Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
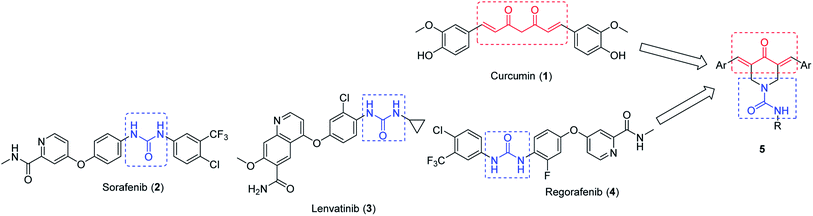
Synthesis, human topoisomerase IIα inhibitory properties and molecular modeling studies of anti-proliferative curcumin mimics†
Nehmedo G. Fawzya,
Siva S. Panda b,
Walid Fayadc,
ElSayed M. Shalaby
b,
Walid Fayadc,
ElSayed M. Shalaby d,
Aladdin M. Srour
d,
Aladdin M. Srour e and
Adel S. Girgis
e and
Adel S. Girgis *a
*a
aDepartment of Pesticide Chemistry, National Research Centre, Dokki, Giza 12622, Egypt. E-mail: girgisas10@yahoo.com
bDepartment of Chemistry & Physics, Augusta University, Augusta, GA 30912, USA
cDrug Bioassay-Cell Culture Laboratory, Pharmacognosy Department, National Research Centre, Dokki, Giza, 12622, Egypt
dX-Ray Crystallography Lab., Physics Division, National Research Centre, Dokki, Giza 12622, Egypt
eDepartment of Therapeutic Chemistry, National Research Centre, Dokki, Giza 12622, Egypt
First published on 21st October 2019
Abstract
3,5-Bis(arylidene)-N-substituted-4-oxo-piperidine-1-carboxamides 24–51 were synthesized as curcumin mimics in a facile pathway through reaction of 3,5-bis(arylidene)-4-piperidones with the appropriate isocyanate in the presence of triethylamine. The 3E,5E′-stereochemical configuration was conclusively supported by single crystal X-ray studies of compounds 25 and 34. Most of the synthesized piperidinecarboxamides showed high anti-proliferative properties with potency higher than that of 5-fluorouracil (clinically approved drug against colon, breast and skin cancers) through in vitro MTT bio-assay. Some of them revealed anti-proliferative properties at sub-micromolar values (IC50 = 0.56–0.70 μM for compounds 29, 30 and 34–38 against HCT116; and IC50 = 0.64 μM for compound 30 against A431 cell lines) with promising inhibitory properties against human DNA topoisomerase IIα. The safe profile of the anti-proliferative active agents against the RPE1 normal cell line may prove their selectivity towards carcinoma cells. Robust molecular models (2D-QSAR, 3D-pharmacophore) supported the SAR and validated the observed bio-properties.
Introduction
Dienone is an attractive chemical motif utilized by many researchers for designing promising biologically/pharmacologically active agents.1–4 Curcumin 1 is one of the most famous dietary natural product dienones isolated from Curcuma longa and used in many Asian countries for its anti-inflammatory and wound healing properties (Ayurvedic medicine).5–7 Curcumin analogues exhibit an extensive broad spectrum of biological properties such as antibacterial,8 anti-tubercular,9 anti-HIV,10 antioxidant,11 antitumor,7 and anti-inflammatory12 activities and exhibit a potential therapeutic effect on Alzheimer's disease.4 Despite the safety profile and broad spectrum biological properties of curcumin, it could not be approved as a therapeutic agent due to its high metabolic instability, low water/plasma solubility and poor systemic bioavailability.13,14 The presence of an active methylene group conjugated with the two β-diketones reduce the stability of curcumin.15–17 Due to these facts, the present study is directed towards the investigation of novel 4-piperidone-1-carboxamides as curcumin mimics (Fig. 1). In other words, a slight modification of the curcumin pharmacophoric skeleton is considered utilizing only one ketonic function conjugated with the two unsaturated olefinic linkages.Interest in the piperidone ring system originate from the diverse biological properties showed by 1,3-diarylidene-4-piperidones as antitumor,18–24 anti-mycobacterial,25 antimalarial26 and acetylcholinesterase inhibitor suggesting the usefulness for Alzheimer's disease treatment.27 The promising properties of 2,4-bis(arylidene)-8-methyl-8-azabicyclo[3.2.1]octan-3-ones against MCF7 (breast) and HepG2 (liver) carcinoma cell lines also prompted the present study.28
Rational for insertion of carboxamide residue at N-1 of the targeted 3,5-bis(arylidene)-4-piperidones is stemmed from the fact that many clinically approved cancer drugs possess 1,3-disubstituted urea function of which, Sorafenib 2, (Nexavar, Bayer Healthcare Pharmaceuticals Inc.) that was approved by the U. S. Food and Drug Administration (US-FDA on November 22, 2013) for the treatment of locally recurrent or metastatic, progressive, differentiated thyroid carcinoma along with its previous approval for the treatment of advanced renal cell carcinoma (2005) and advanced hepatocellular carcinoma (2007).29–31 Lenvatinib 3, was approved by US-FDA (2015) for the treatment of locally recurrent or metastatic, progressive thyroid cancer and recently in 2016 in combination with everolimus for treatment of advanced renal cancer following one prior anti-angiogenic therapy.32 Regorafenib 4, (Stivarga, Bayer HealthCare Pharmaceuticals Inc) approved by US-FDA in 2017 for treatment of hepatocellular carcinoma.33
Generally, rational design of the targeted agents 5 can be recognized as molecular conjugation of pharmacophoric units derived from modified curcuminoid scaffold 1 and uranyl fragment which is the bio-active residue of antitumor drugs sorafenib 2, lenvatinib 3 and regorafenib 4 (Fig. 1).
The targeted 4-piperidone-1-carboxamides 5 are screened against colon, breast and skin human carcinoma cell lines. 5-Fluorouracil (injection) is approved by FDA for clinical treatment of colorectal and breast cancer and topically for skin (basal cell) cancer.34,35 Additionally, pyrimidine scaffold of 5-fluorouracil can be recognized as bio-isosteric form to the targeted skeleton/ring system (piperidine).36,37 For these reasons, 5-fluorouracil is considered as a positive control in the present study.
Results and discussion
Chemistry
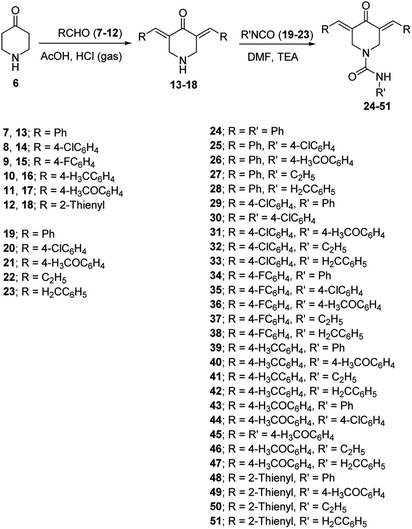
The synthetic pathway for the targeted 3,5-bis(arylidene)-N-substituted-4-oxo-piperidine-1-carboxamides 24–51 is depicted in Scheme 1, through reaction of the appropriate 3,5-bis(arylidene)-4-piperidone 13–18 with the corresponding isocyanate 19–23 in N,N-dimethylformamide (DMF) in the presence of quantitative amount of triethylamine. Spectroscopic (IR, 1H-NMR and 13C-NMR; ESI Fig. S1–S84†) and elemental analysis data support the structure. IR spectrum of compound 24 (representative of the synthesized family), exhibits the ketonic and amidic carbonyls at ν = 1666, 1651 cm−1, respectively. The piperidinyl methylene protons are observed as singlet signal at δH = 4.88 in 1H-NMR spectrum. The exocyclic olefinic protons are revealed as singlet signal at δH = 7.69 supporting the presence of E, E′-configuration.38 13C-NMR spectrum of compound 24 shows the amidic and ketonic carbonyl carbons at δC = 155.1 and 186.8, respectively in addition to the piperidinyl methylene carbon at δC = 45.5. Single crystal X-ray studies of compounds 25 and 34 add conclusive support for the structure.X-ray crystallography
