Open Access Article
Open Access ArticleOligonucleotide-templated lateral flow assays for amplification-free sensing of circulating microRNAs†
Suraj Pavagada a,
Robert B. Channon
a,
Robert B. Channon a,
Jason Y. H. Chang
a,
Jason Y. H. Chang a,
Sung Hye Kimb,
David MacIntyre
a,
Sung Hye Kimb,
David MacIntyre bcd,
Phillip R. Bennettbcd,
Vasso Terzidoubce and
Sylvain Ladame
bcd,
Phillip R. Bennettbcd,
Vasso Terzidoubce and
Sylvain Ladame *ac
*ac
aDepartment of Bioengineering, Imperial College London, South Kensington Campus, London SW7 2AZ, UK. E-mail: sladame@imperial.ac.uk
bParturition Research Group, Institute of Reproductive and Developmental Biology, Imperial College London, London, W12 0NN, UK
cMarch of Dimes European Preterm Birth Research Centre, Imperial College London, London, UK
dQueen Charlotte's Hospital, Imperial College Healthcare NHS Trust, London, W12 0HS, UK
eChelsea & Westminster Hospital, Imperial College Healthcare NHS Trust, London, SW10 9NH, UK
First published on 17th September 2019
Abstract
Herein we demonstrate the first example of oligonucleotide-templated reaction (OTR) performed on paper, using lateral flow to capture and concentrate specific nucleic acid biomarkers on a test line. Quantitative analysis, using a low-cost benchtop fluorescence reader showed very high specificity down to the single nucleotide level and proved sensitive enough for amplification-free, on-chip, detection of endogenous concentrations of miR-150-5p, a recently identified predictive blood biomarker for preterm birth.
Circulating cell-free nucleic acids (cfNAs) in blood have recently emerged as clinically useful and minimally invasive diagnostic and predictive tools for a broad range of pathologies, including cancers1 and prenatal disorders.2 Among them, non-coding microRNAs (or miRNAs) are frequently found to be up-regulated or down-regulated in bodily fluids and have great potential as novel blood-based molecular fingerprints.3 Because of their complexity and cost, current gold-standards for microRNA detection (e.g. RT-qPCR, nCounterTM) remain unsuitable for point-of-care testing.
Sensing strategies based on bio-orthogonal oligonucleotide-templated chemistries have recently emerged as highly promising tools for in vitro or in vivo detection of endogenous nucleic acids (NAs).4 Widespread in nature, oligonucleotide-templated reactions (OTRs) use a DNA or RNA strand as a template to catalyse a highly unfavourable bimolecular chemical reaction by significantly increasing the effective molarity of two molecules, which would otherwise be present in solution at concentrations too low to react with each other.5 NA sensing technologies can be engineered whereby the product of a DNA (or RNA) templated-reaction is detected optically, providing both qualitative (sequence specificity) and quantitative (absolute concentration) information about a given NA target. OTRs based on peptide nucleic acid (PNA) probes6 are particularly powerful for sensing short NAs, such as miRNAs, which are typically challenging to detect using conventional oligonucleotide hybridisation probes such as TaqMan™ probes.7 To date, and despite its proven excellent sensitivity and specificity, OTR sensing has been restricted to laboratory use due to the absence of suitable hosting platforms. Tethering OTR probes onto solid supports (either during the sensing process or post-reaction to facilitate detection) could translate this cumbersome solution-based approach into a portable device format, and pave the way to the next generation of low-cost Nucleic Acid Tests (NATs).
With the introduction of home diagnostic tests for pregnancy, lateral flow assays (LFAs) gained wide popularity as a simple low-cost testing platform for molecular diagnostics.8 A vast number of lateral flow-based serological immunoassays have since been developed towards a wide range of biological and environmental targets.9 Recently, Winssinger et al. reported the design of an LFA for rapid detection of the products of oligonucleotide-templated chemical ligations of two PNA probes.10 It is noteworthy however that the OTR was first carried out in solution before loading the equilibrated mixture onto an LFA strip used here to facilitate detection only. This was successfully applied for sensing specific miRNAs in crude cell lysate using anti-fluorescein isothiocyanate (FITC) antibody labelling.
