Open Access Article
Open Access ArticleIntegrated analysis of lncRNA–mRNA networks associated with an SLA titanium surface reveals the potential role of HIF1A-AS1 in bone remodeling†
Yan Zheng a,
Yunfei Zhengb,
Lingfei Jiacd,
Yu Zhang*a and
Ye Lin*a
a,
Yunfei Zhengb,
Lingfei Jiacd,
Yu Zhang*a and
Ye Lin*a
aDepartment of Oral Implantology, Peking University School and Hospital of Stomatology, 22 Zhongguancun South Avenue, Haidian District, Beijing 100081, China. E-mail: yorcklin@263.net; yuzhang76@bjmu.edu.cn; Fax: +86-10-62173402; Tel: +86-10-62179977 ext. 5344
bDepartment of Orthodontics, Peking University School and Hospital of Stomatology, Beijing 100081, China
cDepartment of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology, Beijing 100081, China
dCentral Laboratory, Peking University School and Hospital of Stomatology, Beijing 100081, China
First published on 2nd June 2020
Abstract
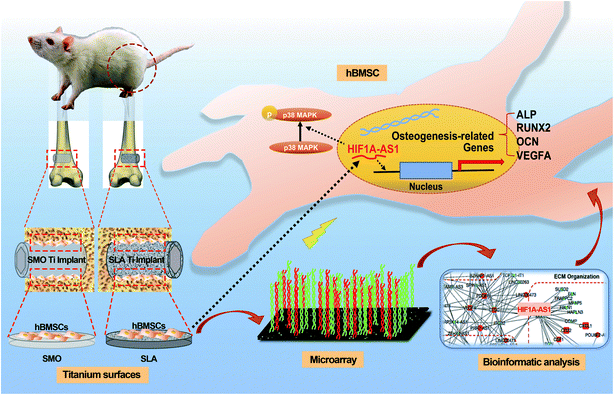
Microstructured titanium surface implants, such as typical sandblasted and acid-etched (SLA) titanium implants, are widely used to promote bone apposition in prosthetic treatment by dental implants following tooth loss. Although there are multiple factors associated with the superior osseointegration of an SLA titanium surface, the molecular mechanisms of long noncoding RNAs (lncRNAs) are still unclear. In this study, we characterized smooth (SMO) and SLA surfaces, and compared the osteoinduction of these surfaces using human bone marrow-derived mesenchymal stem cells (hBMSCs) in vitro and implants in a rat model in vivo. Then, we used microarrays and bioinformatics analysis to investigate the differential expression profiles of mRNAs and lncRNAs on SMO and SLA titanium surfaces. An lncRNA–mRNA network was constructed, which showed an interaction between lncRNA HIF1A antisense RNA 1 (HIF1A-AS1) and vascular endothelial growth factor. We further found that knockdown of HIF1A-AS1 significantly decreased osteogenic differentiation of hBMSCs. This study screened SLA-induced lncRNAs using a systemic strategy and showed that lncRNA HIF1A-AS1 plays a role in promotion of new bone formation in the peri-implant area, providing a novel insight for future surface modifications of implants.
1. Introduction
Successful dental implantation depends on well-established osseointegration that is highly relevant to the design of implants.1 In contrast to earlier smooth (SMO) titanium surfaces, micro-roughened titanium surfaces such as sandblasted and acid-etched (SLA) surfaces consist of micro- and submicro-scale pits on the microscale, which mimic the characteristics of bone. These structures play an important role in promoting the human bone marrow-derived mesenchymal stem cell (hBMSC) response to enhance bone formation and stabilize implants.2,3 Although SLA surfaces have been widely used in the clinic and demonstrated to facilitate osseointegration,4 studies have indicated that osseointegration at the bone-implant interface is still not sufficient to induce ideal biological responses, and some titanium-based surfaces probably induce early pathogenic bacteria adhesion.1,5–7 Thus, knowing the mechanisms of improving osteogenesis and inducing new bone formation at the bone-implant interface with satisfactory surface modification is a pressing health issue. To modify SLA surfaces, previous studies have focused on various bioactive molecules such as growth factors, extracellular matrix (ECM) components, and functional peptides.8–10 Some of them have been loaded onto titanium implants to promote osteogenesis.11–13 However, because the half-life of proteins is short and they are degraded easily, the biological effects appear to be limited. Currently, increasing evidence has shown that non-coding RNAs (ncRNAs), which do not encode proteins but represent the majority of the human transcriptome, play a crucial role in a wide range of biological functions such as gene expression in physiology and development, RNA splicing, chromatin remodeling, and signal transduction.14 Some studies and present references have focused on surfaces functionalized by ncRNAs. Small interfering RNAs (siRNAs) and some microRNAs (miRNAs) can be introduced into cells and act as mediators of osteogenesis-associated biological processes by RNA interference and delivery technologies.12,15,16 However, the studies associated with lncRNA were limited and still deserved to explore.Long non-coding RNAs (lncRNAs), which are commonly defined as ncRNAs of >200 nucleotides and constitute the largest class of ncRNAs, are currently the subject of research. Numerous lncRNAs that mainly exist in chromatin and the nucleus are 3′-polyadenylated, 5′-capped, and multi-exonic and transcribed by RNA polymerase II similarly to mRNA transcripts.17,18 As previous studies revealed, lncRNAs usually act as key regulators synergistically with mRNAs and rarely exert a biological function alone.19 Several lncRNAs, such as H19, MEG3, MIR31HG, and DANCR, modulate the differentiation of osteoblasts and the formation of bone mass by interacting with related mRNAs.20–23 Furthermore, our previous study reported that RNA interference by siRNA targeting lncRNA MIR31HG can be applied to modification of titanium surfaces to improve their osteoconductivity and osseointegration.24 However, it is unknown which lncRNA should be selected for titanium surface biomodification because no studies have systematically examined the differences in lncRNA expression of hBMSCs on SMO and SLA titanium surfaces. We speculated that the different titanium surfaces themselves led to differential expression of lncRNAs and related mRNAs in hBMSC and some of them might be involved in osteogenesis.
LncRNA HIF1A antisense RNA 1 (HIF1A-AS1 or 5′aHIF-1α), located on chromosome 14 (14q23.2) and originating from the 5′ region of the HIF-1α gene, was identified in 2010 by Baranello et al.25 In recent years, a few correlative studies have shown that HIF1A-AS1 can not only be used for cancer and cardiovascular system disease diagnoses, but also has emerging roles in apoptosis and proliferation of vascular smooth muscle cells and osteoblastic differentiation of periodontal ligament cells under hypoxia.26–30 Xu et al. suggested a positive regulatory effect of HIF1A-AS1 on osteogenesis.31 However, the role and mechanism of HIF1A-AS1 in osteogenic differentiation of hBMSCs need to be defined.
Our study investigated lncRNAs that can be used for surface modification in the future. To this end, we compared the expression pattern of lncRNAs associated with osteogenic differentiation of hBMSCs cultured on two titanium implant surfaces (SMO and SLA) and assessed the lncRNA–mRNA interactions on SLA surfaces. Our results demonstrated that the lncRNA HIF1A-AS1 responded to the topography of microstructured titanium implant surfaces and affected the osteogenic differentiation of hBMSCs. The experimental process and related mechanism are schematically illustrated in Scheme 1.
2. Material and methods
2.1 Sample preparation
Titanium specimens were divided into SMO and SLA groups. Pure titanium disks (Wego Jericom Biomaterials Co., Weihai, China) were cut into round sheets with a height of 1 mm and two diameters, 15 and 34 mm, which matched 24-well and 6-well plates, respectively. The SMO disks were polished by a series of silicon carbide sandpapers (from No. 240 to 2000 grits) and then cleaned by acetone, absolute alcohol, and deionized water (dH2O) in sequence. Some SMO disks were further processed by blasting with 0.25–0.50 mm Al2O3 grit, acid etched followed by etching with hot HCl (10–16%)/H2SO4 (68–75%) at 80–90 °C, and then cleaned with nitric acid and deionized water in sequence to obtain SLA surfaces. Cylindrical titanium implants (diameter: 2 mm; length: 4 mm, Wego Jericom Biomaterials Co., Weihai, China) used for in vivo experiments employed these methods as well.2.2 Surface characterization
The morphology of SMO and SLA surfaces was observed by scanning electron microscopy (SEM; S-4800, Hitachi, Tokyo, Japan) at 15.0 kV. The crystallinity of the surfaces was examined by X-ray diffraction (AXS D8 Advanced; Bruker, Karlsruhe, Germany) with a Cu Kα radiation source (λ = 1.541 Å) at 40 kV. The diffraction angles (2θ) ranged from 3° to 90° with an incremental step size of 0.06° s−1. Contact angle (CA) measurements were performed using a Contact Angle Instrument (DSA10, Kruss, Hamburg, Germany) equipped with a camera and SCA20 video software (DataPhysics, Filderstadt, Germany). Deionized water (4 μL) was dropped onto each sample at room temperature (26 ± 1 °C) at least five times. Subsequently, the surface energy was calculated using the associated instrument software (Kino Industry, New York, NY, USA). Surface characterization was performed on more than three titanium specimens in accordance with the grouping.2.3 Cell culture and osteogenic induction
Human bone marrow-derived mesenchymal stem cells (hBMSCs; ScienCell Company, San Diego, CA, USA) were cultured in alpha-minimum essential medium (α-MEM; Gibco, Grand Island, NY, USA) containing 10% (v/v) fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 100 U mL−1 penicillin G (Gibco), and 100 mg mL−1 streptomycin (Gibco) at 37 °C with 5% CO2. Osteogenic medium (OM) consisted of αMEM containing 10% (v/v) FBS, 100 U mL−1 penicillin G, 100 mg mL−1 streptomycin, 10 nM dexamethasone (Sigma-Aldrich, Saint Louis, MO, USA), 10 mM β-glycerophosphate (Sigma-Aldrich), and 50 mg mL−1 L-ascorbic acid (Sigma-Aldrich). Passage 5–7 cells were used for experiments.2.4 Cell adhesion and proliferation assays
Cell attachment analysis was performed on 24-well titanium disks. For SEM observation, hBMSCs were seeded onto SMO and SLA surface titanium disks at 1 × 104 cells per well. After culture for 2, 6, and 24 hours, the cells were washed with phosphate-buffered saline (PBS), fixed with 2.5% glutaraldehyde for 30 minutes at 4 °C, dehydrated through sequential ethanol concentrations, and critical point dried. Prior to the scan by field emission SEM (S-3000N; Hitachi, Tokyo, Japan) at 15.0 kV, samples adhered onto aluminum stubs were sputter-coated with gold.Cell proliferation analysis was performed using Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan), according to the manufacturer's instructions, within the first 6 days on 24-well disks. Briefly, hBMSCs were cultured on the different titanium surfaces. At the scheduled time, cells were incubated in CCK-8 reagent for 2 hours. Subsequently, the culture medium of the reaction was analyzed by a microplate reader (ELX-808 Absorbance Microplate Recorder; BioTek, Winooski, VT, USA) at 450 nm. The optical density absorbance value as units was regarded as the result.
