA12-5, CEA, CY21-1, HE4, PSA and SCCA. A blank control test was conducted by replacing cancer biomarker with PBS solution under the same experimental conditions. As shown in Table S7,† the NS-LFIA exhibited a high specificity in recognition of AFP and ignorable cross reaction with other cancer biomarkers. These results clearly demonstrated the high selectivity of the proposed method for detection of AFP.3.6. Performance evaluation of Nb-SBP-mediated Luminex fluoroimmunoassay
To evaluate the reproducibility of the developed method for AFP, the NS-LFIA was performed to detect AFP in the spiked serum sample. Three spiked serum with low, medium, and high levels of AFP concentrations were prepared for intra- and inter-assay analysis. As displayed in Table 2, the average recoveries for the intra-assay range from 99.7% to 106%, with the relative standard deviation (RSD) from 3.7% to 9.6%. Meanwhile, the inter-assay ranged from 98.2% to 110%, with the RSD from 2.8% to 13.8%, respectively. These results demonstrated the acceptable accuracy and satisfactory repeatability of the developed NS-LFIA for AFP detection in serum samples.| Spiked AFP (ng mL−1) | Intra-assay precisiona | Inter-assay precisionb | ||||
|---|---|---|---|---|---|---|
| Recovered concentration | Recovery (%) | RSD (%) | Recovered concentration | Recovery (%) | RSD (%) | |
| a The assay was carried out in triplicates on the same day.b The assay was performed on three consecutive days. | ||||||
| 2 | 2.12 ± 0.18 | 106 | 8.5 | 2.20 ± 0.15 | 110 | 6.8 |
| 10 | 10.52 ± 0.96 | 105.2 | 9.6 | 10.49 ± 0.11 | 104.9 | 13.8 |
| 50 | 49.73 ± 1.82 | 99.7 | 3.7 | 49.08 ± 0.79 | 98.2 | 2.8 |
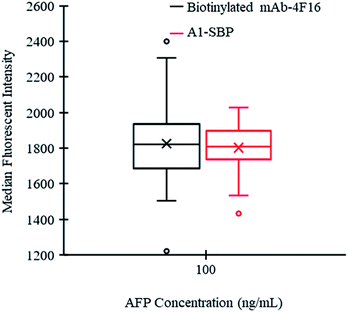
For the developed NS-LFIA, the fluorescence intensity of SA-PE on 50 IMMps was measured and the median value was taken as the result. To evaluate the potential to reduce MMps consumption of the developed method, the data homogeneity of the NS-LFIA was investigated and compared with that of the traditional biotinylated mAb-4F16 (commercial paired antibody for the mAb-5H7)-mediated Luminex fluoroimmunoassay (BM-LFIA). To obtain the comparable fluorescence intensity value, the concentration of biotinylated mAb-4F16 was adjusted when the concentration of target AFP was fixed at 100 ng mL−1. Two sets of data were collected with very close median fluorescence intensity values of 1821.5 for BM-LFIA and 1808.5 for NS-LFIA, respectively. As shown in Fig. 5, the proposed NS-LFIA shows a smaller range of the inter-quartile (from 25th to 75th percentile). Furthermore, the NS-LFIA also shows a lower standard deviation (from 207.4 to 134.3) and RSD (from 11.4% to 7.5%) compared with the BM-LFIA. Hence, these results illustrate that the application of A1-SBP in the Luminex-200 system could significantly improve the homogeneity of the fluorescent signals by comparison with the biotinylated mAb.