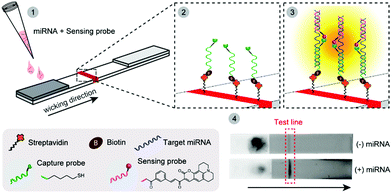
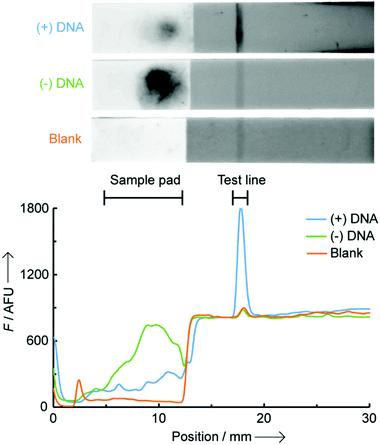
In this work, we report the first example of a fluorogenic OTR carried out directly on paper (a nitrocellulose LFA), which is amenable to incorporation into portable devices and can be analysed quantitatively with a small benchtop reader (Scheme 1). Pre-functionalising the LFA test line with one of the probes minimises unspecific reactions between the two probes that can only interact with each other on paper, at the level of the test line, when simultaneously hybridised to their NA target. As a proof-of-concept study, we developed a NAT for early prediction of preterm birth based on the detection of a recently identified and clinically validated biomarker present in the mother's blood: miR-150-5p.11 The assay was technically validated by analysing clinical plasma samples from pregnant women, and comparison to a reference nCounter™ technology. Building on earlier studies from our group,12 two 7-mer PNA probes were designed and synthesised containing either a butane thiol (at the C-terminus) or a quenched coumarin (at the N-terminus) (ESI† S2). Biotin was also inserted at the N-terminus of the thiol-modified probe to enable its immobilisation on streptavidin-modified LFA test lines. In this new design, the biotinylated PNA thiol was used as a “capture probe” and pre-immobilised onto the streptavidin-coated test line. The PNA coumarin, or “sensing probe” was then added to the sample loading pad, thus spatially separating both probes and preventing their ability to react with each other until the assay is initiated and the sensing probe migrates to the test line. As an initial proof-of-concept, LFAs were performed in the presence and in the absence of a miR-150-5p DNA target. Note, all quoted quantitative analyses herein (i.e. F, fluorescence intensities) represent the integrated peak area obtained from the benchtop reader. A 16-fold increase in fluorescence intensity was observed at the test line when DNA was present ((+) DNA, 0.5 μM), compared to the control without any target ((−) DNA, Fig. 1). It is noteworthy that the residual fluorescence at the test line without DNA (58.1 ± 8.1 AFU, n = 3) was comparable to that observed on blank strips with only buffer added (Blank, 58.4 ± 8.3 AFU, n = 3) suggesting it originates from the strip manufacturing process.
Background noise in solution-phase OTRs typically originates from (i) the residual fluorescence of unreacted probes (in this case from the excess of unreacted PNA coumarin) and (ii) fluorescence from the product of unspecific reactions between probes, in the absence of template. In this LFA setting, no background noise was observed from the excess of the lightly fluorescent PNA coumarin probe, which is washed away during the process, and from unspecific reactions between probes due to the very limited time (<5 min) they are in contact with each other without a template oligonucleotide present. A large background fluorescence at the level of the sample pad is observed on the (−) DNA strip, and a small fluorescence for the (+) DNA strip (Fig. 1). These are likely the result of the highly hydrophobic, and poorly water-soluble, PNA coumarin probe remaining adsorbed onto the nitrocellulose fibres unless it is hybridised to a complementary oligonucleotide. No such effect was observed however with the significantly more water-soluble PNA thiol, enabling its straightforward loading onto the test line.
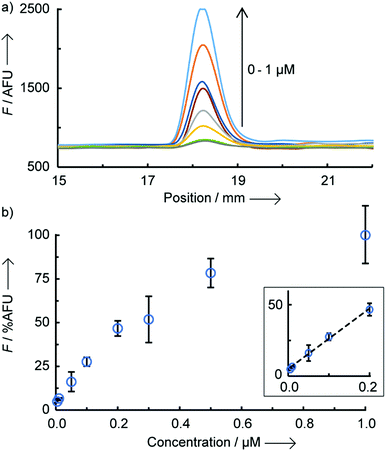
An optimal signal-to-noise ratio (SNR) was obtained with a 1 μM concentration of PNA coumarin (ESI,† S3) and the maximum fluorescence intensity was obtained after complete drying of the LFA strips (150 min passive drying at rt). Importantly, similar fluorescence intensities were achieved when actively drying the LFA strips with a hair dryer for 7 min reducing the time-to-analysis to within 10 min (Fig. S4, ESI†). To assess the assay sensitivity, strips were loaded with increasing concentration of miR-150-5p DNA (0 to 1 μM) whilst keeping all other parameters constant (Fig. 2). A limit of detection of 9 nM was found (3× standard deviation + mean of NTR fluorescence), which is ∼6-fold lower than that previously reported by us for a similar OTR in solution.12a A linear correlation was also observed between fluorescence intensity and DNA concentration in the range of 10–200 nM (Fig. 2b inset and Fig. S5, ESI† for scanned images). Above 200 nM, the fluorescence appears to tail off, which is likely due to the LFA strip test line reaching saturation. A similar effect was also observed when loading increasing concentrations of dual-labelled (3′-biotin, 5′-FITC) DNA oligonucleotides onto the same LFA strips (Fig. S6, ESI†).