2.5 Alizarin red S (ARS) staining
hBMSCs seeded on 6-well disks and cultured in osteogenic differentiation medium for 7 and 21 days were washed with PBS, fixed with 70% cold ethanol for 30 minutes, washed twice with distilled water, and then stained with 2% (w/v) Alizarin Red S (ARS; Sigma-Aldrich), pH 4.3, for 20 minutes at room temperature to evaluate calcium nodules. After staining, the samples were washed with distilled water and PBS. Then, images of the ARS-stained substrates were captured under an optical microscope (M125; Leica, Wetzlar, Germany). This method was also applied to evaluate differentiation after transfection on days 7 and 14.2.6 Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
After hBMSCs were cultured on 6-well disks in proliferation medium overnight and then osteogenically induced for 7 and 21 days, total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and then reverse transcribed to cDNA with a PrimeScript™ RT Reagent Kit (Takara, Tokyo, Japan), according to the manufacturers' instructions. Quantitative PCR was performed with a 7500 Real-Time PCR Detection System (Applied Biosystems, Foster City, CA, USA) using SYBR Green Master Mix (Roche Applied Science, Mannheim, Germany). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. Data were analyzed by the 2−ΔΔCt method. Primer sequences are shown in ESI Table S1.† All experiments were performed in triplicates per group.2.7 Animal experiments
2.8 Microarray and bioinformatics analysis
At days 7 and 21, total RNA of hBMSCs on SMO and SLA titanium disks was extracted with TRIzol reagent as described previously.33 The gene expression profiles of mRNAs and lncRNAs were assessed using a gene expression chip (lncRNA + mRNA Human Gene Expression Microarray V4.0, 4x180K, CapitalBio Technology Inc., Beijing, China). A kit from Agilent with fluorescent probes was used to label the RNA that was then purified using an RNeasy Mini Kit (Qiagen). The purified RNA was hybridized using a Hybridization Kit (Agilent), and the fluorescent intensity was examined using Microarray Scanner and Feature Extraction software (Agilent). Pairwise comparisons of gene expression between SMO and SLA titanium disks at each time point were conducted. The functional classification tool was used to annotate the differentially expressed genes (DEGs) and obtain the enriched gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) categories (http://david.abcc.ncifcrf.gov). LncRNA–mRNA networks and DEGs were built with the correlation interactions of which fold changes were greater than or equal to 2.0 and the p-value was less than 0.05 as described previously.34 A total of 129 transcripts were included in the network map using Cytoscape software (V. 3.2.1).2.9 Cell transfection
Small interfering RNA (siRNA) polyplexes of si-negative control (NC group) and siRNA-targeting lncRNA HIF1A-AS1 (si-HIF1A-AS1 group) were purchased from Suzhou GenePharma Co., Ltd. (Suzhou, China). The siRNA sequences are listed in ESI Table S1.† siRNAs were transfected into hBMSCs using Lipofectamine™ 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Briefly, when cells cultured in 6-well plates reached >50% confluence, the medium was changed from osteogenic medium to Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) before transfection. For comparison, we included a group without small interfering RNA in growth medium for proliferation (PM). To maintain low expression of HIF1A-AS1, the cells were transfected with siRNA every 3 days and expression levels of HIF1A-AS1 were examined for confirmation. After cell transfection for 3, 7, and 14 days, total RNA was extracted and the gene expression of alkaline phosphatase (ALP) and runt-related transcription factor 2 (RUNX2) was examined.2.10 Alkaline phosphatase (ALP) staining
hBMSCs cultured in 6-well plates at 5 × 105 cells per well in proliferation or osteogenic induction medium were evaluated by ALP staining. After transfection with si-NC or si-HIF1A-AS1 for 3 and 7 days, the cells were washed with PBS and fixed in 4% paraformaldehyde for 30 minutes. Subsequently, ALP staining was performed with an NBT/BCIP Staining Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Images were captured under the M125 optical microscope.2.11 Western blot analysis
hBMSCs were divided into si-NC and si-HIF1A-AS1 groups and osteogenically induced for 7 days. Then, total proteins were obtained by lysing the cells in RIPA buffer. Equal amounts of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked in 5% dry skim milk and incubated overnight at 4 °C with primary antibodies. After extensive washing, the membranes were incubated with a secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Cell Signaling Technology) for 1 hour at room temperature and developed with a SuperSignal™ West Femto Substrate Trial Kit. Primary antibodies against RUNX2 (Abcam, Cambridge, UK), osteocalcin (OCN; Abcam), p38 MAPK (Cell Signaling Technology), phosphorylated p38 MAPK (p-p38 MAPK; Cell Signaling Technology), β-actin (Abcam), and VEGFA (Thermo Fisher Scientific) were diluted at 1
000, Cell Signaling Technology) for 1 hour at room temperature and developed with a SuperSignal™ West Femto Substrate Trial Kit. Primary antibodies against RUNX2 (Abcam, Cambridge, UK), osteocalcin (OCN; Abcam), p38 MAPK (Cell Signaling Technology), phosphorylated p38 MAPK (p-p38 MAPK; Cell Signaling Technology), β-actin (Abcam), and VEGFA (Thermo Fisher Scientific) were diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. After normalization to the β-actin band, the intensity of the bands was quantified using ImageJ software (http://rsb.info.nih.gov/ij/).
1000. After normalization to the β-actin band, the intensity of the bands was quantified using ImageJ software (http://rsb.info.nih.gov/ij/).
2.12 Fluorescence in situ hybridization (FISH)
FISH analysis of cultured hBMSCs on sterile glass coverslips was performed with a fluorescence in situ hybridization kit (RiboBio, Guangzhou, China). After preprocessing the sample as described previously,22 hybridization with oligodeoxynucleotide probes for HIF1A-AS1, U6, or 18S was performed overnight at 37 °C while protected from light. The next day, the hBMSCs were counterstained with DAPI. Finally, images were obtained with an LSM 5 EXCITER confocal imaging system (Carl Zeiss, Oberkochen, Germany).2.13 Immunofluorescence staining
Transfected hBMSCs were cultured on glass coverslips. A group without small interfering RNA transfection was included for comparison. The three groups of hBMSCs were cultured in osteogenic induction medium for 24 or 48 hours. After washing with PBS, fixing in 4% paraformaldehyde (Sigma-Aldrich, USA) for 30 minutes, permeabilization with 0.1% Triton X-100 in PBS for 10 minutes at 25 °C, and blocking with 3% bovine serum albumin (Sigma-Aldrich) in PBS for 30 minutes, the cells were incubated with a primary antibody against vascular endothelial growth factor A (VEGFA; Thermo Fisher Scientific, USA) at 4 °C overnight. Then, the samples were incubated with the corresponding secondary antibody for 1 hour. Finally, nuclei were counterstained with DAPI and coverslips were mounted. Then, images were obtained with the LSM 5 EXCITER confocal imaging system.2.14 Statistical analysis
Statistical analysis was performed with SPSS version 20 (IBM, Chicago, IL, USA). Statistical significance was calculated using the two-tailed Student's t-test and one-way analysis of variance with Bonferroni's post hoc correction. All data are presented as the mean ± standard deviation (s.d.) of at least three independent experiments. A difference in values with p < 0.05 was considered as statistically significant.3. Results and discussion
3.1 Physicochemical and biological properties of the prepared SLA and SMO surfaces
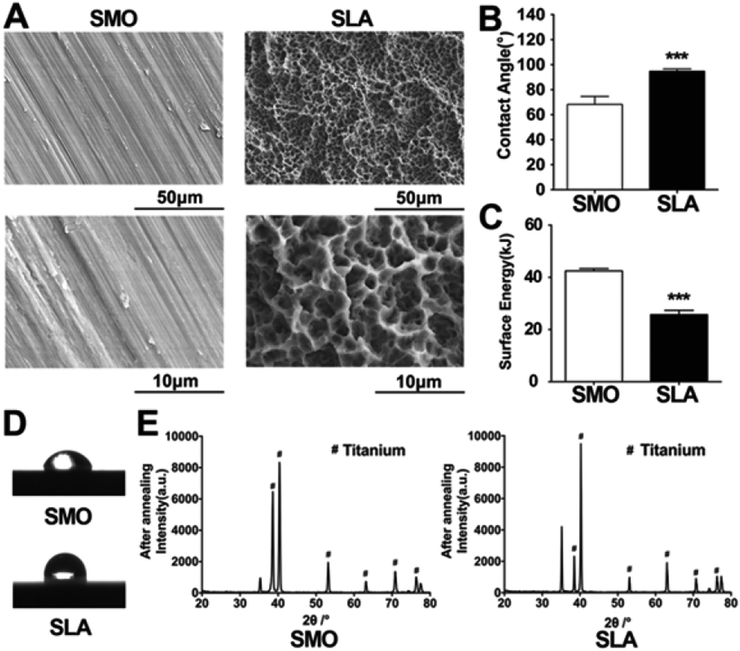
Firstly, the physicochemical properties of the prepared SLA and SMO surfaces were analyzed. SEM was used to distinguish topographical differences between SMO and SLA surfaces. The two surface morphologies of the titanium were quite different. Parallel light scratches were observed on the surface of SMO samples. The SLA surfaces displayed crater-like sags with a diameter of several tens of micrometers and a secondary micropore of micron or sub-micron sizes. The slight waviness of SMO surfaces clearly differed from SLA surfaces (Fig. 1A). We analyzed the surface angle to determine the change in surface wetting. Contact angles (CAs) measured by double distilled water (ddH2O) demonstrated that the SLA surfaces tended to exhibit larger water CAs (Fig. 1B). The SLA surfaces were hydrophobic with a dynamic contact angle (DCA) of <90°, whereas the SMO surfaces were hydrophilic (DCA > 90°) (Fig. 1D). The SMO surfaces were more hydrophilic than SLA surfaces. We quantitatively analyzed the CAs and calculated the surface energy (SFE). The SFE of the SLA surface was lower than that of the SMO surface (Fig. 1C). X-ray diffraction (XRD) patterns of the SLA surface showed that titanium machined by sandblasting with 0.25–0.50 mm grit particles and acid etching with HCl/H2SO4 showed typical titanium peaks similar to SMO surfaces (Fig. 1E).As previous studies using different models have demonstrated that biological reactions such as the cell phenotype and differentiation are altered by surface topographies and chemistry,35,36 hBMSCs adhesion, proliferation, and osteogenic differentiation experiments were also performed in our study on SLA and SMO surfaces in vitro and in vivo.