Compounds 25 and 34 are presented in ESI Fig. S85 and S86,† respectively (ORTEP preview). The two compounds were found to be crystallized in the monoclinic system with space group C2/c for compound 25 and P21/n for compound 34 and with one molecule per asymmetric unit cell. The structures are central 4-oxopiperidine-1-carboxamide heterocycle attached at C17 and C19 to two benzylidenes in compound 25 and two 4-fluorobenzylidines in compound 34 forming E,E′-configuration. The carboxamide group is connected to 4-chlorophenyl and phenyl in compound 25 and 34, respectively. The geometrical parameters of the two compounds were found to be comparable to each other and with similar reported structures.38,39 All aryl rings in both compounds are planar as expected. The piperidine ring adopts a half-chair configuration with maximum deviation of 0.379 (3), 0.373 (2) Å at atom N2 for compounds 25 and 34, respectively (puckering parameters, Q = 0.556 (3), 0.539 (2) Å, Θ = 115.6 (3), 116.1 (3)° and Φ = 166.0 (4), 171.1 (2)° for compounds 25 and 34, respectively). In compound 25, the dihedral angles between the least-square plane of the central piperidinyl heterocycle and the phenyl rings (C1–C6, C10–C15 and C21–C26) are 51.89 (13), 47.92 (12) and 26.65 (14)°, respectively, and for compound 34, these dihedral angles are 54.23 (12), 38.27 (12) and 21.31 (14)°, respectively. Molecules in compound 25 are forming a supramolecular chain via intermolecular C26–H261⋯O1 interaction (ESI Fig. S87 and Table S1†). Similar supramolecular assembly is formed in compound 34 via intermolecular C23–H23⋯O1 interaction (ESI Fig. S88 and Table S2†).Structure optimization studies
The computational studies in the current study are directed to determine the difference(s) between optimized geometric parameters (utilizing DFT, dentistry function theory technique by B3LYP method with 3-21G* basis set, ESI Fig. S89 and S90†) for compounds 25 and 34 and those experimentally observed (single crystal X-ray data). This will identify the effect(s) of lattice form (solid state) on these parameters.40,41 The optimized structures of compounds 25 and 34 reveal bond distances and angles comparable to the experimental values (ESI Tables S3 and S4†). The maximum difference in bond lengths and angles are 0.036 Å, 3.9° and 0.052 Å, 4.2° for compounds 25 and 34, respectively. The root-mean square errors (RMSE) in bond lengths and angles for compounds 25 and 34 are 0.018, 1.602; 0.022 and 1.493, respectively. The central piperidinyl heterocycle of the structures obtained theoretically and experimentally have been superimposed in order to globally compare their conformations (ESI Fig. S91 and S92†). The difference(s) in alignment observed can be explained in terms of crystal packing effects in the solid state which are not present in the gaseous state considered by DFT optimization.Anti-proliferative activity
Anti-proliferative properties of the synthesized piperidinecarboxamides 24–51 were investigated against HCT116 (colon), MCF7 (breast) and A431 (squamous skin) carcinoma cell lines by the standard MTT bio-assay utilizing 5-fluorouracil as a reference standard.28 From the results observed (Table 1, ESI Fig. S93–S95†), it has been noticed that most of the synthesized piperidinecarboxamides show potent anti-proliferative properties higher than that of 5-fluorouracil (clinically approved drug against colon, breast and skin cancers34,35) and curcumin (mimic scaffold). Some of the synthesized compounds reveal anti-proliferative properties at sub-micromolar values (IC50 = 0.56–0.70 μM for compounds 29, 30 and 34–38 against HCT116; and IC50 = 0.64 μM for compound 30 against A431 cell lines). Based on the anti-proliferative properties observed, structure–activity relationships (SAR) could be attained. Attachment of electron-withdrawing function (e.g. chlorine or fluorine) to the phenyl ring of exocyclic olefinic linkages oriented at C-3 and C-5 of piperidinecarboxamides, enhances the anti-proliferative properties relative to the electron-donating functions (methyl or methoxy). Insertion of benzylidene fragment at C-3 and C-5 of the targeted piperidones is important for developing antitumor active agents compared to the 2-thienylidene residue. Fluorine containing-compounds reveal higher anti-proliferative efficacy than the chlorine analogues against HCT116 cell line. Meanwhile, better antitumor properties were shown by the chlorine substituted-compounds compared to their fluorine analogues against MCF7 and A431 cell lines [compound 32 (IC50 = 2.35 μM) is an exception, exhibiting potency close to the corresponding analogue 37 (IC50 = 2.32 μM) against MCF7 cell line. The same observations for compounds 29 and 34 (IC50 = 1.29, 1.27 μM, respectively) against A431 cell line].| ID | Compd | IC50a, μM ± SD (therapeutic index)b | |||
|---|---|---|---|---|---|
| HCT116 | MCF7 | A431 | RPE1 | ||
| a IC50 is the concentration producing 50% inhibition of cell growth relative to the control ± standard deviation (SD).b Therapeutic index is the IC50 in normal cell (RPE1)/IC50 in cancer cell.c 5-FU is 5-fluorouracil (standard reference).d NT is not tested. | |||||
| 1 | 24 | 1.08 ± 0.18 (15.40) | 1.83 ± 0.14 (9.09) | 2.49 ± 0.31 (6.68) | 16.63 ± 1.57 |
| 2 | 25 | 1.03 ± 0.29 (21.43) | 2.09 ± 0.26 (10.56) | 2.51 ± 0.24 (8.79) | 22.07 ± 2.06 |
| 3 | 26 | 1.43 ± 0.20 (12.39) | 2.48 ± 0.31 (7.15) | 2.83 ± 0.36 (6.26) | 17.72 ± 1.98 |
| 4 | 27 | 1.61 ± 0.14 (<31.06) | 2.95 ± 0.24 (<16.95) | 2.56 ± 0.29 (<19.53) | >50.00 ± 3.02 |
| 5 | 28 | 1.31 ± 0.22 (9.13) | 2.58 ± 0.30 (4.64) | 2.58 ± 0.27 (4.64) | 11.96 ± 1.76 |
| 6 | 29 | 0.70 ± 0.06 (13.81) | 1.33 ± 0.24 (7.27) | 1.29 ± 0.15 (7.50) | 9.67 ± 2.03 |
| 7 | 30 | 0.58 ± 0.07 (8.43) | 1.13 ± 0.29 (4.33) | 0.64 ± 0.08 (7.64) | 4.89 ± 1.97 |
| 8 | 31 | 1.03 ± 0.13 (7.28) | 1.44 ± 0.19 (5.21) | 1.24 ± 0.14 (6.05) | 7.50 ± 1.75 |
| 9 | 32 | 1.04 ± 0.11 (11.61) | 2.35 ± 0.23 (5.14) | 1.31 ± 0.09 (9.21) | 12.07 ± 1.76 |
| 10 | 33 | 1.13 ± 0.19 (10.10) | 2.14 ± 0.26 (5.33) | 1.45 ± 0.12 (7.87) | 11.41 ± 1.68 |
| 11 | 34 | 0.56 ± 0.04 (31.25) | 2.34 ± 0.29 (7.48) | 1.27 ± 0.09 (13.78) | 17.50 ± 2.16 |
| 12 | 35 | 0.56 ± 0.06 (14.75) | 1.36 ± 0.17 (6.07) | 1.20 ± 0.14 (6.88) | 8.26 ± 2.33 |
| 13 | 36 | 0.62 ± 0.05 (15.26) | 1.71 ± 0.12 (5.53) | 1.31 ± 0.08 (7.22) | 9.46 ± 1.86 |
| 14 | 37 | 0.57 ± 0.07 (17.16) | 2.32 ± 0.30 (4.22) | 1.18 ± 0.11 (8.29) | 9.78 ± 1.53 |
| 15 | 38 | 0.56 ± 0.03 (19.41) | 2.16 ± 0.15 (5.03) | 1.33 ± 0.17 (8.17) | 10.87 ± 1.42 |
| 16 | 39 | 1.44 ± 0.17 (<34.72) | 3.96 ± 0.29 (<12.63) | 2.57 ± 0.15 (<19.46) | >50.00 ± 3.16 |
| 17 | 40 | 2.73 ± 0.20 (<18.32) | 5.10 ± 0.30 (<9.80) | 4.90 ± 0.34 (<10.20) | >50.00 ± 2.96 |
| 18 | 41 | 2.26 ± 0.23 (8.37) | 4.58 ± 0.28 (4.13) | 2.64 ± 0.27 (7.16) | 18.91 ± 1.33 |
| 19 | 42 | 22.98 ± 1.82 (2.04) | 4.90 ± 0.26 (9.58) | 8.75 ± 0.40 (5.37) | 46.96 ± 2.17 |
| 20 | 43 | 1.23 ± 0.15 (<40.65) | 3.96 ± 0.23 (<12.63) | 2.72 ± 0.20 (<18.38) | >50.00 ± 3.43 |
| 21 | 44 | 1.11 ± 0.09 (<45.05) | 4.17 ± 0.27 (<11.99) | 2.85 ± 0.16 (<17.54) | >50.00 ± 2.76 |
| 22 | 45 | 1.38 ± 0.15 (<36.23) | 5.85 ± 0.31 (<8.55) | 11.35 ± 1.10 (<4.41) | >50.00 ± 3.76 |
| 23 | 46 | 1.50 ± 0.14 (31.81) | 5.73 ± 0.33 (8.33) | 8.83 ± 1.26 (5.40) | 47.72 ± 3.05 |
| 24 | 47 | 1.29 ± 0.19 (<38.76) | 4.89 ± 0.25 (<10.22) | 7.98 ± 1.34 (<6.27) | >50.00 ± 3.99 |
| 25 | 48 | >50.00 ± 1.01 (—) | 33.72 ± 1.38 (<1.48) | >50.00 ± 3.65 (—) | >50.00 ± 4.16 |
| 26 | 49 | 40.83 ± 2.61 (<1.22) | 24.68 ± 1.99 (<2.03) | >50.00 ± 2.08 (—) | >50.00 ± 4.22 |
| 27 | 50 | 20.09 ± 2.44 (2.36) | 21.67 ± 2.78 (2.19) | 40.63 ± 2.88 (1.17) | 47.39 ± 2.66 |
| 28 | 51 | >50.00 ± 2.90 (—) | >50.00 ± 3.01 (—) | >50.00 ± 2.38 (—) | >50.00 ± 2.34 |
| 29 | 5-FUc | 20.43 ± 1.99 | 3.15 ± 0.44 | 23.44 ± 2.09 | NTd |
| 30 | Curcumin | 38.25 ± 2.36 | 16.00 ± 2.04 | NTd | NTd |
Screening the synthesized piperidinecarboxamides 24–51 against normal (non-cancer) RPE1 (human immortalized retinal pigment epithelial cell line) can prove the selectivity towards carcinoma cells. From the results obtained (Table 1, ESI Fig. S96†), it has been noticed that the promising anti-proliferative agents synthesized have IC50 towards RPE1 7–32, 4–11 and 5–14 folds relative to that of HCT116, MCF7 and A431 cell lines, respectively. Additionally, it has been noticed that most of the synthesized agents with methyl/methoxy benzylidenes have high selectivity towards the cancer cell lines relative to the normal/non-cancer cell (therapeutic index).