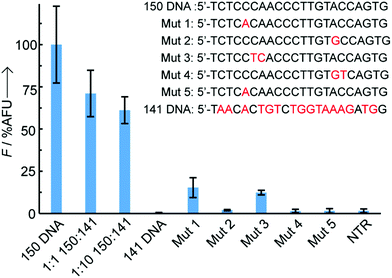
A recurrent challenge when trying to sense short oligonucleotides, such as miRNAs, is the high sequence homology between naturally occurring family members.3 To avoid any risk of false positives it is therefore essential to develop sensing technologies that can discriminate between DNA targets differing by as few as one or two bases. To assess the assay's specificity, probes directed against miR-150-5p were exposed to DNA sequences containing either 1 or 2 base pair mismatches, as well as a non-complementary DNA (Fig. 3). Single point mutations on the DNA sequence complementary to the sensing probe (coumarin) or the capture probe (thiol) led to significant reductions in relative fluorescence intensity of 85% and 98%, respectively, when compared to the signal observed with the perfectly matched target. Double mutations in the DNA target resulted in 88–99% reduction in fluorescence, regardless of their position (side-to-side or split mutations), with absolute intensities similar to background levels observed in the absence of target or in the presence of a non-complementary target (here, miR-141 DNA). Addition of a 1× or 10× excess of competitor miR-141 DNA led to a moderate reduction of fluorescence intensity by 29% and 39% respectively, likely due to unspecific electrostatic interactions between the sensing probe and competitor DNA partially preventing specific hybridisation to the target miRNA. The remarkable overall selectivity can be explained by an on-chip double selection process: (1) the PNA coumarin sensing probes only travel through the paper strips if hybridised to a complementary DNA and (2) the time given to the PNA:DNA binary complex to hybridise to the catch probe at the test line is limited, adding extra selection pressure.
Next, the OTR-LFA approach was applied as a technological solution to a problem that affects 15 million women worldwide every year, namely preterm birth (PTB). PTB is the largest cause of death of children under five worldwide.13 Birth before 26 weeks associates with 80% mortality, 25% severe handicap, and 75% overall morbidity in survivors. The majority of PTBs are to women with no identifiable risk factors, making cases extremely hard to predict at an early stage, when the mother would benefit the most from medical treatment.11 Low-cost, paper-based blood tests for early prediction of PTB could provide a valuable alternative to current approaches which could improve neonatal outcomes.14 Total miRNAs were extracted from the plasma of over 500 pregnant women and profiling analyses were performed using the nCounter nanostring technology.11 The best predictor biomarker for PTB (and cervical shortening) was found to be miR-150-5p, yielding a 100% true positive rate and a 50% false positive rate when measured at 12 weeks. Excess concentrations of this specific miRNA in blood was also shown to rise from 12–15 through 15–18 to 19–22 weeks (3-fold, 6-fold, 10-fold) in women who deliver preterm.11
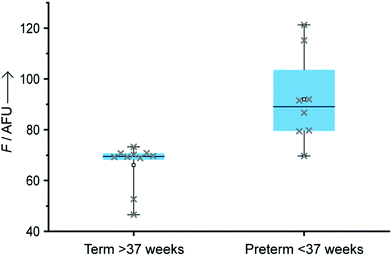
To validate our technology, eighteen total miRNA extracts from plasma samples previously used for a biomarker discovery study and collected between 11th and 19th weeks of pregnancy were also tested through our LFA-OTR approach (Fig. 4). Ten samples were from pregnant women who at the time of collection had low levels of miR-150-5p and delivered at term. Eight others were from women who delivered preterm and had abnormally high levels of miR-150-5p at the time of collection (ESI,† S7). Importantly, all samples tested produced a fluorescent signal above background noise, demonstrating the ability of our technology to detect endogenous concentrations of circulating miR-150-5p, without the need for any amplification steps. As shown in Fig. 4, samples from pre-term patients gave a statistically significantly higher fluorescence than those from term patients (unpaired t-test P value = 0.0006, and Mann–Whitney P value = 0.0021).15 Finally, a good correlation was observed between our approach and a reference method (nCounter) as described in ESI† (Table S1).