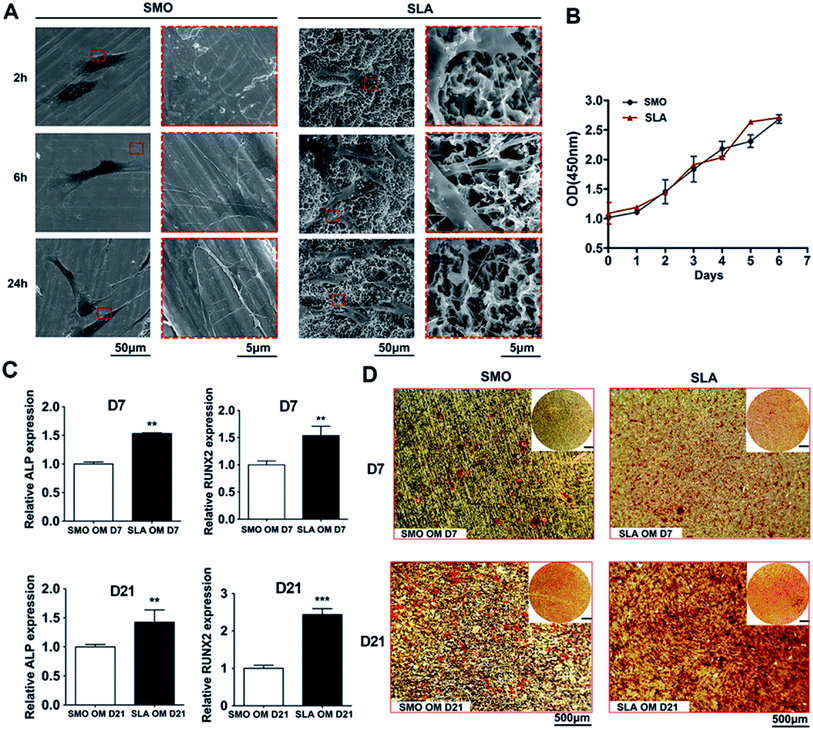
After hBMSCs were seeded on SMO and SLA surfaces, the cellular responses over time were analyzed. SEM with different magnifications was used to observe the morphology and fine structures of adhered cells, such as pseudopodia. At 2 hours after seeding, the cells generally formed a sheet-like expanse (lamellipodia) on SMO surfaces. In contrast, the cells exhibited an obvious web-like extension (filopodia) on SLA surfaces. After 6 hours of culture, cells on SLA surfaces had net-like pseudopodia, whereas cells on SMO surfaces had extended only a few long pseudopodia. After 24 hours of culture, the cells had extended to form a larger polygonal morphology on SLA surfaces with distribution of net-like and strong pseudopodia on the microstructured surfaces. Cells on SMO surfaces had extended to form a much slimmer morphology compared with cells on SLA surfaces (Fig. 2A).
To assess hBMSC proliferation, a logarithmic proliferation curve was constructed for both surfaces, which showed that the cells attached well on these two surfaces and grew with a normal “S” shape growth curve (p > 0.05) (Fig. 2B).
To investigate osteogenic differentiation in vitro, we analyzed the transcriptional responses of hBMSCs to SMO and SLA titanium surfaces that had undergone different surface treatments. For mesenchymal stem cells, surface topography can decide differentiation towards the osteogenic lineage.37 Bone marrow-derived mesenchymal stem cells (BMSCs) as host cells play critical roles in osteoblastogenesis at the bone-implant interface. There are several stages involved in the differentiation of stem cells during osteoblastogenesis and expression of osteoblast-specific genes changes over time.38 Osteoblasts produce a large amount of ALP in the process of synthesizing the secretory bone matrix. ALP interacts with calcium ions to promote deposition of calcium salts to complete the mineralization process. Therefore, ALP is used as an important indicator to detect secretion of extracellular matrix and initial osteogenesis.39 RUNX2 is a major transcription factor involved in osteogenic differentiation. Franceschi et al. found that RUNX2 protein was highly expressed at different stages of bone differentiation and promoted osteogenesis by participating in multiple signal transduction pathways.40 BMP2 as a multifunctional cytokine not only contributes to the proliferation and migration of various cell types, but also strongly induces the formation of mineralized matrix.38,41 Other relevant cell differentiation markers, such as OCN and OPN, have been evaluated to explore the osteogenic potential of MC3T3 cells during mineralization of extracellular matrix.42 Thus, after hBMSCs were cultured in osteogenic medium, gene expression of osteogenic markers, including ALP and RUNX2, was detected. Such gene expression in hBMSCs had increased at days 7 and 21 on SLA surfaces compared with SMO surfaces. At day 7, expression of ALP and RUNX2 showed an obvious increasing trend (p < 0.01). At day 21, relative ALP expression exhibited a constant increasing trend (p < 0.01). Relative RUNX2 expression on SLA surfaces was about two-fold higher than that on SMO surfaces (p < 0.001) (Fig. 2C). Moreover, we detected expression of other osteogenic related genes, such as OCN, OPN, and BMP2, on SLA surfaces compared with SMO surfaces. At day 7, relative OPN and BMP2 expression on SLA surfaces was more than eight-fold higher than that on SMO surfaces (p < 0.001). Expression of OCN also showed an increasing trend (p < 0.01). At day 21, relative OCN and OPN expression on SLA surfaces was more than three-fold higher than that on SMO surfaces (p < 0.001). Relative BMP2 expression still showed an increasing trend (p < 0.05) (Fig. S1†). ARS staining was used to reveal biomineralization of the matrix after 7 and 21 days of osteoinduction. Compared with cells on SMO surfaces, cells on SLA surfaces had enhanced ARS staining at day 7 and mineralized nodules were increased significantly at day 21 (Fig. 2D).
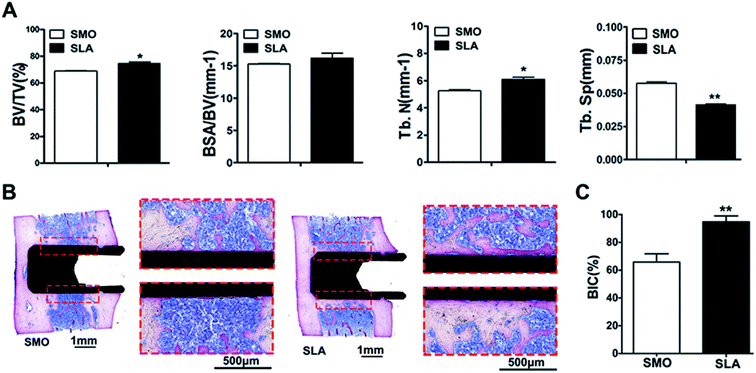
Because previous in vivo studies have shown that the implant interface is completely covered by mature lamellar bone, and osseointegration is completed at 8 weeks,43,44 a rat femur implant model was established to demonstrate osseointegration of SLA and SMO surfaces in vivo. The data showed that SLA surfaces induced more rapid osteogenic responses than SMO surfaces. At 8 weeks after implantation, all implants showed no complications in SMO and SLA surface groups (Fig. S2A†). Qualitative micro-CT two-dimensional images provided information regarding implant osseointegration and the peri-implant trabecular microstructure in different directions. We distinguished the trabecular bone from bone marrow and selected a 1 mm implant bone volume-of-interest by adjusting the threshold value in the software. A total of 1024 slices were used to reconstruct corresponding three-dimensional (3D) models that showed differences in new bone formation around the implants (Fig. S2B†). The SLA surface group showed a tighter structure and promoted peri-implant bone regeneration relative to the SMO surface group. Compared with SMO implants, SLA implants showed a greater ability to increase the bone volume-to-total volume (BV/TV) ratio and trabecular number (Tb.N) (p < 0.05), and reduce trabecular separation (Tb.Sp) (p < 0.01) in the femur. Moreover, a tendency for an increase in the bone surface area fraction (BSA/BV) was found in the SLA group (Fig. 3A). Consistent with these results, histological and histomorphometric analyses also indicated more newly formed bone tissue in the SLA group than in the SMO group. Methylene blue-basic fuchsine staining is the preferred method to observe new bone formation in a peri-implant region. At 8 weeks after implantation, the SLA group formed more direct contact around the implants compared with the SMO group. Additionally, compared with the tiny amounts of new bone on the SMO surface, the SLA group had considerably more new bone formation that tightly adhered to the modified implant surface (Fig. 3B). Thus, a significantly higher bone-to-implant contact (BIC) ratio was also observed in the SLA group (Fig. 3C).