Molecular modeling
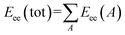 | (1) |
| Entry | Compd | HCT116 | MCF7 | A431 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Observed IC50, μM | Estimated IC50, μM | Errora | Observed IC50, μM | Estimated IC50, μM | Errora | Observed IC50, μM | Estimated IC50, μM | Errora | ||
| a Error is the difference between the observed and estimated bio-activity values. | ||||||||||
| 1 | 24 | 1.08 | 1.17 | −0.09 | 1.83 | 1.39 | 0.44 | 2.49 | 1.94 | 0.55 |
| 2 | 25 | 1.03 | 0.95 | 0.08 | 2.09 | 2.50 | −0.41 | 2.51 | 2.21 | 0.30 |
| 3 | 26 | 1.43 | 1.39 | 0.04 | 2.48 | 2.50 | −0.02 | 2.83 | 2.80 | 0.03 |
| 4 | 27 | 1.61 | 1.29 | 0.32 | 2.95 | 2.62 | 0.33 | 2.56 | 4.25 | −1.69 |
| 5 | 28 | 1.31 | 1.60 | −0.29 | 2.58 | 2.51 | 0.07 | 2.58 | 2.65 | −0.07 |
| 6 | 29 | 0.70 | 0.72 | −0.02 | 1.33 | 1.58 | −0.25 | 1.29 | 1.08 | 0.21 |
| 7 | 30 | 0.58 | 0.60 | −0.02 | 1.13 | 0.89 | 0.24 | 0.64 | 0.92 | −0.28 |
| 8 | 31 | 1.03 | 0.88 | 0.15 | 1.44 | 1.54 | −0.10 | 1.24 | 1.81 | −0.57 |
| 9 | 32 | 1.04 | 0.91 | 0.13 | 2.35 | 2.12 | 0.23 | 1.31 | 1.39 | −0.08 |
| 10 | 33 | 1.13 | 0.93 | 0.20 | 2.14 | 2.00 | 0.14 | 1.45 | 1.76 | −0.31 |
| 11 | 34 | 0.56 | 0.59 | −0.03 | 2.34 | 2.07 | 0.27 | 1.27 | 1.00 | 0.27 |
| 12 | 35 | 0.56 | 0.53 | 0.03 | 1.36 | 1.46 | −0.10 | 1.20 | 1.07 | 0.13 |
| 13 | 36 | 0.62 | 0.69 | −0.07 | 1.71 | 2.18 | −0.47 | 1.31 | 0.98 | 0.33 |
| 14 | 37 | 0.57 | 0.63 | −0.06 | 2.32 | 2.51 | −0.19 | 1.18 | 1.03 | 0.15 |
| 15 | 38 | 0.56 | 0.56 | 0.00 | 2.16 | 2.53 | −0.37 | 1.33 | 1.38 | −0.05 |
| 16 | 39 | 1.44 | 1.36 | 0.08 | 3.96 | 4.46 | −0.50 | 2.57 | 2.79 | −0.22 |
| 17 | 40 | 2.73 | 2.87 | −0.14 | 5.10 | 4.52 | 0.58 | 4.90 | 6.04 | −1.14 |
| 18 | 41 | 2.26 | 3.71 | −1.45 | 4.58 | 4.87 | −0.29 | 2.64 | 3.32 | −0.68 |
| 19 | 42 | 22.98 | 3.43 | 19.55 | 4.90 | 5.10 | −0.20 | 8.75 | 6.78 | 1.97 |
| 20 | 43 | 1.23 | 1.29 | −0.06 | 3.96 | 4.38 | −0.42 | 2.72 | 3.09 | −0.37 |
| 21 | 44 | 1.11 | 0.94 | 0.17 | 4.17 | 3.94 | 0.23 | 2.85 | 3.18 | −0.33 |
| 22 | 45 | 1.38 | 1.55 | −0.17 | 5.85 | 5.82 | 0.03 | 11.35 | 10.62 | 0.73 |
| 23 | 46 | 1.50 | 1.71 | −0.21 | 5.73 | 5.13 | 0.60 | 8.83 | 5.63 | 3.20 |
| 24 | 47 | 1.29 | 1.72 | −0.43 | 4.89 | 4.74 | 0.15 | 7.98 | 6.53 | 1.45 |
Minimum 1-electron reaction index for atom O is an atomic type descriptor (t-criterion = −4.497) participating negatively in the QSAR model (coefficient = −88.6353). Because most of the descriptor values for the tested compounds are with negative sign, the low descriptor value reveals potent anti-proliferative agent as shown for compounds 35 and 41 (descriptor values = −0.00024, −0.00261, respectively). Fukui atomic one-electron reactivity index can be calculated by eqn (2).43
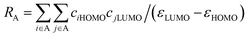 | (2) |
Average information content (order 0) is a topological descriptor. Mean information content index can be calculated by eqn (3).43
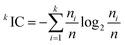 | (3) |
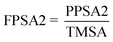 | (4) |
HA dependent HDSA-1 (Zefirov PC) is a charge-related descriptor has a mild power in the QSAR model due to its lowest coefficient value (−0.0318232) among all the descriptors of the attained model. This observation is consistent with the assigned SAR where, attachment of the benzylidene fragment oriented at the C-3 and C-5 of the constructed piperidones with electron-donating function (methyl or methoxy) decreases the anti-proliferative properties (IC50 of compounds 39–47 = 3.96–5.85 μM) relative to the electron-withdrawing function (IC50 of compounds 29–38 with chlorine or fluorine atoms = 1.13–2.35 μM). Hydrogen bonding donor ability of the molecule (HDSA1) can be calculated by eqn (5).43
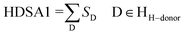 | (5) |
Partial charged surface area (MOPAC PC) for atom C is also a charge-related descriptor. Although this descriptor has the lowest value among all the descriptors of the 2D-QSAR model attained, it observes high effect to the estimated anti-proliferative properties due to its high coefficient value (−866.471) as shown in compounds 30 and 45 (descriptor values = 0.00317, 0.00256, respectively). Atomic charge weighted partial positively charged surface area and atomic charge weighted partial negatively charged surface area can be calculated by eqn (6) and (7), respectively.43
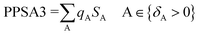 | (6) |
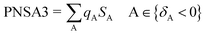 | (7) |
Minimum resonance energy for bond H–C is a semi-empirical descriptor can be calculated by eqn (8).43
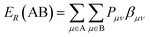 | (8) |
Minimum e–n attraction for atom N is also a semi-empirical descriptor. Nuclear-electron attraction energy for a given atomic species can be determined by eqn (9).43
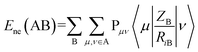 | (9) |
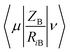 is the electron-nuclear attraction integrals on atomic basis {μν}.
is the electron-nuclear attraction integrals on atomic basis {μν}.
Pharmacophoric model of the anti-proliferative active agents 24–47 against breast (MCF7) carcinoma cell line shows three chemical features [two hydrophobics (H-1, H-2) and one hydrogen bonding acceptor (HBA)] (ESI Fig. S103 and S104†). The substituent of the amidic fragment and aryl group of exocyclic olefinic linkage are fitted with hydrophobics H-1 and H-2, respectively. While the ketonic oxygen gives interaction with HBA for all the tested compounds (ESI Fig. S105†).
Two hydrogen bonding acceptors (HBA-1, HBA-2) and two hydrophobics (H-1, H-2) were viewed by the 3D-pharmacophore of the tested compounds 24–47 against squamous skin (A431) carcinoma cell line (ESI Fig. S106 and S107†). Ketonic and amidic oxygens are fitted with HBA-1 and HBA-2, respectively. However, the exocyclic olefinic linkage and substituent of the amidic fragment are fitted with hydrophobics H-1 and H-2, respectively for all the tested agents (ESI Fig. S108†).
Most of the estimated biological data are correlated with the experimental observations (Table 3). It has been also noticed that all the functions/elements interacted with the pharmacophoric features for all the tested HCT116, MCF7 or A431 cell lines are the controlling parameters governing bio-observations mentioned in SAR. This does not only support the mentioned SAR but also strengthen the assumptions for optimizing more potent anti-proliferative active hits.
| Entry | Compd | HCT116 | MCF7 | A431 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Observed IC50, μM | Estimated IC50, μM | Fit value | Observed IC50, μM | Estimated IC50, μM | Fit value | Observed IC50, μM | Estimated IC50, μM | Fit value | ||
| 1 | 24 | 1.08 | 0.86 | 6.951 | 1.83 | 2.25 | 6.069 | 2.49 | 3.77 | 7.029 |
| 2 | 25 | 1.03 | 1.03 | 6.874 | 2.09 | 2.80 | 5.975 | 2.51 | 1.25 | 7.508 |
| 3 | 26 | 1.43 | 1.00 | 6.885 | 2.48 | 2.62 | 6.002 | 2.83 | 1.21 | 7.524 |
| 4 | 27 | 1.61 | 0.69 | 7.043 | 2.95 | 2.64 | 5.999 | 2.56 | 2.58 | 7.194 |
| 5 | 28 | 1.31 | 1.43 | 6.731 | 2.58 | 2.19 | 6.082 | 2.58 | 3.74 | 7.032 |
| 6 | 29 | 0.70 | 0.93 | 6.917 | 1.33 | 2.93 | 5.954 | 1.29 | 2.31 | 7.242 |
| 7 | 30 | 0.58 | 0.72 | 7.029 | 1.13 | 2.59 | 6.008 | 0.64 | 1.56 | 7.412 |
| 8 | 31 | 1.03 | 1.75 | 6.643 | 1.44 | 2.89 | 5.960 | 1.24 | 2.02 | 7.300 |
| 9 | 32 | 1.04 | 2.31 | 6.521 | 2.35 | 3.14 | 5.924 | 1.31 | 1.65 | 7.387 |
| 10 | 33 | 1.13 | 1.27 | 6.780 | 2.14 | 3.08 | 5.933 | 1.45 | 3.86 | 7.019 |
| 11 | 34 | 0.56 | 0.88 | 6.942 | 2.34 | 2.03 | 6.114 | 1.27 | 2.42 | 7.221 |
| 12 | 35 | 0.56 | 0.70 | 7.040 | 1.36 | 1.82 | 6.162 | 1.2 | 1.46 | 7.442 |
| 13 | 36 | 0.62 | 0.62 | 7.094 | 1.71 | 1.84 | 6.156 | 1.31 | 1.84 | 7.341 |
| 14 | 37 | 0.57 | 0.49 | 7.198 | 2.32 | 2.21 | 6.077 | 1.18 | 1.62 | 7.396 |
| 15 | 38 | 0.56 | 0.48 | 7.203 | 2.16 | 2.46 | 6.030 | 1.33 | 1.46 | 7.440 |
| 16 | 39 | 1.44 | 2.05 | 6.574 | 3.96 | 3.74 | 5.848 | 2.57 | 3.62 | 7.046 |
| 17 | 40 | 2.73 | 1.43 | 6.730 | 5.1 | 3.04 | 5.939 | 4.9 | 1.87 | 7.333 |
| 18 | 41 | 2.26 | 2.02 | 6.581 | 4.58 | 3.85 | 5.836 | 2.64 | 3.58 | 7.052 |
| 19 | 42 | 22.98 | 11.50 | 5.824 | 4.9 | 3.21 | 5.914 | 8.75 | 5.18 | 6.891 |
| 20 | 43 | 1.23 | 1.18 | 6.814 | 3.96 | 2.76 | 5.981 | 2.72 | 2.61 | 7.189 |
| 21 | 44 | 1.11 | 2.43 | 6.500 | 4.17 | 3.45 | 5.883 | 2.85 | 1.60 | 7.401 |
| 22 | 45 | 1.38 | 1.19 | 6.808 | 5.85 | 3.10 | 5.929 | 11.35 | 5.11 | 6.897 |
| 23 | 46 | 1.50 | 1.24 | 6.791 | 5.73 | 3.65 | 5.859 | 8.83 | 5.26 | 6.885 |
| 24 | 47 | 1.29 | 2.98 | 6.411 | 4.89 | 3.72 | 5.850 | 7.98 | 3.07 | 7.118 |
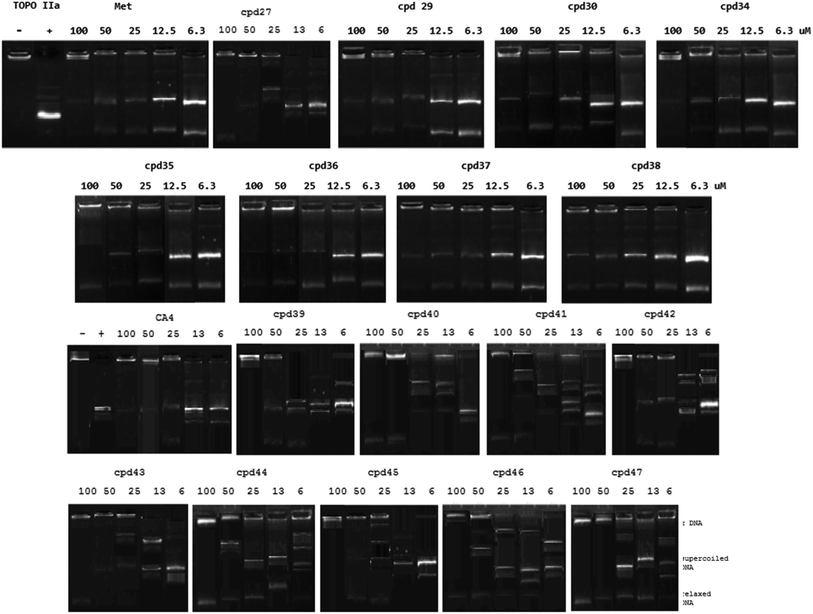
| Entry | Compound | IC50 (μM ± SD)a |
|---|---|---|
| a IC50 is the concentration producing 50% inhibition of the tested enzyme, SD is the standard division. | ||
| 1 | 27 | 33.64 ± 1.30 |
| 2 | 29 | 28.97 ± 1.39 |
| 3 | 30 | 41.79 ± 2.42 |
| 4 | 34 | 30.69 ± 1.32 |
| 5 | 35 | 26.39 ± 1.61 |
| 6 | 36 | 23.04 ± 1.37 |
| 7 | 37 | 27.23 ± 1.72 |
| 8 | 38 | 35.01 ± 2.08 |
| 9 | 39 | 41.30 ± 1.60 |
| 10 | 40 | 40.35 ± 1.56 |
| 11 | 41 | 50.17 ± 1.94 |
| 12 | 42 | 31.49 ± 1.22 |
| 13 | 43 | 28.30 ± 1.09 |
| 14 | 44 | 46.18 ± 1.79 |
| 15 | 45 | 39.31 ± 1.52 |
| 16 | 46 | 56.23 ± 2.18 |
| 17 | 47 | 33.01 ± 1.28 |
| 18 | Methotrexate (Met) | 23.25 ± 1.31 |
| 19 | Combretastatin A-4 (CA-4) | 22.02 ± 0.85 |
Conclusion
(3E,5E)-3,5-Bis(arylidene)-N-substituted-4-oxo-piperidine-1-carboxamides 24–51 were synthesized in excellent yield (80–98%) through reaction of the appropriate 3,5-bis(arylidene)-4-piperidone 13–18 with the corresponding isocyanate 19–23 in DMF in the presence of triethylamine. Single crystal X-ray studies of compounds 25, 34 add good support for the geometrical stereoisomerism. Most of the synthesized piperidinecarboxamides show potent anti-proliferative properties higher than that of 5-fluorouracil (standard reference) through in vitro MTT testing against HCT116 (colon), MCF7 (breast) and A431 (squamous skin) carcinoma cell lines. Promising inhibitory properties were observed against DNA topoisomerase IIα by the tested anti-proliferative agents synthesized. Robust 2D-QSAR and 3D-pharmacophore modeling support the observed anti-proliferative properties. Eventually, it can be concluded that, the series of synthesized compounds may be considered a promising starting point for design of novel highly effective anti-proliferative small molecules based on the high biological activity exhibited towards cancer cells and safety profile against normal cell studied.Experimental section
Melting points were recorded on a Stuart SMP3 melting point apparatus. IR spectra (KBr) were recorded on a Shimadzu FT-IR 8400S spectrophotometer. NMR spectra (DMSO-d6) were taken in a Bruker 500 (1H: 500, 13C: 125 MHz) spectrometer. The starting compounds 13–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46,47 were prepared according to the reported procedures.