In conclusion, this work reports the first example of a direct OTR on paper for detection of endogenous concentrations of circulating microRNAs extracted from blood, using a low-cost benchtop reader and without amplification. This strategy addresses several limitations of solution based OTRs including complex sample handling, high background noise, and expensive detection methodologies. The use of a PNA “capture probe” immobilised at the test line contributes (1) to an increase in specificity by fishing out the biomarker of interest, but also (2) to an increase in sensitivity by concentrating the signal at the test line and reducing background noise resulting from either excess probes (as with solution OTR) or non-templated, unspecific reactions. The background signals are further reduced by the coumarin probe not flowing through the paper unless hybridised to a complementary DNA or RNA.
Our proof-of-concept preterm birth screening test was able to distinguish term and preterm patients based on miR-150 concentrations in plasma. This represents an attractive alternative to cervical length scans or foetal fibronectin tests which are conducted later in pregnancies and are unsuitable for repeated analyses as required for longitudinal monitoring. Work is ongoing to translate our miRNA sensing platform into a portable, low-cost, fully automated device to detect miRNAs directly from whole blood and can be used in a GP's clinic.
This work was supported by an Imperial College Confidence in Concept grant, a Cancer Research UK project grant (C49996/A26141) and by the NIHR Biomedical Research Centre at Imperial.
Conflicts of interest
There are no conflicts to declare.References
- (a) H. Schwarzenbach, Breast Cancer Res., 2013, 15, 211 CrossRef PubMed; (b) A. Sita-Lumsden, C. E. Fletcher, D. A. Dart, G. N. Brooke, J. Waxman and C. L. Bevan, Biomarkers Med., 2013, 7, 867–877 CrossRef CAS PubMed; (c) O. Pös, O. Biró, T. Szemes and B. Nagy, Eur. J. Hum. Genet., 2018, 26, 937–945 CrossRef PubMed.
- (a) L. Hui and D. W. Bianchi, Annu. Rev. Med., 2017, 68, 459–472 CrossRef CAS PubMed; (b) T. T. M. Ngo, M. N. Moufarrej, M. H. Rasmussen, J. Camunas-Soler, W. Pan, J. Okamoto, N. F. Neff, K. Liu, R. J. Wong, K. Downes, R. Tibshirani, G. M. Shaw, L. Skotte, D. K. Stevenson, J. R. Biggio, M. A. Elovitz, M. Melbye and S. R. Quake, Science, 2018, 360, 1133–1136 CrossRef CAS PubMed.
- (a) H. Schwarzenbach, N. Nishida, G. A. Calin and K. Pantel, Nat. Rev. Clin. Oncol., 2014, 11, 145–156 CrossRef CAS PubMed; (b) V. Armand-Labit and A. Pradines, Biomol. Concepts, 2017, 8, 61–81 CAS.
- For recent examples see (a) Y. Huang and J. M. Coull, J. Am. Chem. Soc., 2008, 130, 3238–3239 CrossRef CAS PubMed; (b) R. M. Franzini and E. T. Kool, Org. Lett., 2008, 10, 2935–2938 CrossRef CAS PubMed; (c) Z. Pianowski, K. Gorska, L. Oswald, C. A. Merten and N. Winssinger, J. Am. Chem. Soc., 2009, 131, 6492–6497 CrossRef CAS PubMed; (d) K. Meguellati, G. Koripelly and S. Ladame, Angew. Chem., Int. Ed., 2010, 122, 2798–2802 CrossRef; (e) H. Wu, B. T. Cisneros, C. M. Cole and N. K. Devaraj, J. Am. Chem. Soc., 2014, 136, 17942–17945 CrossRef CAS PubMed; (f) H. Wu, S. C. Alexander, S. Jin and N. K. Devaraj, J. Am. Chem. Soc., 2016, 138, 11429–11432 CrossRef CAS PubMed; (g) O. Zavoiura, U. Resch-Genger and O. Seitz, Bioconjugate Chem., 2018, 29, 1690–1702 CrossRef CAS PubMed; (h) M. Anzola and N. Winssinger, Chem. – Eur. J., 2019, 25, 334–342 CrossRef CAS PubMed; (i) K. Kor, A. P. F. Turner, K. Zaeri, M. Atabati, V. Beni and W. C. Mak, Anal. Bioanal. Chem., 2016, 408, 1475–1485 CrossRef CAS PubMed.
