Open Access Article
Open Access ArticleMachine vision-driven automatic recognition of particle size and morphology in SEM images†
Hyojin
Kim
 *a,
Jinkyu
Han
*a,
Jinkyu
Han
 b and
T. Yong-Jin
Han
b and
T. Yong-Jin
Han
 *b
*b
aCenter for Applied Scientific Computing, Lawrence Livermore National Laboratory, USA. E-mail: hkim@llnl.gov
bMaterials Science Division, Lawrence Livermore National Laboratory, USA. E-mail: han5@llnl.gov
First published on 27th August 2020
Abstract
Scanning Electron Microscopy (SEM) images provide a variety of structural and morphological information of nanomaterials. In the material informatics domain, automatic recognition and quantitative analysis of SEM images in a high-throughput manner are critical, but challenges still remain due to the complexity and the diversity of image configurations in both shape and size. In this paper, we present a generally applicable approach using computer vision and machine learning techniques to quantitatively extract particle size, size distribution and morphology information in SEM images. The proposed pipeline offers automatic, high-throughput measurements even when overlapping nanoparticles, rod shapes, and core–shell nanostructures are present. We demonstrate effectiveness of the proposed approach by performing experiments on SEM images of nanoscale materials and structures with different shapes and sizes. The proposed approach shows promising results (Spearman coefficients of 0.91 and 0.99 using fully automated and semi-automated processes, respectively) when compared with manually measured sizes. The code is made available as open source software at https://github.com/LLNL/LIST.
1. Introduction
The quantitative analysis of SEM images is one of the most important tasks to understand nanomaterial characteristics for a variety of applications. Specifically, visual and structural information such as the particle size, size distribution and morphology of nanomaterials, obtained from SEM images is crucial since desired nanomaterial properties are significantly affected by these properties. Furthermore, the extracted information provides technical and scientific insights into the process of nanomaterial synthesis, fabrication and manufacturing.Despite the importance of the analysis, methods to extract these types of information from SEM images have not been extensively developed. A widely used, conventional approach employs general-purpose image processing tools such as ImageJ.1 Mondini et al.2 proposed a set of methods to measure the morphology and diameter as well as to analyze the extracted information statistically in their user-friendly software environment named Pebbles. This software package allows users to accurately measure nanoparticle diameters manually or semi-automatically. Phromsuwan et al.3 proposed an automated method to analyze the size distribution of nanoparticles in transmission electron microscopy (TEM) images using image processing techniques such as Otsu binarization and Canny edge detectors. Crouzier et al.19 proposed a method to estimate nanoparticle diameters using a remarkable point by varying the electron beam size and profiles. The problems common to these approaches are as follows. First, the entire pipeline to analyze and extract such information from SEM/TEM images requires time-consuming manual intervention, which makes high-throughput measurements infeasible. Furthermore, these methods often fail to measure diameters of overlapping nanoparticles that often appear in most SEM images. Laramy et al.9 proposed a high-throughput algorithm to recognize the structure of core-only nanoparticles in SEM images by estimating heterogeneous nanostructure populations at the bulk level and by using ensemble measurements with an individual nanostructure. Yu et al.6 also presented a high-throughput approach using k-means and computer vision techniques to measure pore parameters such as porosity in EM images. Kopanja et al.7 presented a problem-specific image segmentation method to recognize the shape and aspect ratio of anisotropic magnetic nanochains in TEM images. Wang et al.8 proposed an automated method to determine surface roughness and chemical distribution of nanoparticles from STEM data. The image segmentation method proposed by Le Guen and Paul20 is based on mean-shift clustering and spatially constrained classification.