Here, we found that the SLA surface with a typical complex microtopography providing mechanical stimuli contributed to cell fate decisions. Compared with SMO titanium implant surfaces, the SLA titanium implant surfaces significantly promote osteogenesis in vitro and in vivo, which is consistent with a previous study.43–45
3.2 Gene expression patterns of mRNAs and lncRNAs on SMO and SLA surfaces
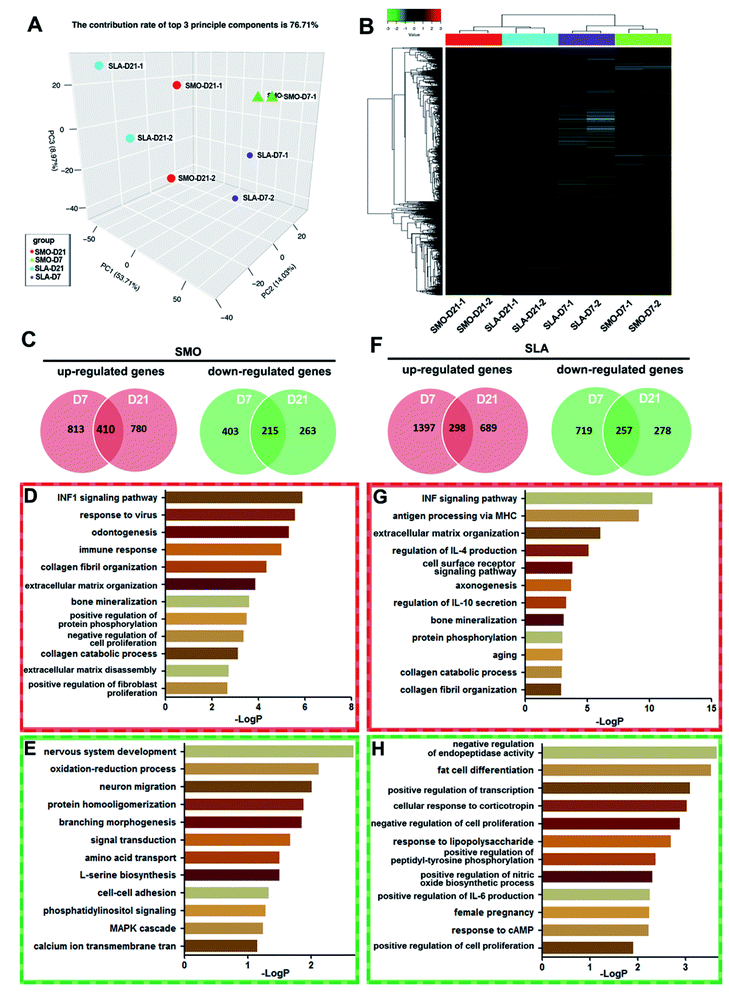
Long noncoding RNAs (lncRNAs), which were regarded as the “noise” of transcription of essentially the entire eukaryotic genome, have been proven to be an important component of the transcriptome.46 Numerous lncRNAs are related to expression of genes that control both physiological and pathological processes such as development and carcinogenesis,47 but only a few have been reported to regulate osteogenic differentiation of stem cells associated with skeletal and dental diseases. Further in-depth studies are urgently needed to reveal the mechanisms relating lncRNAs to osseointegration. In recent years, microarray analysis has been used to investigate the influence of the large quantity of ncRNAs and target mRNAs. Based on the trend of bioinformatics research, scientists have focused on not only chip data management and sharing, but also in-depth information mining and screening of DEGs.48,49 Although microarray analysis has been used to examine the transcription patterns of microRNAs and mRNAs responsible for osteogenic differentiation,50 the role of lncRNAs in the molecular events leading to increased levels of bone formation on titanium surfaces is unclear. With the development of microarray and bioinformatics analyses, unified application of lncRNA and mRNA microarrays has become a mature method to explore the potentially unknown mechanisms of lncRNAs.51 In our study, culturing hBMSCs on SMO and SLA surfaces led to differential expression of thousands of lncRNAs and mRNAs. Because gene expression dynamics have a typical phasic pattern, we selected two time points, i.e., day 7 as the early stage and day 21 as the late stage, and analyzed the expression profiles of genes.38Firstly, we analyzed gene expression patterns of mRNAs between D7 and D21 on SMO and SLA titanium implant surfaces respectively. The levels of affected transcripts in hBMSCs following exposure to the two types of titanium surfaces were investigated using microarrays. hBMSCs cultured on SMO and SLA titanium implant surfaces and osteoinduced for 7 and 21 days in duplicate were collected. Principal component analysis (PCA) was used to analyze cells with similarities in gene expression profiles to cluster together. The PCA analysis showed that the replicates of samples were clustered together. The clustering pattern showed a distinction between cells on SMO and SLA surfaces (Fig. 4A). Hierarchical clustering analysis was based on the similarity in the expression patterns of all genes. hBMSCs were divided into two classes: day 7 and day 21 (D7 and D21, respectively). Each D7 and D21 cluster was further divided into two subclasses according to the surface characteristics, i.e., SMO or SLA (Fig. 4B). In the SMO group and SLA group, differential expression analysis was performed and DEGs (≥two-fold, p < 0.05) at days 7 and 21 were compared. For mRNA transcriptional profiles of hBMSCs on SMO surfaces, 1223 mRNAs (D7) and 1190 mRNAs (D21) were upregulated after osteogenic induction, and 410 of them overlapped in D7 and D21 clusters. In contrast, 618 mRNAs (D7) and 478 mRNAs (D21) were downregulated in BMSCs, and 215 of them overlapped in D7 and D21 clusters (Fig. 4C). GO analysis showed that the upregulated genes were mainly associated with osteogenesis-related processes such as odontogenesis, collagen fibril organization, and bone mineralization (Fig. 4D). The downregulated genes were mainly associated with nervous system development and oxidation–reduction processes. Specifically, the MAPK cascade was involved in the top signaling pathways of hBMSCs (Fig. 4E). For SLA surfaces, 298 genes were commonly upregulated and 257 genes were commonly downregulated in D7 and D21 clusters (Fig. 4F). The upregulated genes were associated with the ECM and mineralization (Fig. 4G), while the downregulated genes were mainly associated with endopeptidase activity and fat cell differentiation (Fig. 4H). Therefore, the mRNA expression of cells seeded on SMO and SLA titanium surfaces showed significant differences on the distinct surfaces.
Here, on both SMO and SLA surfaces, gene expression patterns of mRNAs at day 7 and day 21 were observed respectively. GO analysis of the overlapped mRNA profiles revealed that bone mineralization and collagen catabolic process signal were upregulated from day 7 to 21 on both surfaces. However, most processes were completely different on the distinct surfaces. In particular, we found that the MAPK cascade was downregulated in the SMO group but not in the SLA group, which suggests the SLA surfaces may promote osteogenesis by MAPK signaling pathways.
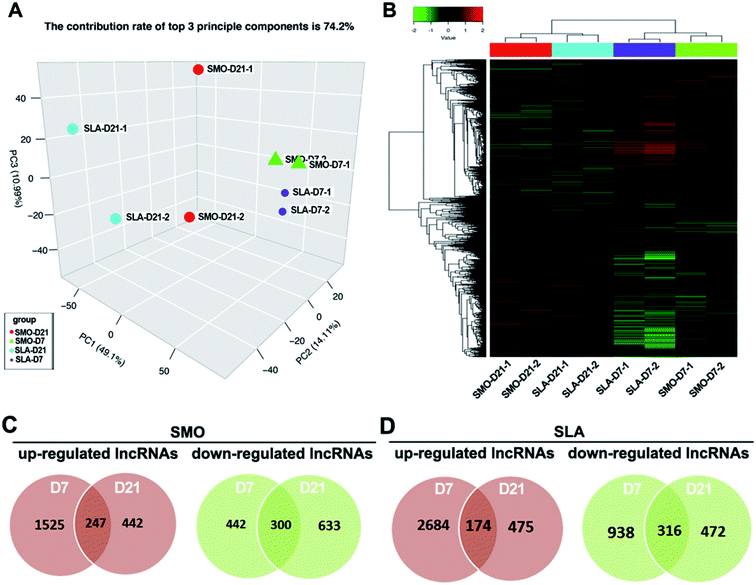
Similarly, gene expression patterns of lncRNAs between D7 and D21 on SMO and SLA implant surfaces also were analyzed respectively. Using LncRNA Human Gene Expression Microarray analysis, we systematically identified lncRNAs related to the differentiation of hBMSCs on SMO and SLA titanium implant surfaces. More than 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lncRNA transcripts were analyzed, including 3433 single-exon and 21
000 lncRNA transcripts were analyzed, including 3433 single-exon and 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 750 multi-exon transcripts derived from 258 unique loci. Of these lncRNAs, 12
750 multi-exon transcripts derived from 258 unique loci. Of these lncRNAs, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 687 intergenic transcripts, 7767 antisense transcripts, 1132 intronic transcripts, and 674 sense lncRNA transcripts were included. PCA showed a distinct separation between cells on SMO and SLA surfaces in terms of the differentially expressed lncRNAs (Fig. 5A). Unsupervised hierarchical clustering analysis showed that samples separated into two main groups, and each D7 and D21 cluster was further divided into two subclasses according to the surface characteristics: SMO and SLA (Fig. 5B). We performed differential expression and Venn diagram analyses to reveal the overlaps between differentially expressed lncRNAs in D7 and D21 clusters (≥two-fold, p < 0.05) of SMO and SLA groups separately. A total of 3589 lncRNAs in the SMO group (2214 upregulated and 1375 downregulated) and 5059 lncRNAs in the SLA group (3333 upregulated and 1726 downregulated) were differentially expressed between D7 and D21 clusters. A total of 247 genes were commonly upregulated, and 300 genes were commonly downregulated on SMO surfaces in D7 and D21 clusters (Fig. 5C). For SLA surfaces, 174 lncRNAs were commonly upregulated and 316 lncRNAs were commonly downregulated in D7 and D21 clusters (Fig. 5D). Therefore, the lncRNA expression of cells seeded on SMO and SLA titanium surfaces showed an obvious difference between the distinct surfaces.
687 intergenic transcripts, 7767 antisense transcripts, 1132 intronic transcripts, and 674 sense lncRNA transcripts were included. PCA showed a distinct separation between cells on SMO and SLA surfaces in terms of the differentially expressed lncRNAs (Fig. 5A). Unsupervised hierarchical clustering analysis showed that samples separated into two main groups, and each D7 and D21 cluster was further divided into two subclasses according to the surface characteristics: SMO and SLA (Fig. 5B). We performed differential expression and Venn diagram analyses to reveal the overlaps between differentially expressed lncRNAs in D7 and D21 clusters (≥two-fold, p < 0.05) of SMO and SLA groups separately. A total of 3589 lncRNAs in the SMO group (2214 upregulated and 1375 downregulated) and 5059 lncRNAs in the SLA group (3333 upregulated and 1726 downregulated) were differentially expressed between D7 and D21 clusters. A total of 247 genes were commonly upregulated, and 300 genes were commonly downregulated on SMO surfaces in D7 and D21 clusters (Fig. 5C). For SLA surfaces, 174 lncRNAs were commonly upregulated and 316 lncRNAs were commonly downregulated in D7 and D21 clusters (Fig. 5D). Therefore, the lncRNA expression of cells seeded on SMO and SLA titanium surfaces showed an obvious difference between the distinct surfaces.