46,47 were prepared according to the reported procedures.
Synthesis of 3,5-bis(arylidene)-N-substituted-4-oxo-piperidine-1-carboxamides 24–51 (general procedure)
A mixture of equimolar amounts of the appropriate 3,5-bis(arylidene)-4-piperidone 13–18 (2.5 mmol) and the corresponding isocyanate 19–23 in N,N-dimethylformamide (10 ml) containing triethylamine (2.5 mmol) was stirred at room temperature (25–30 °C) for the appropriate time (TLC control). The separated solid upon pouring the reaction mixture into water (200 ml) containing sodium chloride (1 g) was collected, washed with water and crystallized from a suitable solvent affording the corresponding 24–51.3,5-Di[(E)-benzylidene]-4-oxo-N-phenylpiperidine-1-carboxamide (24)
Obtained from reaction of 13 and 19. Reaction time 10 h. Yellow microcrystals from n-butanol, mp 179–181 °C, yield 94% (0.93 g). IR: νmax/cm−1 1666, 1651, 1605, 1535. 1H-NMR δ (ppm): 4.88 (s, 4H, 2 NCH2), 6.92 (t, J = 7.0 Hz, 1H, arom. H), 7.18 (t, J = 7.2 Hz, 2H, arom. H), 7.29 (d, J = 7.7 Hz, 2H, arom. H), 7.46–7.53 (m, 6H, arom. H), 7.60 (d, J = 7.1 Hz, 4H, arom. H), 7.69 (s, 2H, 2 olefinic CH), 8.89 (s, 1H, NH). 13C-NMR δ (ppm): 45.5 (NCH2), 119.9, 122.1, 128.3, 128.8, 129.5, 130.7, 133.1, 134.4, 135.8, 140.0 (arom. C + olefinic C), 155.1 (amidic CO), 186.8 (ketonic CO). Elemental analysis: C26H22N2O2 (394.47) required C, 79.17; H, 5.62; N, 7.10, found C, 79.34; H, 5.71; N, 7.29.3,5-Di[(E)-benzylidene]-N-(4-chlorophenyl)-4-oxopiperidine-1-carboxamide (25)
Obtained from reaction of 13 and 20. Reaction time 10 h. Yellow microcrystals from n-butanol, mp 186–188 °C, yield 80% (0.86 g). IR: νmax/cm−1 1674, 1655, 1609, 1578. 1H-NMR δ (ppm): 4.87 (s, 4H, 2 NCH2), 7.22 (d, J = 7.9 Hz, 2H, arom. H), 7.32 (d, J = 7.8 Hz, 2H, arom. H), 7.46–7.53 (m, 6H, arom. H), 7.59 (d, J = 7.0 Hz, 4H, arom. H), 7.67 (s, 2H, 2 olefinic CH), 9.01 (s, 1H, NH). 13C-NMR δ (ppm): 45.5 (NCH2), 121.3, 125.7, 128.1, 128.8, 129.5, 130.7, 133.0, 134.4, 135.8, 139.0 (arom. C + olefinic C), 154.9 (amidic CO), 186.7 (ketonic CO). Elemental analysis: C26H21ClN2O2 (428.92) required C, 72.81; H, 4.94; N, 6.53, found C, 72.89; H, 5.00; N, 6.64.3,5-Di[(E)-benzylidene]-N-(4-methoxyphenyl)-4-oxopiperidine-1-carboxamide (26)
Obtained from reaction of 13 and 21. Reaction time 12 h. Yellow microcrystals from n-butanol, mp 184–186 °C, yield 94% (1.00 g). IR: νmax/cm−1 1670, 1624, 1612, 1531. 1H-NMR δ (ppm): 3.67 (s, 3H, OCH3), 4.85 (s, 4H, 2 NCH2), 6.77 (d, J = 7.9 Hz, 2H, arom. H), 7.16 (d, J = 8.0 Hz, 2H, arom. H), 7.46–7.53 (m, 6H, arom. H), 7.59 (d, J = 7.3 Hz, 4H, arom. H), 7.67 (s, 2H, 2 olefinic CH), 8.71 (s, 1H, NH). 13C-NMR δ (ppm): 45.4 (NCH2), 55.1 (OCH3), 113.5, 122.0, 128.8, 129.5, 130.7, 132.8, 133.2, 134.4, 135.7, 154.7 (arom. C + olefinic C), 155.3 (amidic CO), 186.9 (ketonic CO). Elemental analysis: C27H24N2O3 (424.50) required C, 76.40; H, 5.70; N, 6.60, found C, 76.63; H, 5.79; N, 6.66.3,5-Di[(E)-benzylidene]-N-ethyl-4-oxopiperidine-1-carboxamide (27)
Obtained from reaction of 13 and 22. Reaction time 24 h. Yellow microcrystals from benzene, mp 172–174 °C, yield 92% (0.80 g). IR: νmax/cm−1 1674, 1620, 1535, 1447. 1H-NMR δ (ppm): 0.92 (t, J = 6.8 Hz, 3H, CH3), 2.97 (quintet, J = 6.4 Hz, 2H, NCH2CH3), 4.71 (s, 4H, 2 NCH2), 6.82 (s, 1H, NH), 7.44–7.52 (m, 6H, arom. H), 7.57 (d, J = 7.3 Hz, 4H, arom. H), 7.63 (s, 2H, 2 olefinic CH). 13C-NMR δ (ppm): 15.4 (CH3), 35.0 (NCH2CH3), 45.0 (NCH2), 128.7, 129.4, 130.7, 133.3, 134.5, 135.4 (arom. C + olefinic C), 157.0 (amidic CO), 187.0 (ketonic CO). Elemental analysis: C22H22N2O2 (346.43) required C, 76.28; H, 6.40; N, 8.09, found C, 76.39; H, 6.54; N, 7.95.N-Benzyl-3,5-di[(E)-benzylidene]-4-oxopiperidine-1-carboxamide (28)
Obtained from reaction of 13 and 23. Reaction time 24 h. Pale yellow microcrystals from benzene, mp 163–165 °C, yield 98% (1.00 g). IR: νmax/cm−1 1651, 1605, 1574, 1531. 1H-NMR δ (ppm): 4.16 (s, 2H, PhCH2), 4.78 (s, 4H, 2 NCH2), 7.12–7.52 (m, 12H, 11 arom. H + NH), 7.58 (d, J = 7.2 Hz, 4H, arom. H), 7.65 (s, 2H, 2 olefinic CH). 13C-NMR δ (ppm): 43.4 (PhCH2), 45.2 (NCH2), 126.4, 126.8, 128.0, 128.7, 129.4, 130.7, 133.3, 134.5, 135.6, 140.6 (arom. C + olefinic C), 157.3 (amidic CO), 186.9 (ketonic CO). Elemental analysis: C27H24N2O2 (408.50) required C, 79.39; H, 5.92; N, 6.86, found C, 79.59; H, 6.11; N, 7.03.3,5-Bis[(E)-4-chlorobenzylidene]-4-oxo-N-phenylpiperidine-1-carboxamide (29)
Obtained from reaction of 14 and 19. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 205–207 °C, yield 85% (0.99 g). IR: νmax/cm−1 1651, 1605, 1535, 1489. 1H-NMR δ (ppm): 4.84 (s, 4H, 2 NCH2), 6.92 (t, J = 7.1 Hz, 1H, arom. H), 7.18 (t, J = 7.4 Hz, 2H, arom. H), 7.28 (d, J = 7.7 Hz, 2H, arom. H), 7.50–7.65 (m, 10H, 8 arom. H + 2 olefinic CH), 8.87 (s, 1H, NH). 13C-NMR δ (ppm): 45.5 (NCH2), 120.0, 122.2, 128.3, 128.7, 128.8, 132.2, 132.4, 132.5, 133.3, 133.6, 133.7, 134.2, 134.6, 136.4, 139.9 (arom. C + olefinic C), 155.1 (amidic CO), 186.5 (ketonic CO). Elemental analysis: C26H20Cl2N2O2 (463.36) required C, 67.40; H, 4.35; N, 6.05, found C, 67.27; H, 4.28; N, 6.17.3,5-Bis[(E)-4-chlorobenzylidene]-N-(4-chlorophenyl)-4-oxopiperidine-1-carboxamide (30)
Obtained from reaction of 14 and 20. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 185–187 °C, yield 87% (1.08 g). IR: νmax/cm−1 1655, 1597, 1558, 1516. 1H-NMR δ (ppm): 4.83 (s, 4H, 2 NCH2), 7.23 (d, J = 7.8 Hz, 2H, arom. H), 7.31 (d, J = 7.8 Hz, 2H, arom. H), 7.51–7.65 (m, 10H, 8 arom. H + 2 olefinic CH), 9.00 (s, 1H, NH). 13C-NMR δ (ppm): 45.4 (NCH2), 121.3, 125.8, 128.2, 128.7, 128.8, 132.2, 132.36, 132.45, 133.2, 133.5, 133.69, 133.72, 134.2, 134.6, 136.5, 138.9 (arom. C + olefinic C), 154.9 (amidic CO), 186.4 (ketonic CO). Elemental analysis: C26H19Cl3N2O2 (497.80) required C, 62.73; H, 3.85; N, 5.63, found C, 62.61; H, 3.71; N, 5.47.3,5-Bis[(E)-4-chlorobenzylidene]-N-(4-methoxyphenyl)-4-oxopiperidine-1-carboxamide (31)
Obtained from reaction of 14 and 21. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 210–212 °C, yield 91% (1.12 g). IR: νmax/cm−1 1670, 1636, 1605, 1589. 1H-NMR δ (ppm): 3.67 (s, 3H, OCH3), 4.81 (s, 4H, 2 NCH2), 6.77 (d, J = 7.9 Hz, 2H, arom. H), 7.16 (d, J = 7.9 Hz, 2H, arom. H), 7.51–7.64 (m, 10H, 8 arom. H + 2 olefinic CH), 8.69 (s, 1H, NH). 13C-NMR δ (ppm): 45.4 (NCH2), 55.1 (OCH3), 113.5, 122.1, 128.7, 128.8, 132.2, 132.4, 132.5, 132.7, 133.3, 133.67, 133.72, 134.2, 134.5, 136.5, 154.8 (arom. C + olefinic C), 155.3 (amidic CO), 186.6 (ketonic CO). Elemental analysis: C27H22Cl2N2O3 (493.38) required C, 65.73; H, 4.49; N, 5.68, found C, 65.64; H, 4.38; N, 5.88.3,5-Bis[(E)-4-chlorobenzylidene]-N-ethyl-4-oxopiperidine-1-carboxamide (32)