With the recent drastic success in machine vision and deep learning, several papers have reported methods to automatically recognize morphology and segment region of interest in SEM/TEM images. Xu et al.4 proposed a machine learning-based method to identify key microstructure descriptors in SEM images. Modarres et al.5 introduced a method to employ pre-trained convolutional neural network (CNN)-based models to automatically extract morphology information from SEM images. Ieracitano et al.21 proposed a CNN-based method to classify homogeneous (HNF) and nonhomogeneous nanofibers (NHNF) in SEM images. Azimi et al.22 used a fully convolutional neural network to address the classification of microstructural constituents of low carbon steel in SEM and Light Optical Microscopy (LOM) images. Chen et al.23 proposed to employ the U-Net framework,25 widely used in medical image segmentation, to address SEM image segmentation of shale samples and minerals. CNN-based super-resolution techniques have also been applied to the resolution enhancement of low-quality SEM images.24
Although a number of algorithms to process SEM/TEM images have been developed, a generally applicable, robust measurement of the size and shape of nanomaterials is still a challenge due to the complexity and diversity of nanoparticles in SEM/TEM images. For example, existing methods are somewhat application-specific and are limited to be used for different nanomaterial morphologies such as core/shell and anisotropic structures such as nanorods and nanowires, which are also well-known morphologies in nanomaterials due to their unique material properties. Moreover, an automatic, rapid extraction end-to-end pipeline with little human intervention is crucial in order to facilitate the process of massive SEM/TEM image data.
In this paper, we present a suite of new algorithms to quantitatively analyze SEM images by extracting morphology information and measuring nanoparticle sizes. The proposed approach employing computer vision and machine learning techniques offers fully automated, high-throughput measurements with little user intervention. The distance transform-based size estimation algorithm allows us to extract size information from images containing overlapping regions even when the images are noisy and complex. The proposed size estimation algorithm together with automatic morphology recognition supports core–shell and anisotropic types as well as general nanoparticle types. The automatic extraction of scalebar and text information using the state-of-the-art scene text recognition enables us to automatically convert the size information into an appropriate scale unit. In the case of slightly incorrectly measured sizes using the fully automated pipeline, the proposed approach also offers a semi-automatic process with minimal user intervention by allowing users to choose optimal parameter settings. The proposed algorithm and its implemented GUI software package named Livermore SEM Image Tools (LIST) are publicly available as an open source software code at https://github.com/LLNL/LIST. The main application has been written in C++ with QT.17
The main contribution of this paper is summarized here. To the best of our knowledge, this proposed work is the first attempt to integrate multiple SEM image analysis tasks into a single framework as an open source package. The provided end-to-end pipeline offers great efficiency and effectiveness in the SEM image analysis, which makes high-throughput size and morphology measurement feasible. Unlike the existing application-specific methods, this approach can be used in a wide range of SEM image analyzing applications. Furthermore, the publicly available open source of the proposed work is beneficial to the material science community, where the code can easily be adopted to different SEM/TEM image analysis tasks.
2. Methods
The whole pipeline of LIST consists of three processes: (1) morphology recognition; (2) size measurement; and (3) scalebar and text recognition for scale conversion. The first process is to determine the morphology of the nanoparticles in the input SEM image. We then perform a size measurement process, depending on the input morphology type. Finally, we perform scalebar and text recognition to extract the scalebar and text information embedded on the SEM image to convert the estimated size in pixels into the one in an appropriate unit (e.g., μm or nm). Fig. 1 illustrates the overall pipeline of the proposed approach.2.1. Morphology recognition
The pipeline begins with morphology recognition to classify the nanoparticle shape on the input SEM image. This process enables us to apply each morphology type to its dedicated size measurement algorithm. Defining the number of morphology types of nanoparticles depends on the applications, and we here classify core-only and core–shell nanoparticles. Another goal of this process is to find the correct binary image of the input image, that is, the core regions become white in color where the distance transform can be applied in order to accurately measure the nanoparticle sizes. To accomplish this process, we leverage computer vision-based segmentation algorithms together with morphological image processing techniques. Note that the current version of LIST targets core types including round shape particles (i.e., spherical and oval shape), edged shapes (i.e., cubes and truncated polyhedra), and anisotropic shapes (i.e., rod and needle-like) and their core/shell particles. Triangular edge-shapes and other irregular shapes (e.g., tetrapods and star shapes), where the sizes that are somewhat ill-defined are