3.3 Screening of DEGs between SMO and SLA surfaces at days 7 and 21
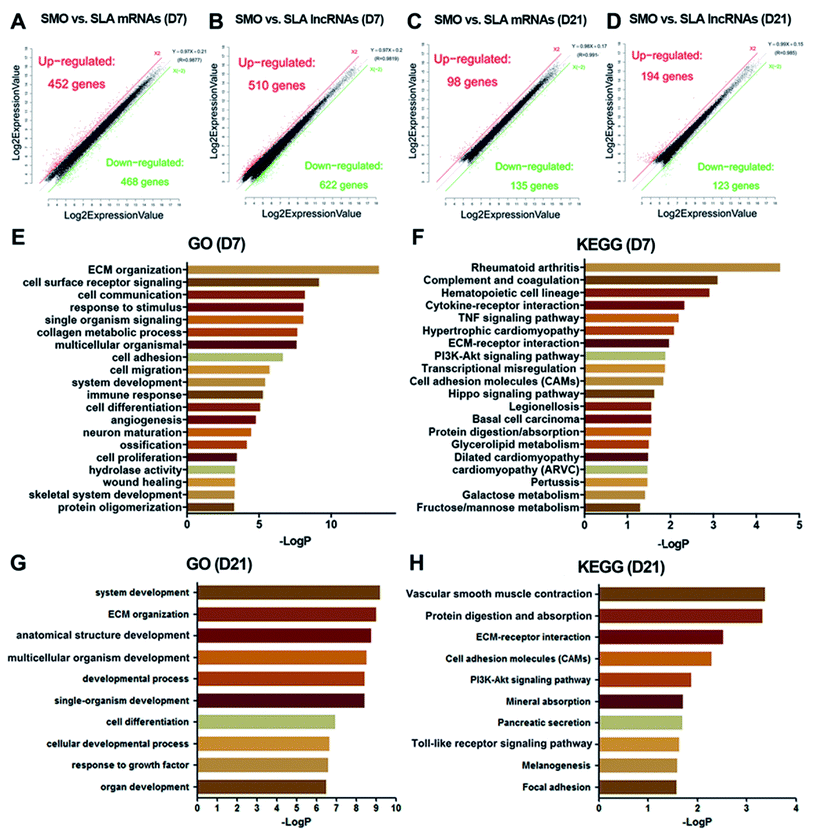
Next, we analyzed the mRNA and lncRNA expression after cells were seeded on the different surfaces at days 7 and 21. At day 7, 920 mRNAs and 1132 lncRNAs on SLA and SMO surfaces were differentially expressed (≥two-fold, p < 0.05). Of these, 452 mRNAs and 510 lncRNAs were upregulated, while 468 mRNAs and 622 lncRNAs were downregulated in the SLA group compared with the SMO group (Fig. 6A and B). The GO categories associated with the DEGs included bone formation-associated processes such as ECM organization, skeletal system development, and ossification (Fig. 6E). The KEGG category of the tumor necrosis factor (TNF) signaling pathway and cell adhesion molecules was over-represented in response to the SLA surface (Fig. 6F). Pivotal genes such as BMP2 and VEGFA were upregulated. At day 21, 98 mRNAs and 194 lncRNAs were upregulated, while 135 mRNAs and 123 lncRNAs were downregulated in the SLA group compared with the SMO group (Fig. 6C and D). The DEGs regulated development-associated processes, ECM organization, and cell differentiation (Fig. 6G). Moreover, KEGG analysis indicated that these DEGs also regulated osteogenesis through ECM–receptor interactions, vascular smooth muscle contraction, mineral absorption, the Toll-like receptor signaling pathway, and vascular smooth muscle contraction (Fig. 6H). The change in transcriptome and the results of osteogenic induction experiments indicated that the regulation of transcription directing osteoblast differentiation occurred mainly at day 7.Differential expression between SMO and SLA surfaces was observed, which showed that the time point was a major factor for mRNA and lncRNA expression during differentiation. Furthermore, on both SMO and SLA surfaces, mRNA and lncRNA expression at day 7 was more active than at day 21, depending on the quantity of DEGs (Fig. 6A–D). GO and KEGG analyses clearly revealed that the highest correlating biological processes and pathways were related to surface topography. SLA surfaces influence the expression of genes in ECM–receptor interactions, the hematopoietic cell lineage, TNF signaling pathways, vascular smooth muscle contraction, and mineral absorption (Fig. 6E–H). Thus, hBMSCs exposed to SLA surfaces had a greater potential for differentiation and revealed candidate genes with a potential osteogenic role in the cellular responses to SLA surfaces especially at the early stage.
3.4 Coexpression analysis of differentially expressed mRNAs and lncRNAs at day 7
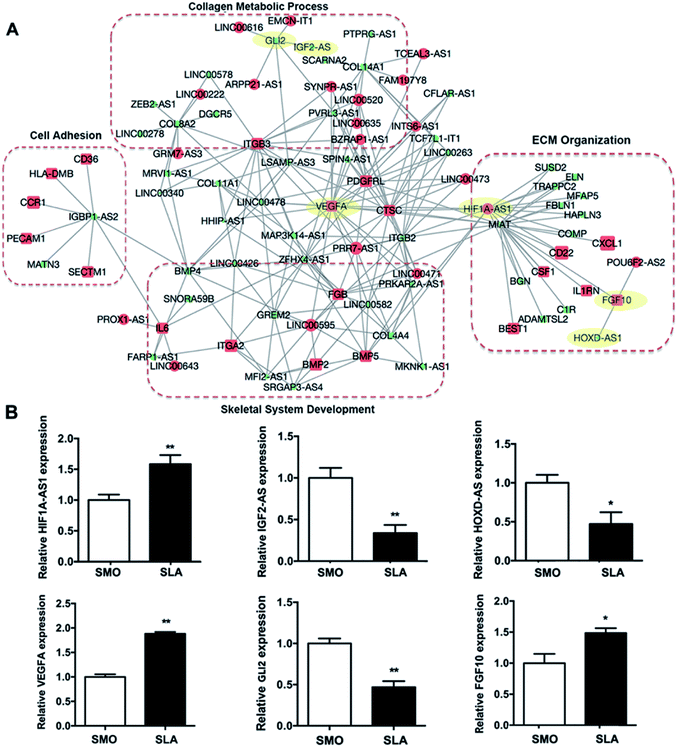
Because lncRNAs may potentially regulate the expression of specific mRNAs, we constructed an lncRNA–mRNA network using a bioinformatics analysis algorithm according to the differential expression at day 7. More than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pieces of interactions were identified with a correlation coefficient of >0.997 and p < 0.01 (Table S2†). GO analysis showed that mRNAs in the network were associated with ECM organization, skeletal development, cell adhesion, and collagen metabolic processes involved in hBMSCs differentiation and bone formation (Fig. 7A). Fifty lncRNAs and 79 mRNAs, which correlated with genes within these four GO categories, were selected to build a simplified network. Each lncRNA was connected to multiple mRNAs and each mRNA may be linked to more than one lncRNA. Some mRNAs of osteogenic genes showed the highest degree of interaction, including BMP2, VEGFA, ITGA2, ITGB3, and GLI2. We also validated the expression of three types of lncRNAs and coexpressed mRNAs (i.e., lncRNA upregulated and mRNA upregulated; lncRNA downregulated and mRNA downregulated; lncRNA downregulated and mRNA upregulated) by qRT-PCR. Consistently, the expression of HIF1A-AS1/VEGFA, IGF2-AS/GLI2, and HOXD-AS1/FGF10 was correlated (Fig. 7B).
000 pieces of interactions were identified with a correlation coefficient of >0.997 and p < 0.01 (Table S2†). GO analysis showed that mRNAs in the network were associated with ECM organization, skeletal development, cell adhesion, and collagen metabolic processes involved in hBMSCs differentiation and bone formation (Fig. 7A). Fifty lncRNAs and 79 mRNAs, which correlated with genes within these four GO categories, were selected to build a simplified network. Each lncRNA was connected to multiple mRNAs and each mRNA may be linked to more than one lncRNA. Some mRNAs of osteogenic genes showed the highest degree of interaction, including BMP2, VEGFA, ITGA2, ITGB3, and GLI2. We also validated the expression of three types of lncRNAs and coexpressed mRNAs (i.e., lncRNA upregulated and mRNA upregulated; lncRNA downregulated and mRNA downregulated; lncRNA downregulated and mRNA upregulated) by qRT-PCR. Consistently, the expression of HIF1A-AS1/VEGFA, IGF2-AS/GLI2, and HOXD-AS1/FGF10 was correlated (Fig. 7B).
Recent studies have shown that lncRNAs regulate biological functions at the mRNA expression level. Among their complex effects on the body, the discovery has expanded our knowledge of the regulatory mechanisms involved in biological processes.52 To obtain useful information, an lncRNA–mRNA coexpression network at day 7 was constructed to understand the roles of the differentially expressed lncRNAs. The coexpressed mRNAs were associated with certain functions such as ECM organization, metabolic processes, cell adhesion, and skeletal system development. The considerable number of differentially expressed osteogenesis-associated lncRNAs of the SLA group reflected the complexity of the bone regeneration process that occurs on titanium surfaces with varied topography (Fig. 7A). In the lncRNA–mRNA coexpression network, we also found that genes such as platelet-derived growth factor receptor-like (PDGFRL) and BMP2 genes, were upregulated in hBMSCs on SLA surfaces. While the lncRNAs, such as MIAT and HOXD-AS1 were downregulated. The PDGFRL gene has been regarded as a tumor suppressor gene. However, a recent study indicated that PDGFRL also plays an important role in chondrocytes.53 In our study, we first found that PDGFRL expressed in hBMSCs seeded on micro-roughened surfaces might play a positive role in osteogenic differentiation. BMP2 is used clinically to improve osseointegration and peri-implant bone formation. An SLA implant with a coating of BMP2 was shown to promote osseointegration and bone regeneration in a pilot study.54 However, a recent study showed that the use of BMP2 with SLA implants might increase inflammation and possibly delay bone formation depending on the dose.55 Our results showed that the SLA surface itself increased the mRNA level of BMP2. Thus, the proper dose of BMP2 for clinical applications during implant insertion may need to be re-evaluated. SLA surfaces also suppressed the expression of HOXD-AS1. HOXD-AS1 acts as an oncogene and is associated with the development of multiple cancers such as lung and liver cancers.56,57 Although the role of HOXD-AS1 in osteoblastic differentiation is unknown, knockdown of HOXD-AS1 leads to differential expression of genes involved in angiogenesis.58 Moreover, knockdown of HOXD-AS1 inactivates the JAK2/STAT3 pathway,59 which may suppress transcriptional induction of RUNX2.60 As reported in our previous study, knockdown of MIAT was found to act as a role of promoting human adipose-derived stem cells' osteogenic differentiation and reverse the negative effects of inflammation on osteoblastic differentiation.61 Thus, the downregulation of MIAT in hBMSCs which cultured on the SLA surfaces may contribute to enhanced osteogenesis. Here, three randomly selected lncRNA–mRNA interactions (i.e., HIF1A-AS1/VEGFA, IGF2-AS/GLI2, and HOXD-AS1/FGF10) matching to the lncRNA–mRNA coexpression network confirm its validity. Among them, HIF1A-AS1/VEGFA genes were found as the greatest differentially expressed lncRNA–mRNA interaction (Fig. 7B).