Obtained from reaction of 14 and 22. Reaction time 24 h. Yellow microcrystals from n-butanol, mp 211–213 °C, yield 88% (0.92 g). IR: νmax/cm−1 1670, 1628, 1612, 1535. 1H-NMR δ (ppm): 0.91 (t, J = 6.7 Hz, 3H, CH3), 2.95 (quintet, J = 6.0 Hz, 2H, NCH2CH3), 4.67 (s, 4H, 2 NCH2), 6.80 (s, 1H, NH), 7.55–7.60 (m, 10H, 8 arom. H + 2 olefinic CH). 13C-NMR δ (ppm): 15.4 (CH3), 34.9 (NCH2CH3), 44.9 (NCH2), 128.8, 132.4, 133.3, 133.8, 134.1, 134.3 (arom. C + olefinic C), 156.9 (amidic CO), 186.8 (ketonic CO). Elemental analysis: C22H20Cl2N2O2 (415.31) required C, 63.62; H, 4.85; N, 6.75, found C, 63.81; H, 4.93; N, 6.64.N-Benzyl-3,5-bis[(E)-4-chlorobenzylidene]-4-oxopiperidine-1-carboxamide (33)
Obtained from reaction of 14 and 23. Reaction time 24 h. Yellow microcrystals from n-butanol, mp 199–201 °C, yield 92% (1.10 g). IR: νmax/cm−1 1674, 1628, 1612, 1528. 1H-NMR δ (ppm): 4.15 (s, 2H, PhCH2), 4.74 (s, 4H, 2 NCH2), 7.12 (d, J = 7.2 Hz, 2H, arom. H), 7.17 (t, J = 7.0 Hz, 1H, arom. H), 7.24 (t, J = 7.1 Hz, 2H, arom. H), 7.40 (br s, 1H, NH), 7.54 (d, J = 7.9 Hz, 4H, arom. H), 7.59 (d, J = 7.9 Hz, 4H, arom. H), 7.63 (s, 2H, 2 olefinic CH). 13C-NMR δ (ppm): 43.4 (PhCH2), 45.2 (NCH2), 126.4, 126.7, 128.0, 128.7, 132.4, 133.3, 133.8, 134.1, 134.4, 140.5 (arom. C + olefinic C), 157.2 (amidic CO), 185.6 (ketonic CO). Elemental analysis: C27H22Cl2N2O2 (477.39) required C, 67.93; H, 4.65; N, 5.87, found C, 68.00; H, 4.56; N, 5.73.3,5-Bis[(E)-4-fluorobenzylidene]-4-oxo-N-phenylpiperidine-1-carboxamide (34)
Obtained from reaction of 15 and 19. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 191–193 °C, yield 87% (0.94 g). IR: νmax/cm−1 1651, 1597, 1566, 1535. 1H-NMR δ (ppm): 4.85 (s, 4H, 2 NCH2), 6.92 (t, J = 7.0 Hz, 1H, arom. H), 7.18 (t, J = 7.4 Hz, 2H, arom. H), 7.29–7.36 (m, 6H, arom. H), 7.67 (br s, 6H, 4 arom. H + 2 olefinic CH), 8.88 (s, 1H, NH). 13C-NMR δ (ppm): 45.4 (NCH2), 115.7, 115.9, 120.0, 122.2, 128.3, 131.0, 132.8, 133.0, 133.1, 134.7, 139.9, 161.5, 163.5 (arom. C + olefinic C), 155.1 (amidic CO), 186.6 (ketonic CO). Elemental analysis: C26H20F2N2O2 (430.45) required C, 72.55; H, 4.68; N, 6.51, found: C, 72.41; H, 4.75; N, 6.59.N-(4-Chlorophenyl)-3,5-bis[(E)-4-fluorobenzylidene]-4-oxopiperidine-1-carboxamide (35)
Obtained from reaction of 15 and 20. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 192–194 °C, yield 95% (1.10 g). IR: νmax/cm−1 1655, 1600, 1574, 1504. 1H-NMR δ (ppm): 4.85 (s, 4H, 2 NCH2), 7.22–7.37 (m, 7H, arom. H), 7.55–7.58 (br d, 2H, arom. H), 7.66 (br s, 5H, 3 arom. H + 2 olefinic CH), 9.00 (s, 1H, NH). 13C-NMR δ (ppm): 45.4 (NCH2), 115.6, 115.7, 115.9, 119.8, 121.3, 125.7, 128.2, 131.0, 132.6, 132.67, 132.72, 133.1, 134.8, 139.0, 161.5, 163.5 (arom. C + olefinic C), 154.9 (amidic CO), 186.5 (ketonic CO). Elemental analysis: C26H19ClF2N2O2 (464.90) required C, 67.17; H, 4.12; N, 6.03, found C, 67.33; H, 4.17; N, 6.07.3,5-Bis[(E)-4-fluorobenzylidene]-N-(4-methoxyphenyl)-4-oxopiperidine-1-carboxamide (36)
Obtained from reaction of 15 and 21. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 189–191 °C, yield 97% (1.12 g). IR: νmax/cm−1 1670, 1628, 1597, 1504. 1H-NMR δ (ppm): 3.67 (s, 3H, OCH3), 4.82 (s, 4H, 2 NCH2), 6.78 (d, J = 7.8 Hz, 2H, arom. H), 7.17 (d, J = 7.8 Hz, 2H, arom. H), 7.34 (t, J = 8.1 Hz, 4H, arom. H) 7.66 (br s, 6H, 4 arom. H + 2 olefinic CH), 8.71 (s, 1H, NH). 13C-NMR δ (ppm): 45.3 (NCH2), 55.1 (OCH3), 113.5, 115.7, 115.9, 122.1, 131.02, 131.04, 132.8, 132.9, 133.0, 133.1, 134.6, 154.8, 161.5, 163.5 (arom. C + olefinic C), 155.3 (amidic CO), 186.7 (ketonic CO). Elemental analysis: for C27H22F2N2O3 (460.48) required C, 70.43; H, 4.82; N, 6.08, found C, 70.49; H, 4.94; N, 6.01.N-Ethyl-3,5-bis[(E)-4-fluorobenzylidene]-4-oxopiperidine-1-carboxamide (37)
Obtained from reaction of 15 and 22. Reaction time 24 h. Pale yellow microcrystals from benzene, mp 180–182 °C, yield 94% (0.90 g). IR: νmax/cm−1 1674, 1620, 1582, 1543. 1H-NMR δ (ppm): 0.92 (t, J = 6.9 Hz, 3H, CH3), 2.96 (quintet, J = 6.2 Hz, 2H, NCH2CH3), 4.68 (s, 4H, 2 NCH2), 6.81 (s, 1H, NH), 7.33 (t, J = 8.3 Hz, 4H, arom. H) 7.62 (br d, 6H, 4 arom. H + 2 olefinic CH). 13C-NMR δ (ppm): 15.4 (CH3), 34.9 (NCH2CH3), 44.9 (NCH2), 115.9, 131.1, 133.0, 134.4, 161.5, 163.5 (arom. C + olefinic C), 156.9 (amidic CO), 186.8 (ketonic CO). Elemental analysis: C22H20F2N2O2 (382.41) required C, 69.10; H, 5.27; N, 7.33, found C, 69.15; H, 5.36; N, 7.19.N-Benzyl-3,5-bis[(E)-4-fluorobenzylidene]-4-oxopiperidine-1-carboxamide (38)
Obtained from reaction of 15 and 23. Reaction time 24 h. Pale yellow microcrystals from n-butanol, mp 178–180 °C, yield 97% (1.02 g). IR: νmax/cm−1 1674, 1606, 1582, 1543. 1H-NMR δ (ppm): 4.17 (s, 2H, PhCH2), 4.75 (s, 4H, 2 NCH2), 7.13 (d, J = 7.2 Hz, 2H, arom. H), 7.17 (t, J = 7.1 Hz, 1H, arom. H), 7.24 (t, J = 7.2 Hz, 2H, arom. H), 7.33 (t, J = 8.3 Hz, 4H, arom. H), 7.41 (br s, 1H, NH), 7.64 (br s, 6H, 4 arom. H + 2 olefinic CH). 13C-NMR δ (ppm): 43.4 (PhCH2), 45.1 (NCH2), 115.7, 115.8, 126.4, 126.8, 128.0, 131.0, 131.1, 132.99, 133.0, 133.04, 133.1, 134.5, 140.6, 161.5, 163.5 (arom. C + olefinic C), 157.2 (amidic CO), 186.7 (ketonic CO). Elemental analysis: C27H22F2N2O2 (444.48) required C, 72.96; H, 4.99; N, 6.30, found C, 73.12; H, 4.92; N, 6.10.3,5-Bis[(E)-4-methylbenzylidene]-4-oxo-N-phenylpiperidine-1-carboxamide (39)
Obtained from reaction of 16 and 19. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 221–223 °C, yield 87% (0.92 g). IR: νmax/cm−1 1670, 1643, 1601, 1535. 1H-NMR δ (ppm): 2.37 (s, 6H, 2 ArCH3), 4.85 (s, 4H, 2 NCH2), 6.91 (t, J = 7.1 Hz, 1H, arom. H), 7.18 (t, J = 7.3 Hz, 2H, arom. H), 7.28–7.33 (m, 6H, arom. H), 7.49 (d, J = 7.5 Hz, 4H, arom. H), 7.64 (s, 2H, olefinic CH), 8.88 (s, 1H, NH). 13C-NMR δ (ppm): 21.0 (ArCH3), 45.5 (NCH2), 119.9, 122.1, 128.3, 129.4, 130.8, 131.7, 132.3, 135.7, 139.4, 140.0 (arom. C + olefinic C), 155.1 (amidic CO), 186.6 (ketonic CO). Elemental analysis: C28H26N2O2 (422.53) required C, 79.59; H, 6.20; N, 6.63, found C, 79.66; H, 6.31; N, 6.60.N-(4-Methoxyphenyl)-3,5-bis[(E)-4-methylbenzylidene]-4-oxopiperidine-1-carboxamide (40)