outside the scope of this paper and the current version of LIST.To find the correct binary image, we first apply contrast limited adaptive histogram equalization26 to enhance low contrast between the background and the particle cores or shell regions. This step also makes the dynamic range consistent over the input images. Then, we binarize the image by performing Otsu's image thresholding14 to adaptively divide the image into foreground and background regions. To obtain the correct binary image, we perform two binarization steps, each of which is done with and without inverting the input image. In each binary image, we perform multiple erosion processes to separate nanoparticle cores as much as possible. This step allows us not only to effectively separate core regions but also to precisely extract particle centers, even when they are heavily adjacent or cluttered. Then, we find isolated core regions and their center locations, that is, any region that has a single adjacency to another segment region. Among the isolated regions, we down-select valid core regions by checking the solidity between the region and its convex hull region, that is, any core region that is close to its convex hull is marked as a valid region. Between two binary images, we pick the correct binary image that has more valid core regions. The core centers are also extracted by computing the centroid of each valid core region. To examine the existence of the shell regions in the binary image, we check whether (1) the aspect ratios between the core and the outer region are similar and (2) the centroid of the outer region of each core is located inside the core region. The detailed algorithm description is shown in Algorithm 1 in the ESI.†
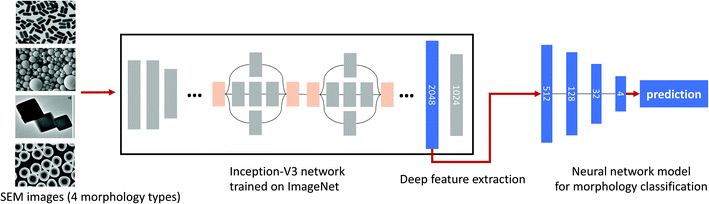
Alternatively, we propose to leverage a convolutional neural network (CNN)-driven image classifier to automatically determine the morphology type. In the case of a small variation in training sample size and image distribution, the state-of-the-art deep CNN models trained on a large number of natural images are known to provide a more distinctive feature representation, compared to shallow CNNs directly trained on the target dataset from scratch. Furthermore, collecting a large number of labeled SEM images for training is almost infeasible. For these reasons, the proposed classifier employs deep features using a pre-trained deep residual network known as the Google inception network.15 In the case of more complex or sequential images (e.g., 3D medical CT images), one can apply more advanced CNN architectures (e.g., DenseNet28 and ENAS29) or Recurrent Neural Network (RNN).30–32 Having said that, we observed that the residual inception network is sufficient to accomplish our morphology recognition task.
The features extracted from the pre-trained inception model are trained by the second neural network model comprising 4 fully connected layers, as shown in Fig. 2. Given an input SEM image, we first run the inception model to extract its feature, which is then fed into the second neural network for training. Note that this approach was separately implemented using python and tensorflow16 as an alternative solution since we observed that the method described above is sufficient for our task to separate core-only and core–shell nanoparticles with the correct binary image extraction.
2.2. Size measurements
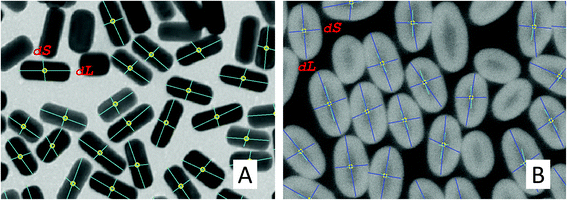
We present two different algorithms to measure the nanoparticle sizes in the input SEM image, depending on the morphology type. In the case of the core–shell type, the shell sizes need to be measured in addition to the core sizes. To this end, we perform an additional process to measure the shell sizes for core–shell nanoparticles.2.3. Scalebar and text recognition
This subsection describes the scalebar and text recognition to identify a scalebar together with a scale number and a unit embedded on the SEM images. The goal of this process is to convert the measured core and shell sizes in pixels into a proper unit (μm or nm) to acquire the physical sizes of the nanoparticles. To localize and extract the scale information, we first perform a scalebar detection process. The scalebar detection is accomplished by a threshold-based segmentation to segment the input image into multiple small segments, each of which is then examined to identify out whether it is a scalebar, i.e., whether the segment is a horizontal-shaped rectangle. To determine whether a segment is a scalebar, we examine its shape feature such as length, size, location and completeness.Once candidate scalebar segments are collected, their neighboring regions are examined to find out whether there is text within each region. This step allows us to detect and localize the text region more efficiently than detecting the entire image region. For precise text region detection, we propose to use a deep CNN-driven algorithm known as an efficient and accurate scene text detector (EAST).11 The EAST algorithm uses a fully convolutional network-based pipeline trained on ICDAR 2015 images12 to make dense per-pixel predictions. The non-maximum suppression is then used to yield possible text regions. We observed that incorporating the EAST detector into the following text recognition process clearly outperforms a naïve text recognition method with randomly selected candidate text regions since performance of the text recognition algorithm is heavily affected by the accuracy of the input text region. Fig. 4 shows a detected scalebar from the segmentation and text regions surrounding the scalebar, detected by the EAST algorithm.