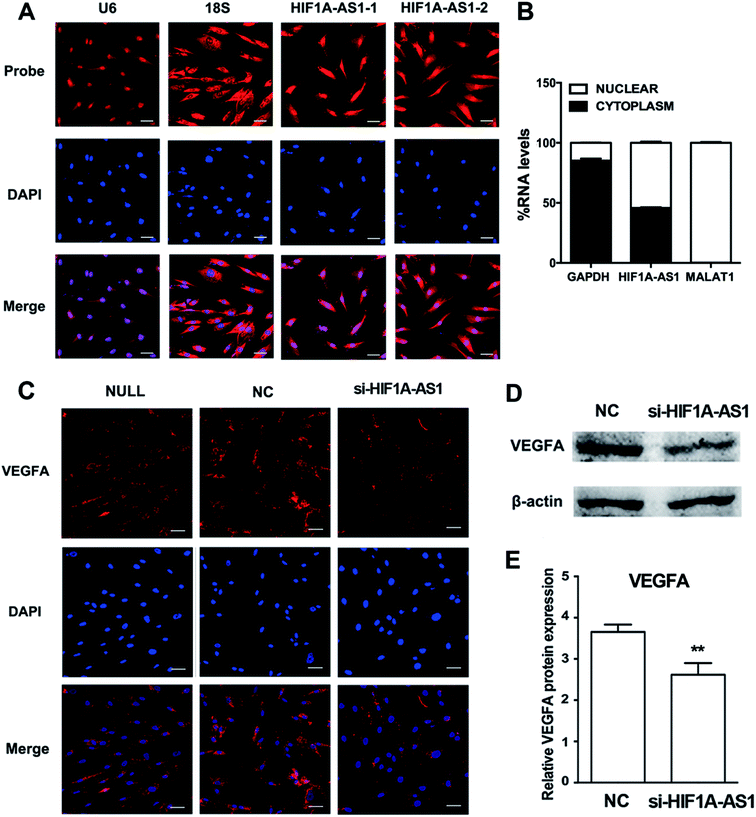
3.5 Knockdown of lncRNA HIF1A-AS1 inhibits VEGFA expression
We first analyzed the distribution of HIF1A-AS1 by FISH and confocal microscopy after 24 hours of osteogenic induction. Using two probes for HIF1A-AS1, we found that HIF1A-AS1 was predominantly located in the nucleus and peri-nuclear compartment (Fig. 8A), which was confirmed quantitatively by nuclear/cytoplasm fractionation (Fig. 8B). Thus, for inducing gene silencing of HIF1A-AS1 in hBMSCs, siRNA transfection can be considered as an effective method.According to the coexpression network, HIF1A-AS1 and VEGFA were connected and had a role in ECM organization of hBMSCs. To investigate the effect of HIF1A-AS1 on VEGFA expression, we knocked down endogenous HIF1A-AS1 in hBMSCs and cultured the hBMSCs in osteogenic medium. Immunofluorescence staining revealed that VEGFA was localized predominantly in the cytoplasm after knockdown of lncRNA HIF1A-AS1 in the si-HIF1A-AS1 group similarly to the si-NC group. However, compared with null and si-NC groups, the fluorescent density and intensity of VEGFA protein in the si-HIF1A-AS1 group were much lower at both 24 and 48 hours (Fig. S3† and 8C).
After that, we examined the effect of HIF1A-AS1 of VEGFA expression in hBMSCs after osteogenic induction for 7 days. When HIF1A-AS1 was knocked down, western blotting showed inhibition of VEGFA protein expression at day 7 (p < 0.01) (Fig. 8D and E).
Although Xu et al. suggested that HIF1A-AS1 interferes with acetylation of HOXD10 and subsequently results in improvement of osteoblastic differentiation,31 there is no additional evidence to reveal the function of HIF1A-AS1 in hBMSCs. HIF1A-AS1 was significantly upregulated in hBMSCs on SLA surfaces. In our study, FISH was used to detect the location of HIF1A-AS1 in hBMSCs (Fig. 8A). The results revealed that HIF1A-AS1 accumulated in not only the nucleus, but also in the perinuclear cellular compartment, which is different from its expression in other human cell types and tissues.30,62 Interestingly, based on the lncRNA–mRNA crosstalk mentioned above, we confirmed that VEGFA as a core gene involved in ECM organization, skeletal development, cell adhesion, and collagen metabolic processes had a strong connection with HIF1A-AS1. As a regulator of angiogenesis, VEGFA promotes endothelial cell proliferation, migration, and survival.63 Additionally, VEGFA plays an essential role in stimulating intramembranous ossification and in endochondral bone formation during development of the craniofacial skeletal system.64,65All of these results confirmed our hypothesis that HIF1A-AS1 may mediate osteogenic differentiation through upregulated VEGFA expression.
3.6 Knockdown of lncRNA HIF1A-AS1 inhibits osteogenic differentiation of hBMSCs
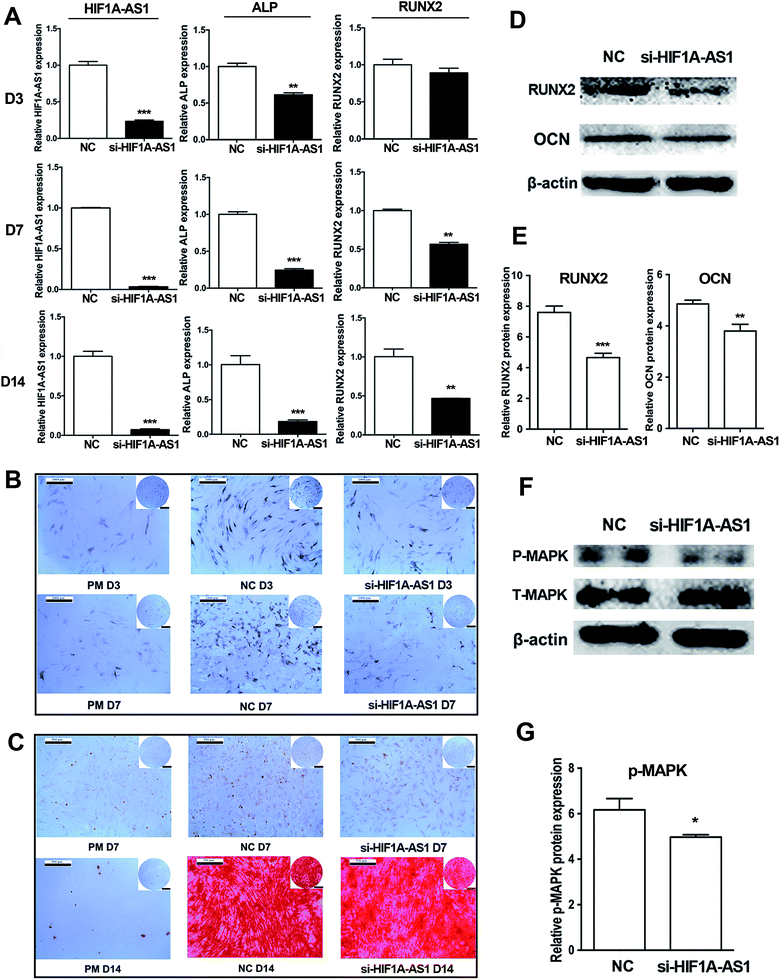
Next, we investigated the effect of HIF1A-AS1 on osteogenic differentiation of hBMSCs. After osteogenic induction of hBMSCs for 7 days, qRT-PCR showed that expression of HIF1A-AS1 was decreased from 70% to 90% in the HIF1A-AS1 knockdown group (p < 0.01 at day 3, p < 0.001 at days 7 and 14). Similarly, gene expression of osteogenic related marker ALP was also significantly downregulated in the si-HIF1A-AS1 group compared with the NC group at days 3, 7, and 14 (p < 0.01 at day 3, p < 0.001 at days 7 and 14). In addition, expression of RUNX2 was downregulated in the si-HIF1A-AS1 group, especially at days 7 and 14 (p < 0.01) (Fig. 9A). Compared with cells in the negative control group (NC group) at days 3 and 7, ALP activity in the si-HIF1A-AS1 group was decreased significantly after endogenous HIF1A-AS1 was knocked down in hBMSCs (Fig. 9B). ARS staining was used to reveal biomineralization of the matrix at days 7 and 14 of osteoinduction. Compared with the NC group, mineralized nodules in the si-HIF1A-AS1 group were decreased at both days 7 and 14 (Fig. 9C). Western blotting confirmed that expression of osteogenic related markers RUNX2 and OCN was reduced in HIF1A-AS1 knockdown cells at day 7 (p < 0.01) (Fig. 9D and E), indicating that HIF1A-AS1 silencing delayed osteoblastic differentiation.According to the results of GO analysis, the MAPK cascade signaling pathways involved in modulating the osteogenesis. Zhou et al. also found that a p38 MAPK-related signaling pathway was involved in regulating osteogenic differentiation of hypoxia-simulated osteogenesis.66 To confirm the effect of HIF1A-AS1, we analyzed protein expression of p38 MAPK protein. In our present data, synchronizing with the expression of HIF1A-AS1, western blotting showed that phosphorylated p38 MAPK was decreased in the HIF1A-AS1 knockdown group at day 7 (p < 0.05) (Fig. 9F and G), which suggested that HIF1A-AS1 influenced expression of the p38 MAPK signaling pathway during osteoinduction.
Another study indicated that enhanced expression of RUNX2, OCN, VEGF, and PDGF-β in BMSCs after osteogenic induction may be mediated by activation of MAPK signaling.67 Instead, inhibition of the p38 MAPK signaling pathway not only decreases homing of BMSCs, but also local vascularization, which highly correlate with secretion of VEGFA.68 In our present study, Fig. 8 and 9 showed that knockdown of HIF1A-AS1 inhibited the VEGFA expression and osteogenic differentiation. Moreover, we confirmed that HIF1A-AS1 can regulate the MAPK cascade by modulating p38 MAPK phosphorylation. Thus, we revealed an lncRNA–mRNA coexpression network involving HIF1A-AS1, VEGFA, and MAPK cascade signaling pathways. LncRNA HIF1A-AS1 can be considered as potential biologic modifiers to improve the osteogenic function of implant materials.
4. Conclusion
In our study, two kinds of titanium surfaces (SMO and SLA) were manufactured with different roughness, and we demonstrated that SLA titanium surfaces markedly enhanced the differentiation of hBMSCs in vitro and in vivo compared with SMO titanium surfaces. The SLA surfaces resulted in unique profiles of mRNAs and lncRNAs that can be potentially used to modify titanium surfaces. Notably, we revealed an lncRNA–mRNA coexpression network involving HIF1A-AS1, VEGFA, and MAPK cascade signaling pathways. The hBMSCs cultured on SLA disks displayed a higher level of HIF1A-AS1 and VEGFA expression. Knockdown of HIF1A-AS1 inhibited the osteogenic differentiation of hBMSCs by regulating p38 MAPK cascade proteins. Because titanium surfaces functionalized with miRNAs have been created successfully, lncRNAs may be used for modification of titanium-based materials. Thus, HIF1A-AS1 may be used to regulate the osteogenic property of titanium implants, although further investigation is needed.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We wish to thank Xiyuan Ge (Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology) and Muyang Sun (Department of Biomedical Engineering, College of Engineering, Peking University) for their valuable support and technical assistance. This work was supported by grants from the National Key Research and Development Program of China (No. 2016YFC1102705 and No. 2018YFC1105302); the National Natural Science Foundation of China (No. 81700938); and the Science Foundation of Peking University School and Hospital of Stomatology (PKUSS20150106 and PKUSS20140104).References
- L. Le Guehennec, A. Soueidan, P. Layrolle and Y. Amouriq, Surface treatments of titanium dental implants for rapid osseointegration, Dent. Mater., 2007, 23(7), 844–854 CrossRef CAS PubMed.