3.7. Practicability of Nb-SBP-mediated Luminex fluoroimmunoassay
Considering the good selectivity, satisfactory repeatability and excellent sensitivity of the developed strategy toward AFP, the practicability of this proposed method was further explored in real samples according to the general assay procedures. In total forty clinical serum samples were analyzed by both the NS-LFIA and the well-known Cobas E601 electrochemiluminescence immunoassay analyzer. As shown in Fig. 6, a good correlation was obtained between the two methods (R2 = 0.9894), which indicates the potential of the NS-LFIA for early and accurate detection of AFP and the rest of disease biomarkers in real clinical samples.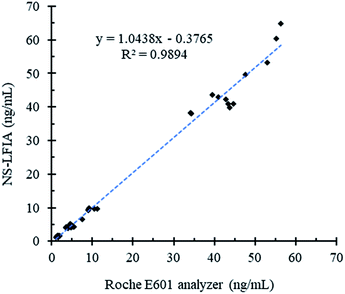 | ||
| Fig. 6 Correlation analysis between the Cobas E601 chemiluminescence immunoassay analyzer and the proposed NS-LFIA in quantifying AFP from 40 clinical serum samples. | ||
4. Conclusion
Herein we constructed a phage-displayed Nb library from an alpaca immunized with AFP and two unique sequences of AFP-specific Nbs (A1 and A2) were isolated after three rounds of biopanning. Two Nbs tagged with SBP (A1-SBP and A2-SBP) were constructed through simple genetic manipulation and a Luminex fluoroimmunoassay using the A1-SBP (NS-LFIA) was developed for detecting AFP in serum. Due to the application of the Luminex-200 system, the NS-LFIA can handle a large number of serum samples without matrix interference and implement simultaneous detection of multiple targets. This study illustrates the value of Nbs in analyzing macromolecules. Nbs have the advantages of monoclonal antibody and are more low-cost and time-saving to develop and produce. Moreover, the single domain property of Nbs makes it very easy for genetic manipulation as demonstrated here. We also demonstrated that the SBP tag fusion is superb in macromolecule analysis. Not only does the SBP tag fusion technology yield streptavidin-binding molecules with defined analyte/reporter molar ratio, but the Nb-SBP can significantly improve the homogeneity of the fluorescent signals in NS-LFIA compared with the traditional biotinylated mAb, which can reduce MMps consumption and test cost.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was funded by the Natural Science Foundation of Hainan Province (grant number 219QN149 and 2019RC119) and the National Natural Science Foundation of China (grant number 31760493 and 31901800). Partial support was provided by NIEHS/Superfund Research Program (grant number P42 ES004699) and NIEHS RIVER Award (grant number R35ES030443). The APC was funded by the Scientific Research Foundation of Hainan University [grant number KYQD1631 and KYQD(ZR)1957].References
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- W. Chen, R. Zheng, P. D. Baade, S. Zhang, H. Zeng, F. Bray, A. Jemal, X. Q. Yu and J. He, Ca-Cancer J. Clin., 2016, 66, 115–132 CrossRef PubMed.
- Y. Shieh, M. Eklund, G. F. Sawaya, W. C. Black, B. S. Kramer and L. J. Esserman, Nat. Rev. Clin. Oncol., 2016, 13, 550–565 CrossRef CAS PubMed.
- A. Wöckel, U. S. Albert, W. Janni, A. Scharl, R. Kreienberg and T. Stüber, Dtsch Arztebl Int, 2018, 115, 316–323 Search PubMed.
- J. Bruix, M. Reig and M. Sherman, Gastroenterology, 2016, 150, 835–853 CrossRef PubMed.
- L. Ping, P. Yin, H. Rui, Y. Tan, Z. Li, G. Qiu, Z. Yin, X. Xie, X. Wang and W. Chen, Hepatology, 2018, 67, 662–675 CrossRef PubMed.
- J. Peng, Z. Guoxin, K. H. Kee, H. Seon-Ah, Z. Jin, S. Qiao, Z. Cui, L. Gong, Y. Jinah and K. Sanghee, Dis. Markers, 2011, 30, 307–315 CrossRef CAS.
- Y. Leng, K. Sun, X. Chen and W. Li, Chem. Soc. Rev., 2015, 44, 5552–5595 RSC.