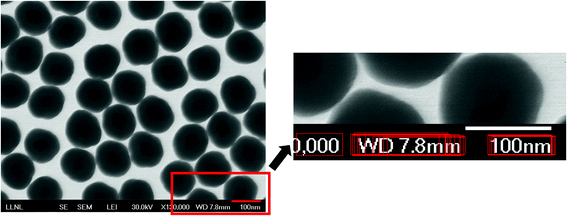 | ||
| Fig. 4 A detected scalebar region and several text regions detected by EAST.11 | ||
Given candidate text regions, we now perform text recognition to check if there is a text to present a scale number with a scale unit. For this, we employ an open source optical character recognition (OCR) algorithm known as Tesseract.13 If the text contains numbers followed by a distance unit (μm or nm), we extract the scale number and the unit. With the width of the scalebar region in pixels with the number and the actual unit, we finally convert all the detected core and shell sizes in pixels to a proper unit.
2.4. High-throughput processes and interactive user interfaces
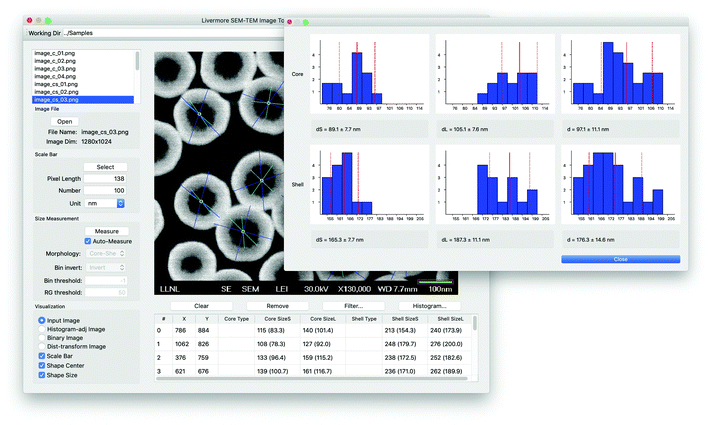
We aim for two different directions in the implementation of the proposed approach. First, the size measurement process can be performed in a high-throughput manner with little user intervention. In this case, all of the processes including the measurement of nanoparticle shapes and sizes are performed through batch processing with default or user-defined parameter settings. This high-throughput measurement enables the automatic processing of a large collection of SEM images. The second direction is to provide a user-friendly interface for more sophisticated manual intervention and visualization, as shown in Fig. 5. This graphical user interface (GUI) based interactive framework enables users to improve the size measurement by changing the thresholds or removing outliers, especially when the nanoparticle sizes appear to be incorrectly measured using the current parameter settings. Moreover, the proposed GUI provides visual results and summary of the size statistics interactively. Through the GUI, users are allowed to perform the following manual intervention tasks: (a) editing scale bar and text information; (b) nanoparticle shape selection; (c) core detection outlier removal by mouse selection or size statistics; (d) intermediate result visualization and other parameter settings; (e) individual nanoparticle information and size statistics visualization. The main code was implemented using C++ and QT,17 together with EAST,11 Tesseract13 and OpenCV.183. Results and discussion
We first report performance evaluation of the CNN-based morphology classification algorithm proposed as an alternative method, described in section 2.1. Note that the main size measurement pipeline uses its own shape recognition to distinguish between core only and core–shell types. Although the shape recognition in the main pipeline is sufficient for the following nanoparticle size measurement task, this deep feature-based CNN method can be used as an alternative solution for improved shape recognition in future implementation. For this experiment, we used 4 different morphology types: circular-shaped core-only, cube-shaped core-only, rod-shaped core-only and core–shell. We split 274 SEM images containing all 4 types into 191 samples for training and the remaining samples for testing. Due to the small number of images to be trained, we augmented data samples by cropping each image into multiple 128 × 128 image patches. The numbers of training and testing image patches are 