- R. A. Gittens, R. Olivares-Navarrete, Z. Schwartz and B. D. Boyan, Implant osseointegration and the role of microroughness and nanostructures: lessons for spine implants, Acta Biomater., 2014, 10(8), 3363–3371 CrossRef PubMed.
- R. A. Gittens, T. McLachlan, R. Olivares-Navarrete, Y. Cai, S. Berner, R. Tannenbaum, Z. Schwartz, K. H. Sandhage and B. D. Boyan, The effects of combined micron-/submicron-scale surface roughness and nanoscale features on cell proliferation and differentiation, Biomaterials, 2011, 32(13), 3395–3403 CrossRef CAS PubMed.
- G. Gao, D. Lange, K. Hilpert, J. Kindrachuk, Y. Zou, J. T. Cheng, M. Kazemzadeh-Narbat, K. Yu, R. Wang, S. K. Straus, D. E. Brooks, B. H. Chew, R. E. Hancock and J. N. Kizhakkedathu, The biocompatibility and biofilm resistance of implant coatings based on hydrophilic polymer brushes conjugated with antimicrobial peptides, Biomaterials, 2011, 32(16), 3899–3909 CrossRef CAS PubMed.
- H. J. Busscher, H. C. van der Mei, G. Subbiahdoss, P. C. Jutte, J. J. van den Dungen, S. A. Zaat, M. J. Schultz and D. W. Grainger, Biomaterial-associated infection: locating the finish line in the race for the surface, Sci. Transl. Med., 2012, 4(153), 153rv10 Search PubMed.
- J. P. Albouy, I. Abrahamsson, L. G. Persson and T. Berglundh, Implant surface characteristics influence the outcome of treatment of peri-implantitis: an experimental study in dogs, J. Clin. Periodontol., 2011, 38(1), 58–64 CrossRef PubMed.
- G. Pan, S. Sun, W. Zhang, R. Zhao, W. Cui, F. He, L. Huang, S. H. Lee, K. J. Shea, Q. Shi and H. Yang, Biomimetic Design of Mussel-Derived Bioactive Peptides for Dual-Functionalization of Titanium-Based Biomaterials, J. Am. Chem. Soc., 2016, 138(45), 15078–15086 CrossRef CAS PubMed.
- D. H. Yang, S. W. Moon and D. W. Lee, Surface Modification of Titanium with BMP-2/GDF-5 by a Heparin Linker and Its Efficacy as a Dental Implant, Int. J. Mol. Sci., 2017, 18(1), 229 CrossRef PubMed.
- M. R. Khan, N. Donos, V. Salih and P. M. Brett, The enhanced modulation of key bone matrix components by modified titanium implant surfaces, Bone, 2012, 50(1), 1–8 CrossRef CAS PubMed.
- M. Heller, V. V. Kumar, A. Pabst, J. Brieger, B. Al-Nawas and P. W. Kammerer, Osseous response on linear and cyclic RGD-peptides immobilized on titanium surfaces in vitro and in vivo, J. Biomed. Mater. Res., Part A, 2018, 106(2), 419–427 CrossRef CAS PubMed.
- H. Cheng, K. Yue, M. Kazemzadeh-Narbat, Y. Liu, A. Khalilpour, B. Li, Y. S. Zhang, N. Annabi and A. Khademhosseini, Mussel-Inspired Multifunctional Hydrogel Coating for Prevention of Infections and Enhanced Osteogenesis, ACS Appl. Mater. Interfaces, 2017, 9(13), 11428–11439 CrossRef CAS PubMed.
- J. Shin, J. H. Cho, Y. Jin, K. Yang, J. S. Lee, H. J. Park, H. S. Han, J. Lee, H. Jeon, H. Shin and S. W. Cho, Mussel Adhesion-Inspired Reverse Transfection Platform Enhances Osteogenic Differentiation and Bone Formation of Human Adipose-Derived Stem Cells, Small, 2016, 12(45), 6266–6278 CrossRef CAS PubMed.
- S. J. Lee, D. Lee, T. R. Yoon, H. K. Kim, H. H. Jo, J. S. Park, J. H. Lee, W. D. Kim, I. K. Kwon and S. A. Park, Surface modification of 3D-printed porous scaffolds via mussel-inspired polydopamine and effective immobilization of rhBMP-2 to promote osteogenic differentiation for bone tissue engineering, Acta Biomater., 2016, 40, 182–191 CrossRef CAS PubMed.
- P. Kapranov, J. Cheng, S. Dike, D. A. Nix, R. Duttagupta, A. T. Willingham, P. F. Stadler, J. Hertel, J. Hackermuller, I. L. Hofacker, I. Bell, E. Cheung, J. Drenkow, E. Dumais, S. Patel, G. Helt, M. Ganesh, S. Ghosh, A. Piccolboni, V. Sementchenko, H. Tammana and T. R. Gingeras, RNA maps reveal new RNA classes and a possible function for pervasive transcription, Science, 2007, 316(5830), 1484–1488 CrossRef CAS PubMed.
- Z. Wang, G. Wu, Z. Feng, S. Bai, Y. Dong, G. Wu and Y. Zhao, Microarc-oxidized titanium surfaces functionalized with microRNA-21-loaded chitosan/hyaluronic acid nanoparticles promote the osteogenic differentiation of human bone marrow mesenchymal stem cells, Int. J. Nanomed., 2015, 10, 6675–6687 CAS.
- X. Zhang, Y. Li, Y. E. Chen, J. Chen and P. X. Ma, Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects, Nat. Commun., 2016, 7, 10376 CrossRef CAS PubMed.
- V. A. Moran, R. J. Perera and A. M. Khalil, Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs, Nucleic Acids Res., 2012, 40(14), 6391–6400 CrossRef CAS PubMed.
- A. E. Kornienko, P. M. Guenzl, D. P. Barlow and F. M. Pauler, Gene regulation by the act of long non-coding RNA transcription, BMC Biol., 2013, 11, 59 CrossRef PubMed.
- J. J. Quinn and H. Y. Chang, Unique features of long non-coding RNA biogenesis and function, Nat. Rev. Genet., 2016, 17(1), 47–62 CrossRef CAS PubMed.
- Y. Huang, Y. Zheng, L. Jia and W. Li, Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-beta1/Smad3/HDAC Signaling Pathway by Deriving miR-675, Stem Cells, 2015, 33(12), 3481–3492 CrossRef CAS PubMed.
- W. Zhuang, X. Ge, S. Yang, M. Huang, W. Zhuang, P. Chen, X. Zhang, J. Fu, J. Qu and B. Li, Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription, Stem Cells, 2015, 33(6), 1985–1997 CrossRef CAS PubMed.
- C. Jin, L. Jia, Y. Huang, Y. Zheng, N. Du, Y. Liu and Y. Zhou, Inhibition of lncRNA MIR31HG Promotes Osteogenic Differentiation of Human Adipose-Derived Stem Cells, Stem Cells, 2016, 34(11), 2707–2720 CrossRef CAS PubMed.
- J. Zhang, Z. Tao and Y. Wang, Long noncoding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway, Int. J. Mol. Med., 2018, 41(1), 213–219 CAS.
- Y. P. Huang, Y. F. Zheng, Y. X. Xu, X. B. Li, Y. Zheng, L. F. Jia and W. R. Li, Titanium Surfaces Functionalized with siMIR31HG Promote Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells, ACS Biomater. Sci. Eng., 2018, 4(8), 2986–2993 CrossRef CAS.
- L. Baranello, D. Bertozzi, M. V. Fogli, Y. Pommier and G. Capranico, DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1alpha gene locus, Nucleic Acids Res., 2010, 38(1), 159–171 CrossRef CAS PubMed.
- J. Tantai, D. Hu, Y. Yang and J. Geng, Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer, Int. J. Clin. Exp. Pathol., 2015, 8(7), 7887–7895 CAS.
- J. Gao, R. Cao and H. Mu, Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer, Int. J. Clin. Exp. Pathol., 2015, 8(10), 12936–12942 CAS.
- Y. B. Zhao, G. X. Feng, Y. Z. Wang, Y. H. Yue and W. C. Zhao, Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: implications for TAA pathogenesis, Int. J. Clin. Exp. Pathol., 2014, 7(11), 7643–7652 Search PubMed.
- S. Wang, X. Zhang, Y. Yuan, M. Tan, L. Zhang, X. Xue, Y. Yan, L. Han and Z. Xu, BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro, Eur. J. Cardiothorac. Surg., 2015, 47(3), 439–446 CrossRef PubMed.
- D. Chen, L. Wu, L. Liu, Q. Gong, J. Zheng, C. Peng and J. Deng, Comparison of HIF1AAS1 and HIF1AAS2 in regulating HIF1alpha and the osteogenic differentiation of PDLCs under hypoxia, Int. J. Mol. Med., 2017, 40(5), 1529–1536 CrossRef CAS PubMed.
- Y. Xu, S. Wang, C. Tang and W. Chen, Upregulation of long non-coding RNA HIF 1alpha-anti-sense 1 induced by transforming growth factor-beta-mediated targeting of sirtuin 1 promotes osteoblastic differentiation of human bone marrow stromal cells, Mol. Med. Rep., 2015, 12(5), 7233–7238 CrossRef CAS PubMed.
- C. Yi, K. Y. Hao, T. Ma, Y. Lin, X. Y. Ge and Y. Zhang, Inhibition of cathepsin K promotes osseointegration of titanium implants in ovariectomised rats, Sci. Rep., 2017, 7, 44682 CrossRef PubMed.
- Y. Zheng, J. Cai, A. P. Hutchins, L. Jia, P. Liu, D. Yang, S. Chen, L. Ge, D. Pei and S. Wei, Remission for Loss of Odontogenic Potential in a New Micromilieu In Vitro, PLoS One, 2016, 11(4), e0152893 CrossRef PubMed.