- (a) A. P. Silverman and E. T. Kool, Chem. Rev., 2006, 106, 3775–3789 CrossRef CAS PubMed; (b) T. N. Grossmann and O. Seitz, Chem. – Eur. J., 2009, 15, 6723–6730 CrossRef CAS PubMed; (c) A. Shibata, H. Abe and Y. Ito, Molecules, 2012, 17, 2446–2463 CrossRef CAS PubMed; (d) C. Percivalle, J.-F. Bartolo and S. Ladame, Org. Biomol. Chem., 2013, 11, 16–26 RSC; (e) K. Gorska and N. Winssinger, Angew. Chem., Int. Ed., 2013, 52, 6820–6843 CrossRef CAS PubMed; (f) M. Di Pisa and O. Seitz, ChemMedChem, 2017, 12, 872–882 CrossRef CAS PubMed; (g) S. Pavagada and S. Ladame, Chimia, 2018, 72, 809–814 CrossRef CAS PubMed.
- (a) Y. Choi, G. Metcalf, M. H. Sleiman, D. Vair-Turnbull and S. Ladame, Bioorg. Med. Chem., 2014, 22, 4395–4398 CrossRef CAS PubMed; (b) S. Barluenga and N. Winssinger, Acc. Chem. Res., 2015, 48, 1319–1331 CrossRef CAS PubMed.
- C. C. Pritchard, H. H. Cheng and M. Tewari, Nat. Rev. Genet., 2012, 13, 358–369 CrossRef CAS PubMed.
- (a) A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC; (b) K. M. Koczula and A. Gallotta, Essays Biochem., 2016, 60(1), 111–120 CrossRef PubMed; (c) C. Dincer, R. Bruch, E. Costa-Rama, M. T. Fernández-Abedul, A. Merkoçi, A. Manz, G. A. Urban and F. Güder, Adv. Mater., 2019, 1806739 CrossRef PubMed.
- (a) G. A. Posthuma-Trumpie, J. Korf and J. A. van Amerongen, Anal. Bioanal. Chem., 2009, 393(2), 569–582 CrossRef CAS PubMed; (b) D. R. Hristov, C. Rodriguez-Quijada, J. Gomez-Marquez and K. Hamad-Schifferli, Sensors, 2019, 19, E554 CrossRef PubMed.
- J. Sayers, R. J. Payne and N. Winssinger, Chem. Sci., 2017, 9, 896–903 RSC.
- (a) P. R. Bennett, J. Cook, D. A. MacIntyre and V. Terzidou, WO2016/142688, 2016; (b) J. Cook, P. R. Bennett, S. H. Kim, T. G. Teoh, L. Sykes, L. M. Kindinger, A. Grant, R. Binkhamis, D. A. MacIntyre and V. Terzidou, Sci. Rep., 2019, 9, 5861 CrossRef PubMed.
- (a) G. A. D. Metcalf, A. Shibakawa, H. Patel, A. Sita-Lumsden, A. Zivi, N. Rama, C. L. Bevan and S. Ladame, Anal. Chem., 2016, 88, 8091–8098 CrossRef CAS PubMed; (b) D. Al Sulaiman, J. Y. H. Chang and S. Ladame, Angew. Chem., Int. Ed., 2017, 56, 5247–5251 CrossRef CAS PubMed.
- March of Dimes, PMNCH, Save the Children, WHO, Born Too Soon: The Global Action Report on Preterm Birth, ed. H. Blencowe, S. Cousens, D. Chou, M. Oestergaard, L. Say, A. B. Moller, M. V. Kinney and J. E. Lawn, and the Born Too Soon Preterm Birth Action Group, WHO, Geneva, 2012 Search PubMed.
- Institute of Medicine (US) Committee on Understanding Premature Birth and Assuring Healthy Outcomes, Preterm birth: causes, consequences, and prevention, ed. R. E. Behrman and A. S. Butler, National Academies Press, 2007 Search PubMed.
- J. N. Miller and J. C. Miller, Statistics and Chemometrics for Analytical Chemistry, 6th edn, Pearson Education Ltd, London, 2005 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Including experimental information, PNA synthesis, coumarin probe concentration, optimization of the LFA parameters and information on the clinical samples. See DOI: 10.1039/c9cc05607f |
| This journal is © The Royal Society of Chemistry 2019 |