4488 and 1688, respectively. We compared the proposed deep feature-based deep network with a baseline method, similar to AlexNet,27 composed of a typical CNN architecture without using pre-trained deep features (4 convolutional layers with 3 max pooling layers followed by 4 fully connected layers). We used the standard multi-class cross-entropy as the main loss, a learning rate of 0.004 with the Adam optimizer. The number of epochs and the mini-batch size are 20 and 50, respectively. The accuracy, precision, recall and f1-score of the proposed method are 87%, 0.88, 0.87, 0.87, respectively. Compared to the results of the baseline approach (56%, 0.55, 0.56, 0.54), the deep features extracted from the pre-trained CNNs significantly improve the overall performance.We herein report performance evaluation of the main size measurement pipeline using a SEM image dataset containing both our custom and publicly available images. The image set has 66 SEM images in total, consisting of 20 core-only circular, 6 core-only rod, 32 core–shell circular and 8 core–shell rod types. Fig. 6 shows several examples of size measured images of core-only, rod-shape, and their core/shell particles and there are more successful examples shown in Fig. S1.†
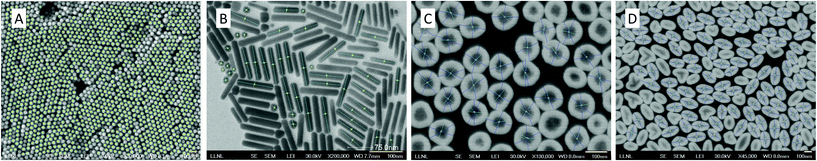 | ||
| Fig. 6 Examples of successfully measured nanoparticles with different shapes. (A) Nanoparticles; (B) rod-shape; (C) core–shell; and (D) rod-shaped core–shell structures. | ||
Due to the lack of ground truth size information, we compared our results with manually measured sizes using the scale bars to show general performance and effectiveness of the proposed automatic and semi-automatic processes with minimal manual intervention. In the case of rod shapes, we report both nanoparticle sizes (dS and dL) to compute the error between the proposed and the manual measurement, whereas the averaged sizes (dM) of the two sizes were used in the circular shapes. The automatic process refers to the proposed pipeline with a pre-defined parameter setting (no user intervention), while the semi-automatic process refers to the same pipeline with a minimal user intervened parameter selection. The user intervention includes (1) changing the morphology type (when the structure is incorrectly classified), (2) selecting different segmentation parameters (when the measured sizes appear to be somewhat incorrect), and (3) removing outliers by visually selecting them on the image or by using the size histograms (when the outliers are not properly removed).
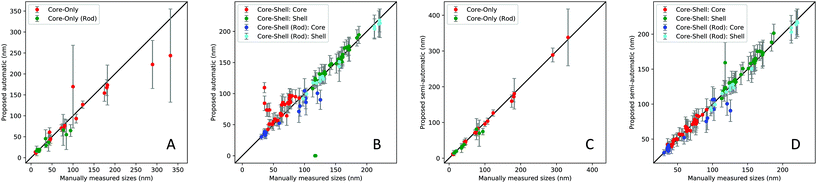
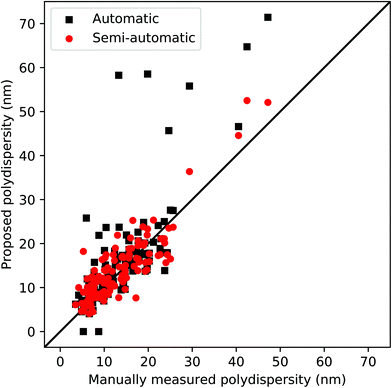
Table 1 summarizes the overall performance of the proposed automatic (auto) and semi-automatic processes (semi). We report the accuracy between the manually measured sizes and the algorithm generated sizes using the binary logistic regression-based accuracy (Logit-Acc) and Spearman coefficients to evaluate the overall performance. Note that the performance results in Table 1 do not include the cases where the automatic process failed to extract scalebar information (3 failure cases out of 66 images). Fig. 7 shows the scatter plots between the manually measured sizes and sizes using two proposed approaches.