- S. F. Parsa, A. Vafajoo, A. Rostami, R. Salarian, M. Rabiee, N. Rabiee, G. Rabiee, M. Tahriri, A. Yadegari, D. Vashaee, L. Tayebi and M. R. Hamblin, Anal. Chim. Acta, 2018, 1032, 1–17 CrossRef CAS PubMed.
- C. Yuan, Y. Deng, X. Li, C. Li, Z. Xiao and Z. Liu, Anal. Chem., 2018, 90, 8178–8187 CrossRef CAS PubMed.
- M. Lowe, A. Spiro, Y.-Z. Zhang and R. Getts, Cytometry A, 2004, 60A, 135–144 CrossRef CAS PubMed.
- H. H. Gorris and O. S. Wolfbeis, Angew. Chem. Int. Ed., 2013, 52, 3584–3600 CrossRef CAS PubMed.
- T. Sheng, Z. Xie, P. Liu, J. Chen and D. Deng, Adv. Mater. Interfaces, 2018, 5, 1870096 CrossRef.
- X. Qu, F. Bian, Q. Guo, Q. Ge, Q. Sun and X. Huang, Anal. Chem., 2018, 90, 12051–12058 CrossRef CAS PubMed.
- F. Song, P. S. Tang, H. Durst, D. T. Cramb and W. C. Chan, Angew. Chem. Int. Ed., 2012, 51, 8773–8777 CrossRef CAS PubMed.
- V. Biju, Chem. Soc. Rev., 2014, 43, 744–764 RSC.
- V. Crivianu-Gaita, A. Romaschin and M. Thompson, Biochem. Biophys. Rep., 2015, 2, 23–28 Search PubMed.
- S. Farajnia, V. Ahmadzadeh, A. Tanomand, K. Veisi, S. A. Khosroshahi and L. Rahbarnia, Immunopharm. Immunot., 2014, 36, 297–308 CrossRef CAS PubMed.
- F. Khodabakhsh, M. Behdani, A. Rami and F. Kazemi-Lomedasht, Int. Rev. Immunol., 2018, 37, 316–322 CrossRef CAS PubMed.
- J.-P. Salvador, L. Vilaplana and M.-P. Marco, Anal. Bioanal. Chem., 2019, 411, 1703–1713 CrossRef CAS PubMed.
- V. Crivianu-Gaita and M. Thompson, Biosens. Bioelectron., 2016, 85, 32–45 CrossRef CAS PubMed.
- H. English, J. Hong and M. Ho, Antib. Ther., 2020, 3, 1–9 Search PubMed.
- Z. Sun, J. Lv, X. Liu, Z. Tang, X. Wang, Y. Xu and B. D. Hammock, Anal. Chem., 2018, 90, 10628–10634 CrossRef CAS PubMed.
- X. Liu, Y. Xu, Y.-h. Xiong, Z. Tu, Y.-p. Li, Z.-y. He, Y.-l. Qiu, J.-h. Fu, S. J. Gee and B. D. Hammock, Anal. Chem., 2014, 86, 7471–7477 CrossRef CAS PubMed.
- X. Wang, Q. Chen, Z. Sun, Y. Wang, B. Su, C. Zhang, H. Cao and X. Liu, Int. J. Biol. Macromol., 2020, 151, 312–321 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1: the validation of monoclonal antibody-conjugated magnetic microspheres, Table S1: the 1st round PCR system, Table S2: the 2nd round PCR system, Table S3: the biopanning conditions for the selection of AFP-specific VHH phages, Table S4: the enrichment of phage in three rounds biopanning, Table S5: the screening of paired antibodies based on Luminex-200 system, Table S6: the orthogonal test for parameter optimization of the NS-LFIA, Table S7: the selectivity of the NS-LFIA, Table S8: the LOD of the NS-LFIA. See DOI: 10.1039/d0ra04210b |
| This journal is © The Royal Society of Chemistry 2020 |