- Y. Zheng, X. Li, Y. Huang, L. Jia and W. Li, The Circular RNA Landscape of Periodontal Ligament Stem Cells During Osteogenesis, J. Periodontol., 2017, 88(9), 906–914 CrossRef CAS PubMed.
- J. Y. Martin, Z. Schwartz, T. W. Hummert, D. M. Schraub, J. Simpson, J. Lankford Jr, D. D. Dean, D. L. Cochran and B. D. Boyan, Effect of titanium surface roughness on proliferation, differentiation, and protein synthesis of human osteoblast-like cells (MG63), J. Biomed. Mater. Res., 1995, 29(3), 389–401 CrossRef CAS PubMed.
- R. J. McMurray, N. Gadegaard, P. M. Tsimbouri, K. V. Burgess, L. E. McNamara, R. Tare, K. Murawski, E. Kingham, R. O. Oreffo and M. J. Dalby, Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency, Nat. Mater., 2011, 10(8), 637–644 CrossRef CAS PubMed.
- G. Abagnale, M. Steger, V. H. Nguyen, N. Hersch, A. Sechi, S. Joussen, B. Denecke, R. Merkel, B. Hoffmann, A. Dreser, U. Schnakenberg, A. Gillner and W. Wagner, Surface topography enhances differentiation of mesenchymal stem cells towards osteogenic and adipogenic lineages, Biomaterials, 2015, 61, 316–326 CrossRef CAS PubMed.
- E. Karner, C. M. Backesjo, J. Cedervall, R. V. Sugars, L. Ahrlund-Richter and M. Wendel, Dynamics of gene expression during bone matrix formation in osteogenic cultures derived from human embryonic stem cells in vitro, Biochim. Biophys. Acta, 2009, 1790(2), 110–118 CrossRef PubMed.
- T. Yoshikawa, H. Ohgushi and S. Tamai, Immediate bone forming capability of prefabricated osteogenic hydroxyapatite, J. Biomed. Mater. Res., 1996, 32(3), 481–492 CrossRef CAS PubMed.
- R. T. Franceschi, C. Ge, G. Xiao, H. Roca and D. Jiang, Transcriptional regulation of osteoblasts, Ann. N. Y. Acad. Sci., 2007, 1116, 196–207 CrossRef CAS PubMed.
- V. S. Salazar, L. W. Gamer and V. Rosen, BMP signalling in skeletal development, disease and repair, Nat. Rev. Endocrinol., 2016, 12(4), 203–221 CrossRef CAS PubMed.
- H. Zhou, M. Nabiyouni, B. Lin and S. B. Bhaduri, Fabrication of novel poly(lactic acid)/amorphous magnesium phosphate bionanocomposite fibers for tissue engineering applications via electrospinning, Mater. Sci. Eng., C, 2013, 33(4), 2302–2310 CrossRef CAS PubMed.
- C. Colnot, D. M. Romero, S. Huang, J. Rahman, J. A. Currey, A. Nanci, J. B. Brunski and J. A. Helms, Molecular analysis of healing at a bone-implant interface, J. Dent. Res., 2007, 86(9), 862–867 CrossRef CAS PubMed.
- T. Berglundh, I. Abrahamsson, N. P. Lang and J. Lindhe, De novo alveolar bone formation adjacent to endosseous implants, Clin. Oral Implants Res., 2003, 14(3), 251–262 CrossRef PubMed.
- G. Zhao, A. L. Raines, M. Wieland, Z. Schwartz and B. D. Boyan, Requirement for both micron- and submicron scale structure for synergistic responses of osteoblasts to substrate surface energy and topography, Biomaterials, 2007, 28(18), 2821–2829 CrossRef CAS PubMed.
- C. P. Ponting, P. L. Oliver and W. Reik, Evolution and functions of long noncoding RNAs, Cell, 2009, 136(4), 629–641 CrossRef CAS PubMed.
- P. J. Batista and H. Y. Chang, Long noncoding RNAs: cellular address codes in development and disease, Cell, 2013, 152(6), 1298–1307 CrossRef CAS PubMed.
- C. J. Stoeckert Jr, H. C. Causton and C. A. Ball, Microarray databases: standards and ontologies, Nat. Genet., 2002, 32(Suppl.), 469–473 CrossRef PubMed.
- F. Cordero, M. Botta and R. A. Calogero, Microarray data analysis and mining approaches, Briefings Funct. Genomics Proteomics, 2007, 6(4), 265–281 CrossRef CAS PubMed.
- N. Chakravorty, S. Ivanovski, I. Prasadam, R. Crawford, A. Oloyede and Y. Xiao, The microRNA expression signature on modified titanium implant surfaces influences genetic mechanisms leading to osteogenic differentiation, Acta Biomater., 2012, 8(9), 3516–3523 CrossRef CAS PubMed.
- Q. Li, C. Wu, G. Song, H. Zhang, B. Shan, Y. Duan and Y. Wang, Genome-Wide Analysis of Long Noncoding RNA Expression Profiles in Human Xuanwei Lung Cancer, Clin. Lab., 2015, 61(10), 1515–1523 CAS.
- Y. Yan, L. Zhang, Y. Jiang, T. Xu, Q. Mei, H. Wang, R. Qin, Y. Zou, G. Hu, J. Chen and Y. Lu, LncRNA and mRNA interaction study based on transcriptome profiles reveals potential core genes in the pathogenesis of human glioblastoma multiforme, J. Cancer Res. Clin. Oncol., 2015, 141(5), 827–838 CrossRef CAS PubMed.
- K. Kawata, S. Kubota, T. Eguchi, E. Aoyama, N. H. Moritani, M. Oka, H. Kawaki and M. Takigawa, A Tumor Suppressor Gene Product, Platelet-Derived Growth Factor Receptor-Like Protein Controls Chondrocyte Proliferation and Differentiation, J. Cell. Biochem., 2017, 118(11), 4033–4044 CrossRef CAS PubMed.
- N. H. Kim, S. H. Lee, J. J. Ryu, K. H. Choi and J. B. Huh, Effects of rhBMP-2 on Sandblasted and Acid Etched Titanium Implant Surfaces on Bone Regeneration and Osseointegration: Spilt-Mouth Designed Pilot Study, BioMed Res. Int., 2015, 2015, 459393 Search PubMed.
- S. L. Hyzy, R. Olivares-Navarrete, S. Ortman, B. D. Boyan and Z. Schwartz, Bone Morphogenetic Protein 2 Alters Osteogenesis and Anti-Inflammatory Profiles of Mesenchymal Stem Cells Induced by Microtextured Titanium In Vitro, Tissue Eng., Part A, 2017, 23(19–20), 1132–1141 CrossRef CAS PubMed.
- H. Wang, X. Huo, X. R. Yang, J. He, L. Cheng, N. Wang, X. Deng, H. Jin, N. Wang, C. Wang, F. Zhao, J. Fang, M. Yao, J. Fan and W. Qin, STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4, Mol. Cancer, 2017, 16(1), 136 CrossRef PubMed.
- Q. Wang, S. Jiang, A. Song, S. Hou, Q. Wu, L. Qi and X. Gao, HOXD-AS1 functions as an oncogenic ceRNA to promote NSCLC cell progression by sequestering miR-147a, OncoTargets Ther., 2017, 10, 4753–4763 CrossRef PubMed.
- A. A. Yarmishyn, A. O. Batagov, J. Z. Tan, G. M. Sundaram, P. Sampath, V. A. Kuznetsov and I. V. Kurochkin, HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome, BMC Genomics, 2014, 15(Suppl. 9), S7 CrossRef PubMed.
- L. Zheng, J. Chen, Z. Zhou and Z. He, Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway, Tumor Biol., 2017, 39(5), 1010428317705335 CrossRef PubMed.
- G. Dalagiorgou, C. Piperi, C. Adamopoulos, U. Georgopoulou, A. N. Gargalionis, A. Spyropoulou, I. Zoi, M. Nokhbehsaim, A. Damanaki, J. Deschner, E. K. Basdra and A. G. Papavassiliou, Mechanosensor polycystin-1 potentiates differentiation of human osteoblastic cells by upregulating Runx2 expression via induction of JAK2/STAT3 signaling axis, Cell. Mol. Life Sci., 2017, 74(5), 921–936 CrossRef CAS PubMed.
- C. Jin, Y. Zheng, Y. Huang, Y. Liu, L. Jia and Y. Zhou, Long non-coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose-derived stem cells, Cell Biol. Int., 2017, 41(1), 33–41 CrossRef CAS PubMed.
- D. Bertozzi, R. Iurlaro, O. Sordet, J. Marinello, N. Zaffaroni and G. Capranico, Characterization of novel antisense HIF-1alpha transcripts in human cancers, Cell Cycle, 2011, 10(18), 3189–3197 CrossRef CAS PubMed.
- P. Carmeliet and R. K. Jain, Molecular mechanisms and clinical applications of angiogenesis, Nature, 2011, 473(7347), 298–307 CrossRef CAS PubMed.
- A. Ben Shoham, C. Rot, T. Stern, S. Krief, A. Akiva, T. Dadosh, H. Sabany, Y. Lu, K. E. Kadler and E. Zelzer, Deposition of collagen type I onto skeletal endothelium reveals a new role for blood vessels in regulating bone morphology, Development, 2016, 143(21), 3933–3943 CrossRef CAS PubMed.
- X. Duan, S. R. Bradbury, B. R. Olsen and A. D. Berendsen, VEGF stimulates intramembranous bone formation during craniofacial skeletal development, Matrix Biol., 2016, 52–54, 127–140 CrossRef CAS PubMed.
- Y. Zhou, X. Guan, H. Wang, Z. Zhu, C. Li, S. Wu and H. Yu, Hypoxia induces osteogenic/angiogenic responses of bone marrow-derived mesenchymal stromal cells seeded on bone-derived scaffolds via ERK1/2 and p38 pathways, Biotechnol. Bioeng., 2013, 110(6), 1794–1804 CrossRef CAS PubMed.
- W. You, H. Gao, L. Fan, D. Duan, C. Wang and K. Wang, Foxc2 regulates osteogenesis and angiogenesis of bone marrow mesenchymal stem cells, BMC Musculoskeletal Disord., 2013, 14, 199 CrossRef CAS PubMed.
- J. L. Sun, K. Jiao, Q. Song, C. F. Ma, C. Ma, F. R. Tay, L. N. Niu and J. H. Chen, Intrafibrillar silicified collagen scaffold promotes in situ bone regeneration by activating the monocyte p38 signaling pathway, Acta Biomater., 2018, 67, 354–365 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Tables S1, S2 and Fig. S1–S3. See DOI: 10.1039/d0ra01242d |
| This journal is © The Royal Society of Chemistry 2020 |