| Morphology type | Logit-Acc | Spearman | ||
|---|---|---|---|---|
| auto | semi | auto | semi | |
| Core-only | 13/18 | 17/18 | 0.97 | 0.99 |
| Core-only rod | 6/10 | 9/10 | 0.89 | 0.96 |
Core–shell![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) core core |
18/32 | 32/32 | 0.60 | 0.97 |
Core–shell![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) shell shell |
30/32 | 29/32 | 0.98 | 0.95 |
Core–shell rod![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) core core |
13/16 | 14/16 | 0.86 | 0.90 |
Core–shell rod![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) shell shell |
14/16 | 14/16 | 0.98 | 0.98 |
| Total | 94/124 | 115/124 | 0.91 | 0.99 |
First, we estimate the accuracy by computing the number of correctly measured images as a binary logistic regression problem. The cut-off criterion for the logistic regression is based on the standard deviation of the measured sizes of the nanoparticle structure in each SEM image, thus, 1 if |siM − siP| ≤ σi, 0 otherwise, where siM, siP and σi are the mean of the manually measured sizes, the mean of the proposed sizes and the standard deviation of the proposed sizes of image i, respectively. Note that we count two nanoparticle sizes independently for the rod types. As expected, the semi-automatic process shows improved accuracy compared to the fully automated process. More specifically, the average percentage to meet the above criteria is around 76% (94 out of 124) when the automatic process is used, while the semi-automatic process yields 93% (115 out of 124), as shown in Table 1. Since this logistic accuracy heavily depends on the cut-off value, only exhibiting true (1) or false (0), this evaluation is not sufficient to represent the actual performance of the proposed method. For example, the standard deviation of the manually measured sizes are not taken into consideration and it is likely to be categorized into false cases when the particles have a smaller standard deviation.
Due to the continuous values in the size measurement, we evaluate Spearman coefficients between the manually measured sizes and the algorithm generated sizes, as shown in Table 1. The coefficients show that both automatic and semi-automatic methods are highly correlated with the manual measurement, except for the core–shell![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) core cases with the automated pipeline (coefficient of 0.6). The averaged coefficients of the automatic and semi-automatic processes are 0.91 and 0.99, respectively. We observed that several outliers in the results of the automatic approach are due to the incorrect shape recognition or improper segmentation thresholds which degrades the performance of the particle size estimation, especially for core–shell
core cases with the automated pipeline (coefficient of 0.6). The averaged coefficients of the automatic and semi-automatic processes are 0.91 and 0.99, respectively. We observed that several outliers in the results of the automatic approach are due to the incorrect shape recognition or improper segmentation thresholds which degrades the performance of the particle size estimation, especially for core–shell![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) core cases and some core-only particles. Note that the failure or poorly measured cases are related to (1) incorrect morphology recognition due to ambiguity of the core or shell structures, (2) incorrect segmentation thresholds for less contrast of the SEM images, and (3) scalebar and text detection error, e.g., the scale bar or text was unable to be detected due to blurriness or noise. See several of the failure cases in the ESI (Fig. S2 and S3†). To address such cases, the semi-automatic method with a minimum level of user intervention is applied to improve the accuracy. The results show that the semi-automatic method marginally outperforms the fully automated one. The coefficients of all morphology types using the semi-automatic approach are in the range of 0.9 to 0.99.
core cases and some core-only particles. Note that the failure or poorly measured cases are related to (1) incorrect morphology recognition due to ambiguity of the core or shell structures, (2) incorrect segmentation thresholds for less contrast of the SEM images, and (3) scalebar and text detection error, e.g., the scale bar or text was unable to be detected due to blurriness or noise. See several of the failure cases in the ESI (Fig. S2 and S3†). To address such cases, the semi-automatic method with a minimum level of user intervention is applied to improve the accuracy. The results show that the semi-automatic method marginally outperforms the fully automated one. The coefficients of all morphology types using the semi-automatic approach are in the range of 0.9 to 0.99.