Obtained from reaction of 16 and 21. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 230–232 °C, yield 98% (1.11 g). IR: νmax/cm−1 1674, 1628, 1605, 1582. 1H-NMR δ (ppm): 2.36 (s, 6H, 2 ArCH3), 3.67 (s, 3H, OCH3), 4.83 (s, 4H, 2 NCH2), 6.77 (d, J = 7.7 Hz, 2H, arom. H), 7.18 (d, J = 7.7 Hz, 2H, arom. H), 7.31 (d, J = 7.0 Hz, 4H, arom. H) 7.48 (d, J = 7.1 Hz, 4H, rom. H), 7.63 (s, 2H, 2 olefinic CH), 8.71 (s, 1H, NH). 13C-NMR δ (ppm): 21.0 (ArCH3), 45.5 (NCH2), 55.1 (OCH3), 113.5, 122.0, 129.4, 130.8, 131.7, 132.4, 132.9, 135.6, 139.4, 154.7 (arom. C + olefinic C), 155.3 (amidic CO), 186.7 (ketonic CO). Elemental analysis: C29H28N2O3 (452.55) required C, 76.97; H, 6.24; N, 6.19, found C, 76.81; H, 6.05; N, 6.10.N-Ethyl-3,5-bis[(E)-4-methylbenzylidene]-4-oxopiperidine-1-carboxamide (41)
Obtained from reaction of 16 and 22. Reaction time 24 h. Yellow microcrystals from benzene, mp 174–176 °C, yield 88% (0.82 g). IR: νmax/cm−1 1670, 1628, 1605, 1535. 1H-NMR δ (ppm): 0.92 (t, J = 6.6 Hz, 3H, CH3), 2.37 (s, 6H, 2 ArCH3), 2.96 (quintet, J = 6.3 Hz, 2H, NCH2CH3), 4.68 (s, 4H, 2 NCH2), 6.79 (s, 1H, NH), 7.31 (d, J = 7.2 Hz, 4H, arom. H) 7.45 (d, J = 7.3 Hz, 4H, arom. H), 7.58 (s, 2H, 2 olefinic CH). 13C-NMR δ (ppm): 15.4 (NCH2CH3), 21.0 (ArCH3), 34.9 (NCH2CH3), 45.0 (NCH2), 129.4, 130.8, 131.8, 132.5, 135.4, 139.3 (arom. C + olefinic C), 157.0 (amidic CO), 186.9 (ketonic CO). Elemental analysis: C24H26N2O2 (374.48) required C, 76.98; H, 7.00; N, 7.48, found C, 77.06; H, 6.94; N, 7.60.N-Benzyl-3,5-bis[(E)-4-methylbenzylidene]-4-oxopiperidine-1-carboxamide (42)
Obtained from reaction of 16 and 23. Reaction time 24 h. Yellow microcrystals from methanol, mp 166–168 °C, yield 92% (1.00 g). IR: νmax/cm−1 1651, 1601, 1566, 1512. 1H-NMR δ (ppm): 2.36 (s, 6H, 2 ArCH3), 4.17 (s, 2H, PhCH2), 4.75 (s, 4H, 2 NCH2), 7.14 (d, J = 7.2 Hz, 2H, arom. H), 7.17 (d, J = 7.1 Hz, 1H, arom. H), 7.24 (t, J = 7.2 Hz, 2H, arom. H), 7.31 (d, J = 7.5 Hz, 4H, arom. H), 7.40 (br s, 1H, NH), 7.46 (d, J = 7.5 Hz, 4H, arom. H), 7.62 (s, 2H, 2 olefinic CH). 13C-NMR δ (ppm): 21.0 (ArCH3), 43.5 (PhCH2), 45.2 (NCH2), 126.4, 126.8, 128.0, 129.4, 130.8, 131.7, 132.5, 135.5, 139.3, 140.6 (arom. C + olefinic C), 157.2 (amidic CO), 186.7 (ketonic CO). Elemental analysis: C29H28N2O2 (436.56) required C, 79.79; H, 6.47; N, 6.42, found C, 79.92; H, 6.61; N, 6.48.3,5-Bis[(E)-4-methoxybenzylidene]-4-oxo-N-phenylpiperidine-1-carboxamide (43)
Obtained from reaction of 17 and 19. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 190–192 °C, yield 93% (1.05 g). IR: νmax/cm−1 1670, 1643, 1597, 1558. 1H-NMR δ (ppm): 3.83 (s, 6H, 2 OCH3), 4.84 (s, 4H, 2 NCH2), 6.91 (t, J = 6.5 Hz, 1H, arom. H), 7.07 (d, J = 7.6 Hz, 4H, arom. H), 7.18 (t, J = 7.0 Hz, 2H, arom. H), 7.30 (d, J = 7.7 Hz, 2H, arom. H), 7.56 (d, J = 7.7 Hz, 4H, arom. H), 7.62 (s, 2H, olefinic CH), 8.88 (s, 1H, NH). 13C-NMR δ (ppm): 45.5 (NCH2), 55.4 (OCH3), 114.4, 119.9, 122.1, 127.1, 128.3, 131.0, 132.7, 135.4, 140.1, 160.3 (arom. C + olefinic C), 155.2 (amidic CO), 186.4 (ketonic CO). Elemental analysis: C28H26N2O4 (454.53) required C, 73.99; H, 5.77; N, 6.16, found C, 73.80; H, 5.82; N, 6.19.N-(4-Chlorophenyl)-3,5-bis[(E)-4-methoxybenzylidene]-4-oxopiperidine-1-carboxamide (44)
Obtained from reaction of 17 and 20. Reaction time 12 h. Pale yellow microcrystals from n-butanol, mp 185–187 °C, yield 84% (1.02 g). IR: νmax/cm−1 1663, 1636, 1597, 1566. 1H-NMR δ (ppm): 3.83 (s, 6H, 2 OCH3), 4.84 (s, 4H, 2 NCH2), 7.07 (br d, 4H, arom. H), 7.24 (br s, 2H, arom. H), 7.34 (br s, 2H, arom. H) 7.55 (br s, 4H, rom. H), 7.62 (s, 2H, 2 olefinic CH), 9.01 (s, 1H, NH). 13C-NMR δ (ppm): 45.5 (NCH2), 55.3 (OCH3), 114.4, 121.3, 125.6, 127.1, 128.1, 130.8, 132.7, 135.5, 139.1, 160.3 (arom. C + olefinic C), 154.9 (amidic CO), 186.3 (ketonic CO). Elemental analysis: C28H25ClN2O4 (488.97) required C, 68.78; H, 5.15; N, 5.73, found C, 68.89; H, 5.07; N, 5.92.3,5-Bis[(E)-4-methoxybenzylidene]-N-(4-methoxyphenyl)-4-oxopiperidine-1-carboxamide (45)
Obtained from reaction of 17 and 21. Reaction time 12 h. Yellow microcrystals from n-butanol, mp 212–214 °C, yield 80% (0.97 g). IR: νmax/cm−1 1670, 1628, 1597, 1558. 1H-NMR δ (ppm): 3.67 (s, 3H, OCH3), 3.83 (s, 6H, 2 OCH3), 4.82 (s, 4H, 2 NCH2), 6.77 (d, J = 8.0 Hz, 2H, arom. H), 7.06 (d, J = 7.7 Hz, 4H, arom. H), 7.19 (d, J = 8.1 Hz, 2H, arom. H) 7.56 (d, J = 8.0 Hz, 4H, rom. H), 7.61 (s, 2H, 2 olefinic CH), 8.71 (s, 1H, NH). 13C-NMR δ (ppm): 45.4 (NCH2), 55.1 (OCH3), 55.3 (OCH3), 113.5, 114.3, 122.0, 127.1, 131.0, 132.7, 135.3, 155.3, 160.3 (arom. C + olefinic C), 154.7 (amidic CO), 186.5 (ketonic CO). Elemental analysis: C29H28N2O5 (484.55) required C, 71.88; H, 5.82; N, 5.78, found C, 71.94; H, 5.95; N, 5.86.N-Ethyl-3,5-bis[(E)-4-methoxybenzylidene]-4-oxopiperidine-1-carboxamide (46)
Obtained from reaction of 17 and 22. Reaction time 24 h. Yellow microcrystals from benzene, mp 173–175 °C, yield 98% (0.99 g). IR: νmax/cm−1 1665, 1620, 1597, 1566. 1H-NMR δ (ppm): 0.93 (t, J = 6.4 Hz, 3H, CH3), 2.98 (br s, 2H, NCH2CH3), 3.82 (s, 6H, 2 OCH3), 4.67 (s, 4H, 2 NCH2), 6.81 (s, 1H, NH), 7.05 (d, J = 7.9 Hz, 4H, arom. H) 7.52 (d, J = 7.9 Hz, 4H, arom. H), 7.57 (s, 2H, 2 olefinic CH). 13C-NMR δ (ppm): 15.5 (CH3), 35.0 (NCH2CH3), 44.9 (NCH2), 55.3 (OCH3), 114.3, 127.2, 131.2, 132.7, 135.1, 160.2 (arom. C + olefinic C), 157.0 (amidic CO), 186.6 (ketonic CO). Elemental analysis: C24H26N2O4 (406.48) required C, 70.92; H, 6.45; N, 6.89, found C, 71.06; H, 6.51; N, 6.92.N-Benzyl-3,5-bis[(E)-4-methoxybenzylidene]-4-oxopiperidine-1-carboxamide (47)
Obtained from reaction of 17 and 23. Reaction time 24 h. Yellow microcrystals from n-butanol, mp 177–179 °C, yield 85% (1.00 g). IR: νmax/cm−1 1670, 1628, 1597, 1535. 1H-NMR δ (ppm): 3.82 (s, 6H, 2 OCH3), 4.18 (s, 2H, PhCH2), 4.74 (s, 4H, 2 NCH2), 7.05 (d, J = 7.9 Hz, 4H, arom. H), 7.14–7.18 (m, 3H, arom. H), 7.24 (t, J = 7.2 Hz, 2H, arom. H), 7.41 (br s, 1H, NH), 7.54 (d, J = 7.9 Hz, 4H, arom. H), 7.60 (s, 2H, 2 olefinic CH). 13C-NMR δ (ppm): 43.5 (PhCH2), 45.2 (NCH2), 55.3 (OCH3), 114.3, 126.4, 126.8, 127.1, 128.0, 131.2, 132.7, 135.2, 140.7, 160.2 (arom. C + olefinic C), 157.2 (amidic CO), 186.5 (ketonic CO). Elemental analysis: C29H28N2O4 (468.55) required C, 74.34; H, 6.02; N, 5.98, found C, 74.55; H, 6.06; N, 5.96.(3E,5E)-4-Oxo-N-phenyl-3,5-bis(thiophen-2-ylmethylene)piperidine-1-carboxamide (48)
Obtained from reaction of 18 and 19. Reaction time 12 h. Yellow microcrystals from n-butanol, mp 188–190 °C, yield 87% (0.88 g). IR: νmax/cm−1 1670, 1647, 1589, 1535. 1H-NMR δ (ppm): 4.86 (s, 4H, 2 NCH2), 6.94 (t, J = 7.0 Hz, 1H, arom. H), 7.21 (t, J = 7.4 Hz, 2H, arom. H), 7.29 (br s, 2H, arom. H), 7.36 (d, J = 7.6 Hz, 2H, arom. H), 7.64 (s, 2H, olefinic CH), 7.86 (br s, 2H, arom. H), 7.96 (br s, 2H, arom. H), 9.08 (s, 1H, NH). 13C-NMR δ (ppm): 46.0 (NCH2), 120.2, 122.6, 128.4, 128.8, 129.0, 130.3, 132.7, 134.7, 138.2, 140.6 (arom. C + olefinic C), 156.0 (amidic CO), 185.9 (ketonic CO). Elemental analysis: C22H18N2O2S2 (406.52) required C, 65.00; H, 4.46; N, 6.89, found C, 64.93; H, 4.56; N, 6.95.(3E,5E)-N-(4-Methoxyphenyl)-4-oxo-3,5-bis(thiophen-2-ylmethylene)piperidine-1-carboxamide (49)
Obtained from reaction of 18 and 21. Reaction time 12 h. Yellow microcrystals from n-butanol, mp 194–196 °C, yield 84% (0.92 g). IR: νmax/cm−1 1667, 1651, 1593, 1535. 1H-NMR δ (ppm): 3.68 (s, 3H, OCH3), 4.83 (s, 4H, 2 NCH2), 6.80 (d, J = 7.9 Hz, 2H, arom. H), 7.24 (d, J = 7.7 Hz, 2H, arom. H), 7.28 (br s, 2H, arom. H), 7.64 (s, 2H, 2 olefinic CH), 7.84 (br s, 2H, arom. H), 7.96 (br s, 2H, arom. H), 8.90 (s, 1H, NH). 13C-NMR δ (ppm): 45.4 (NCH2), 55.1 (OCH3), 113.5, 121.7, 127.9, 128.6, 129.9, 132.2, 133.0, 134.2, 137.7, 154.7 (arom. C + olefinic C), 155.7 (amidic CO), 185.5 (ketonic CO). Elemental analysis: C23H20N2O3S2 (436.54) required C, 63.28; H, 4.62; N, 6.42, found C, 63.12; H, 4.53; N, 6.56.(3E,5E)-N-Ethyl-4-oxo-3,5-bis(thiophen-2-ylmethylene)piperidine-1-carboxamide (50)
Obtained from reaction of 18 and 22. Reaction time 24 h. Yellow microcrystals from n-butanol, mp 197–199 °C, yield 96% (0.86 g). IR: νmax/cm−1 1670, 1636, 1589, 1551. 1H-NMR δ (ppm): 0.97 (t, J = 6.5 Hz, 3H, CH3), 3.02 (br s, 2H, NCH2CH3), 4.69 (s, 4H, 2 NCH2), 6.91 (s, 1H, NH), 7.28 (d, J = 3.1 Hz, 2H, arom. H), 7.60 (s, 2H, 2 olefinic CH), 7.79 (br s, 2H, arom. H), 7.95 (br s, 2H, arom. H). 13C-NMR δ (ppm): 15.4 (CH3), 35.1 (NCH2CH3), 44.9 (NCH2), 127.6, 128.5, 130.2, 132.1, 134.0, 137.8 (arom. C + olefinic C), 157.3 (amidic CO), 185.7 (ketonic CO). Elemental analysis: C18H18N2O2S2 (358.47) required C, 60.31; H, 5.06; N, 7.81, found C, 60.23; H, 5.21; N, 7.98.(3E,5E)-N-Benzyl-4-oxo-3,5-bis(thiophen-2-ylmethylene)piperidine-1-carboxamide (51)
Obtained from reaction of 18 and 23. Reaction time 24 h. Yellow microcrystals from n-butanol, mp 175–177 °C, yield 95% (1.00 g). IR: νmax/cm−1 1663, 1620, 1597, 1566. 1H-NMR δ (ppm): 4.24 (s, 2H, PhCH2), 4.76 (s, 4H, 2 NCH2), 7.20–7.28 (m, 7H, arom. H), 7.55 (br s, 1H, NH), 7.61 (s, 2H, 2 olefinic CH), 7.83 (br s, 2H, arom. H), 7.94 (br s, 2H, arom. H). 13C-NMR δ (ppm): 43.7 (PhCH2), 45.1 (NCH2), 126.4, 126.9, 127.8, 128.1, 128.5, 130.1, 132.1, 134.0, 137.8, 140.6 (arom. C + olefinic C), 157.5 (amidic CO), 185.6 (ketonic CO). Elemental analysis: C23H20N2O2S2 (420.55) required C, 65.69; H, 4.79; N, 6.66, found C, 65.81; H, 4.85; N, 6.52.X-ray crystallography
Experimental part of X-ray crystallography is reported in the ESI.†In vitro antitumor screening
Experimental part of the in vitro antitumor screening is reported in the ESI.†2D-QSAR studies
Experimental part of the 2D-QSAR studies is reported in the ESI.†Human DNA topoisomerase IIα inhibitory properties
Experimental part of the Human DNA topoisomerase IIα inhibitory properties determination is reported in the ESI.†Conflicts of interest
There is no conflict to declare.Acknowledgements
This work was supported financially by National Research Centre, Egypt, Project ID: 11010341. Thanks also to Dr Nicola Demitri (Elettra-Sincrotrone Trieste S. C. p. A., Trieste, Italy) due to his help in X-ray measument.Notes and references
- Y. Zhang, Z. Liu, J. Wu, B. Bai, H. Chen, Z. Xiao, L. Chen, Y. Zhao, H. Lum, Y. Wang, H. Zhang and G. Liang, New MD2 inhibitors derived from curcumin with improved anti-inflammatory activity, Eur. J. Med. Chem., 2018, 148, 291–305 CrossRef CAS.
- P. V. S. Ramya, L. Guntuku, S. Angapelly, C. S. Digwal, U. J. Lakshmi, D. K. Sigalapalli, B. N. Babu, V. G. M. Naidu and A. Kamal, Synthesis and biological evaluation of curcumin inspired imidazo[1,2-a]pyridine analogues as tubulin polymerization inhibitors, Eur. J. Med. Chem., 2018, 143, 216–231 CrossRef CAS.
- Z.-S. Tu, Q. Wang, D.-D. Sun, F. Dai and B. Zhou, Design, synthesis, and evaluation of curcumin derivatives as Nrf2 activators and cytoprotectors against oxidative death, Eur. J. Med. Chem., 2017, 134, 72–85 CrossRef CAS.
- Z. Liu, L. Fang, H. Zhang, S. Gou and L. Chen, Design, synthesis and biological evaluation of multifunctional tacrine-curcumin hybrids as new cholinesterase inhibitors with metal ions-chelating and neuroprotective property, Bioorg. Med. Chem., 2017, 25, 2387–2398 CrossRef CAS.
- D. J. Newman, G. M. Cragg and K. M. Snader, Natural products as sources of new drugs over the period 1981-2002, J. Nat. Prod., 2003, 66, 1022–1037 CrossRef CAS.
- D. Tunc, E. Dere, D. Karakas, B. Cevatemre, V. T. Yilmaz and E. Ulukaya, Cytotoxic and apoptotic effects of the combination of palladium (II) 5,5-diethylbarbiturate complex with bis(2-pyridylmethyl)amine and curcumin on non small lung cancer cell lines, Bioorg. Med. Chem., 2017, 25, 1717–1723 CrossRef CAS.
- V. Lopes-Rodrigues, A. Oliveira, M. Correia-da-Silva, M. Pinto, R. T. Lima, E. Sousa and M. H. Vasconcelos, A novel curcumin derivative which inhibits P-glycoprotein, arrests cell cycle and induces apoptosis in multidrug resistance cells, Bioorg. Med. Chem., 2017, 25, 581–596 CrossRef CAS.
- C. H. Liu and H. Y. Huang, Antimicrobial activity of curcumin-loaded myristic acid microemulsions against Staphylococcus epidermidis, Chem. Pharm. Bull., 2012, 60, 1118–1124 CrossRef CAS.
- D. K. Agrawal, D. Saikia, R. Tiwari, S. Ojha, K. Shanker, J. K. Kumar, A. K. Gupta, S. Tandon, A. S. Negi and S. P. S. Khanuja, Demethoxycurcumin and its semisynthetic analogues as antitubercular agents, Planta Med., 2008, 74, 1828–1831 CrossRef CAS.
- A. Mazumder, N. Neamati, S. Sunder, J. Schulz, H. Pertz, E. Eich and Y. Pommier, Curcumin analogs with altered potencies against HIV-1 integrase as probes for biochemical mechanisms of drug action, J. Med. Chem., 1997, 40, 3057–3063 CrossRef CAS.
- Q. Li, J. Chen, S. Luo, J. Xu, Q. Huang and T. Liu, Synthesis and assessment of the antioxidant and antitumor properties of asymmetric curcumin analogues, Eur. J. Med. Chem., 2015, 93, 461–469 CrossRef CAS.
- N. Paulino, A. S. Paulino, S. N. Diniz, S. de Mendonça, I. D. Gonçalves, F. F. Flores, R. P. Santos, C. Rodrigues, P. C. Pardi and J. A. Q. Suarez, Evaluation of the anti-inflammatory action of curcumin analog (DM1): Effect on iNOS and COX-2 gene expression and autophagy pathways, Bioorg. Med. Chem., 2016, 24, 1927–1935 CrossRef CAS PubMed.