We also report the particle size distribution between the proposed automatic pipeline and the semi-automatic process, known as poly-dispersity (i.e. standard deviation/average size) in Fig. 8. This evaluation provides another critical nanomaterial characteristic obtained from SEM images, to comprehensively evaluate the different sizes and units of nanoparticles in the entire SEM image dataset. As shown in Fig. 8, both processes yield reasonably accurate poly-dispersity, while the semi-automatic process improves the overall accuracy, compared to the fully automatic pipeline. The execution time of the entire size measurement process depends on the size of the image and the number of detected nanoparticles. Typically, the execution time for a single SEM image is 1–3 s when the fully automatic pipeline is used, thereby enabling us to handle over 3500 images in an hour. For SEM images where there is a need for user intervention, additional time is required to select correct thresholds and other parameters for the segmentation process and outlier removal.
4. Conclusions
This paper presented a novel approach to measure the sizes and shapes of the nanoparticles in SEM images in a high-throughput manner. The proposed algorithm is widely applicable to different morphology types including core–shell and rod nanostructures. The automatic size measurement pipeline in a high-throughput manner will also enable the processing of a large number of SEM images with little manual intervention. In the case of failure or poorly measured sizes, the proposed approach offers a semi-automatic process which allows users to interactively intervene the measurement process to improve the accuracy. The experimental results show that both approaches yield reasonably accurate sizes, while the semi-automatic approach slightly outperforms the fully automated one. These two methods are complementary; the automatic pipeline offers rapid size measurements without user intervention and the semi-automatic process enables flexibility with marginally improved accuracy. The proposed approach also offers quantitative analysis of size information such as detailed size information and histograms. We believe that the proposed system will be a powerful software tool especially for material science and chemistry researchers and their communities to effectively accelerate and improve nanoscale size and morphology estimation, thereby potentially characterizing their desired properties. The proposed algorithm has been implemented in a GUI software package, which is publicly available as open source software known as LIST.Disclaimer
This document was prepared as an account of work sponsored by an agency of the United States government. Neither the United States government nor Lawrence Livermore National Security, LLC, nor any of their employees make any warranty, expressed or implied, or assume any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represent that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States government or Lawrence Livermore National Security, LLC. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States government or Lawrence Livermore National Security, LLC, and shall not be used for advertising or product endorsement purposes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Laboratory Directed Research and Development (LDRD) program at Lawrence Livermore National Laboratory (16-ERD-019 and 19-SI-001). Lawrence Livermore National Laboratory is operated by Lawrence Livermore National Security, LLC, for the U.S. Department of Energy, National Nuclear Security Administration under Contract DEAC52-07NA27344. LLNL-JRNL-809488.References
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS.
- S. Mondini, A. M. Ferretti, A. Puglisi and A. Ponti, Nanoscale, 2012, 4, 5356–5372 RSC.
- U. Phromsuwan, C. Sirisathitkul, Y. Sirisathitkul, B. Uyyanonvara and P. Muneesawang, J. Magn., 2013, 18(3), 311–316 CrossRef.
- H. Xu, R. Liu, A. Choudhary and W. Chen, J. Mech. Des., 2015, 137(5), 051403 CrossRef.
- M. H. Modarres, R. Aversa, S. Cozzini, R. Ciancio, A. Leto and G. P. Brandino, Sci. Rep., 2017, 7(1), 13282 CrossRef.
- Z. X. Yu, S. C. Wei, J. W. Zhang, B. Wang, Y. J. Wang, Y. Liang and H. L. Tian, Comput. Mater. Sci., 2020, 171, 109216 CrossRef CAS.
- L. Kopanja, M. Tadic, S. Kralj and J. Žunić, Ceram. Int., 2018, 44(11), 12340–12351 CrossRef CAS.
- Y. C. Wang, T. J. A. Slater, T. S. Rodrigues, P. H. C. Camargo and S. J. Haigh, J. Phys.: Conf. Ser., 2017, 902, 012018 CrossRef.
- C. R. Laramy, K. A. Brown, M. N. O'Brien and C. A. Mirkin, ACS Nano, 2015, 9(12), 12488–12495 CrossRef CAS.
- A. S. Kornilov and I. V. Safonov, J. Imaging, 2018, 4(10), 123 CrossRef.
- X. Zhou, C. Yao, H. Wen, Y. Wang, S. Zhou, W. He and J. Liang, CVPR, 2017, pp. 2642–2651, DOI:10.1109/CVPR.2017.283.
- D. Karatzas, L. Gomez, A. Nicolaou, S. Ghosh, A. Bagdanov, M. Iwamura, J. Matas, L. Neumann, V. Chandrasekhar, S. Lu, F. Shafait, S. Uchida and E. Valveny, ICDAR 2015 competition on Robust Reading, 2015, pp. 1156–1160, DOI:10.1109/ICDAR.2015.7333942.
- R. Smith, ICDAR, 2007, 2, pp. 629–633, DOI:10.1109/ICDAR.2007.4376991.
- M. Sezgin and B. Sankur, J. Electron. Imaging, 2004, 13(1), 146–168 CrossRef.
- C. Szegedy, V. Vanhoucke, S. Ioffe and J. Shlens, CVPR, 2016, DOI:10.1109/CVPR.2016.308.
- M. Abadi, A. Agarwal, P. Barham, E. Brevdo, Z. Chen, C. Citro, G. Corrado, A. Davis, J. Dean, M. Devin, S. Ghemawat, I. Goodfellow, A. Harp, G. Irving, M. Isard, Y. Jia, L. Kaiser, M. Kudlur, J. Levenberg and X. Zheng, TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems, tensorflow.org, 2015 Search PubMed.
- Qt, Cross-platform software development for embedded & desktop http://www.qt.io.
- G. Bradski, OpenCV Library, Dr. Dobb's Journal of Software Tools, 2000 Search PubMed.
- L. Crouzier, A. Delvallee, S. Ducourtieux, L. Devoille, C. Tromas and N. Feltin, Ultramicroscopy, 2019, 207, 112847 CrossRef CAS.
- V. L. Guen and N. Paul, ICIP, 2014, pp. 4447–4451, DOI:10.1109/ICIP.2014.7025902.
- C. Ieracitano, F. Pantó, N. Mammone, A. Paviglianiti, P. Frontera and F. C. Morabito, in Neural Approaches to Dynamics of Signal Exchanges, ed. A. Esposito, M. Faundez-Zanuy, F. Morabito and E. Pasero, Springer, 2020, vol. 151, DOI:10.1007/978-981-13-8950-4_7.
- S. Azimi, D. Britz, M. Engstler, M. Fritz and F. Mücklich, Sci. Rep., 2018, 8, 2128 CrossRef.
- Z. Chen, X. Liu, J. Yang, E. Little and Y. Zhou, Comput. Geosci., 2020, 138, 104450 CrossRef.
- K. Haan, Z. Ballard, Y. Rivenson, Y. Wu and A. Ozcan, Sci. Rep., 2019, 9, 12050 CrossRef.
- O. Ronneberger, P. Fischer and T. Brox, LNCS, 2015, vol. 9351, pp. 234–241.
- K. Zuiderveld, in Graphics Gems IV, ed. P. Heckbert, Academic Press, 1994, pp. 474–485 Search PubMed.
- A. Krizhevsky, I. Sutskever and G. E. Hinton, NIPS, 2012, p. 25, DOI:10.1145/3065386.
- G. Huang, Z. Liu, L. Van Der Maaten and K. Q. Weinberger, CVPR, 2017, pp. 2261–2269, DOI:10.1109/CVPR.2017.243.
- H. Pham, M. Guan, B. Zoph, Q. Le and J. Dean, ICML, 2018, pp. 4095–4104.
- M. Hesamian, W. Jia, X. He and P. Kennedy, J. Digital Imaging, 2019, 32(4), 582–596 CrossRef.
- J. Chen, L. Yang, Y. Zhang, M. Alber and D. Z. Chen, NIPS, 2016, pp. 3044–3052.
- A. Chakravarty and J. Sivaswamy, IEEE J. Biomed. Health Inf., 2019, 23(3), 1151–1162 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr04140h |
| This journal is © The Royal Society of Chemistry 2020 |