- R. A. Sharma, H. R. McLelland, K. A. Hill, C. R. Ireson, S. A. Euden, M. M. Manson, M. Pirmohamed, L. J. Marnett, A. J. Gescher and W. P. Steward, Pharmacodynamic and pharmacokinetic study of oral curcuma extract in patients with colorectal cancer, Clin. Cancer Res., 2001, 7, 1894–1900 CAS.
- P. V. S. Ramya, S. Angapelly, L. Guntuku, C. S. Digwal, B. N. Babu, V. G. M. Naidu and A. Kamal, Synthesis and biological evaluation of curcumin inspired indole analogues as tubulin polymerization inhibitors, Eur. J. Med. Chem., 2017, 127, 100–114 CrossRef.
- P. V. S. Ramya, L. Guntuku, S. Angapelly, S. Karri, C. S. Digwal, B. N. Babu, V. G. M. Naidu and A. Kamal, Curcumin inspired 2-chloro/phenoxy quinoline analogues: Synthesis and biological evaluation as potential anticancer agents, Bioorg. Med. Chem. Lett., 2018, 28, 892–898 CrossRef.
- C. Qiu, Y. Hu, K. Wu, K. Yang, N. Wang, Y. Ma, H. Zhu, Y. Zhang, Y. Zhou, C. Chen, S. Li, L. Fu, X. Zhang and Z. Liu, Synthesis and biological evaluation of allylated mono-carbonyl analogues of curcumin (MACs) as anti-cancer agents for cholangiocarcinoma, Bioorg. Med. Chem. Lett., 2016, 26, 5971–5976 CrossRef CAS.
- P. K. Sahu, Design, structure activity relationship, cytotoxicity and evaluation of antioxidant activity of curcumin derivatives/analogues, Eur. J. Med. Chem., 2016, 121, 510–516 CrossRef CAS.
- E. Addala, H. Rafiei, S. Das, B. Bandy, U. Das, S. S. Karki and J. R. Dimmock, 3,5-Bis(3-dimethylaminomethyl-4-hydroxybenzylidene)-4-piperidone and related compounds induce glutathione oxidation and mitochondria-mediated cell death in HCT-116 colon cancer cells, Bioorg. Med. Chem. Lett., 2017, 27, 3669–3673 CrossRef CAS.
- M. Hossain, U. Das, N. Umemura, H. Sakagami, J. Balzarini, E. De Clercq, M. Kawase and J. R. Dimmock, Tumour-specific cytotoxicity and structure–activity relationships of novel 1-[3-(2-methoxyethylthio)propionyl]-3,5-bis(benzylidene)-4-piperidones, Bioorg. Med. Chem., 2016, 24, 2206–2214 CrossRef CAS.
- S. S. Karki, U. Das, N. Umemura, H. Sakagami, S. Iwamoto, M. Kawase, J. Balzarini, E. De Clercq, S. G. Dimmock and J. R. Dimmock, 3,5-Bis(3-alkylaminomethyl-4-hydroxybenzylidene)-4-piperidones: A novel class of potent tumor-selective cytotoxins, J. Med. Chem., 2016, 59, 763–769 CrossRef CAS PubMed.
- A. Jha, K. M. Duffield, M. R. Ness, S. Ravoori, G. Andrews, K. S. Bhullar, H. P. V. Rupasinghe and J. Balzarini, Curcumin-inspired cytotoxic 3,5-bis(arylmethylene)-1-(N-(ortho-substituted aryl)maleamoyl)-4-piperidones: A novel group of topoisomerase II alpha inhibitors, Bioorg. Med. Chem., 2015, 23, 6404–6417 CrossRef CAS.
- E. Potter, M. Jha, K. S. Bhullar, H. P. V. Rupasinghe, J. Balzarini and A. Jha, Investigation of fatty acid conjugates of 3,5-bisarylmethylene-4-piperidone derivatives as antitumor agents and human topoisomerase-IIα inhibitors, Bioorg. Med. Chem., 2015, 23, 411–421 CrossRef CAS.
- Y. Santiago-Vazquez, S. Das, U. Das, E. Robles-Escajeda, N. M. Ortega, C. Lema, A. Varela-Ramírez, R. J. Aguilera, J. Balzarini, E. De Clercq, S. G. Dimmock, D. K. J. Gorecki and J. R. Dimmock, Novel 3,5-bis(arylidene)-4-oxo-1-piperidinyl dimers: Structure–activity relationships and potent antileukemic and antilymphoma cytotoxicity, Eur. J. Med. Chem., 2014, 77, 315–322 CrossRef CAS.
- N. K. Paul, M. Jha, K. S. Bhullar, H. P. V. Rupasinghe, J. Balzarini and A. Jha, All trans 1-(3-arylacryloyl)-3,5-bis(pyridin-4-ylmethylene)piperidin-4-ones as curcumin-inspired antineoplastics, Eur. J. Med. Chem., 2014, 87, 461–470 CrossRef CAS.
- U. Das, S. Das, B. Bandy, J. P. Stables and J. R. Dimmock, N-Aroyl-3,5-bis(benzylidene)-4-piperidones: Anovel class of antimycobacterial agents, Bioorg. Med. Chem., 2008, 16, 3602–3607 CrossRef CAS PubMed.
- R. S. P. Singh, U. Das, J. M. Auschwitz, S. E. Leed, M. R. Hickman, J. R. Dimmock and J. Alcorn, From a cytotoxic agent to the discovery of a novel antimalarial agent, Bioorg. Med. Chem. Lett., 2013, 23, 584–587 CrossRef CAS PubMed.
- G.-F. Zha, C.-P. Zhang, H.-L. Qin, I. Jantan, M. Sher, M. W. Amjad, M. A. Hussain, Z. Hussain and S. N. A. Bukhari, Biological evaluation of synthetic α,β-unsaturated carbonyl based cyclohexanone derivatives as neuroprotective novel inhibitors of acetylcholinesterase, butyrylcholinesterase and amyloid-β aggregation, Bioorg. Med. Chem., 2016, 24, 2352–2359 CrossRef CAS.
- N. S. M. Ismail, R. F. George, R. A. T. Serya, F. N. Baselious, M. El-Manawaty, E. M. Shalaby and A. S. Girgis, Rational design, synthesis and 2D-QSAR studies of antiproliferative tropane-based compounds, RSC Adv., 2016, 6, 101911–101923 RSC.
- http://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm376547.htm.
- https://www.cancer.gov/about-cancer/treatment/drugs/sorafenibtosylate.
- https://www.cancer.gov/about-cancer/treatment/drugs/fda-sorafenib-tosylate.
- https://www.cancer.gov/about-cancer/treatment/drugs/lenvatinibmesylate.
- https://www.cancer.gov/about-cancer/treatment/drugs/fda-regorafenib.
- https://www.cancer.gov/about-cancer/treatment/drugs/fluorouracil.
- https://www.cancer.gov/about-cancer/treatment/drugs/fluorouracil-topical.
- G. A. Patani and E. J. LaVoie, Bioisosterism: A Rational Approach in Drug Design, Chem. Rev., 1996, 96, 3147–3176 CrossRef CAS.
- N. G. Fawzy, S. S. Panda, W. Fayad, M. A. El-Manawaty, A. M. Srour and A. S. Girgis, Novel curcumin inspired antineoplastic 1-sulfonyl-4-piperidones: design, synthesis and molecular modeling studies, Anti-Cancer Agents Med. Chem., 2019, 19, 1069–1078 CrossRef.
- A. S. Girgis, S. S. Panda, M. N. Aziz, P. J. Steel, C. D. Hall and A. R. Katritzky, Rational design, synthesis, and 2D-QSAR study of anti-oncological alkaloids against hepatoma and cervical carcinoma, RSC Adv., 2015, 5, 28554–28569 RSC.
- M. S. Nandhini, N. Srinivasan, R. Ranjithkumar, S. Perumal and R. V. Krishnakumar, 3,5-Bis[(E)-4-methylbenzylidene]-4-oxopiperidine-1-carbonitrile, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2007, 63, o3749 CrossRef CAS.
- M. N. Aziz, S. S. Panda, E. M. Shalaby, N. G. Fawzy and A. S. Girgis, Facile synthetic approach towards vasorelaxant active 4-hydroxyquinazoline-4-carboxamides, RSC Adv., 2019, 9, 28534–28540 RSC.
- E. M. Shalaby, A. M. Srour, S. S. Panda, R. F. George, A. N. Fitch and A. S. Girgis, Synthesis, X-ray powder diffraction and DFT-d studies of indole-based compounds, Z. Kristallogr. - Cryst. Mater., 2018, 233, 421–428 CAS.
- E. A. Soliman, S. S. Panda, M. N. Aziz, E. M. Shalaby, N. Mishriky, F. M. Asaad and A. S. Girgis, Synthesis, molecular modeling studies and bronchodilation properties of nicotinonitrile containing-compounds, Eur. J. Med. Chem., 2017, 138, 920–931 CrossRef CAS.
- A. R. Katritzky, R. Petrukhin, I. Petrukhina, A. Lomaka, D. B. Tatham and M. Karelson, CODESSA-Pro software manual, 2005, pp. 42, 54, 55, 57, 61, 69, 72, 74, 76 Search PubMed.
- M. A. Ibrahim, S. S. Panda, A. A. Oliferenko, P. V. Oliferenko, A. S. Girgis, M. Elagawany, Z. Küçükbay, C. S. Panda, G. G. Pillai, A. Samir, K. Tämm, C. D. Hall and A. R. Katritzky, Macrocyclic peptidomimetics with antimicrobial activity: synthesis, bioassay, and molecular modeling studies, Org. Biomol. Chem., 2015, 13, 9492–9503 RSC.
- P. Li, W. Zhang, Ho. Jiang, Y. Li, C. Dong, H. Chen, K. Zhangad and Z. Du, Design, synthesis and biological evaluation of benzimidazole-rhodanine conjugates as potent topoisomerase II inhibitors, Med. Chem. Commun., 2018, 9, 1194–1205 RSC.
- J. R. Dimmock, M. P. Padmanilayam, R. N. Puthucode, A. J. Nazarali, N. L. Motaganahalli, G. A. Zello, J. W. Quail, E. O. Oloo, H.-B. Kraatz, J. S. Prisciak, T. M. Allen, C. L. Santos, J. Balzarini, E. De Clercq and E. K. Manavathu, A conformational and structure–activity relationship study of cytotoxic 3,5-bis(arylidene)-4-piperidones and related N-acryloyl analogues, J. Med. Chem., 2001, 44, 586–593 CrossRef CAS.
- A.-M. Katsori, M. Chatzopoulou, K. Dimas, C. Kontogiorgis, A. Patsilinakos, T. Trangas and D. Hadjipavlou-Litina, Curcumin analogues as possible anti-proliferative & anti-inflammatory agents, Eur. J. Med. Chem., 2011, 46, 2722–2735 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1855508, 1855509. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra05661k |
| This journal is © The Royal Society of Chemistry 2019 |