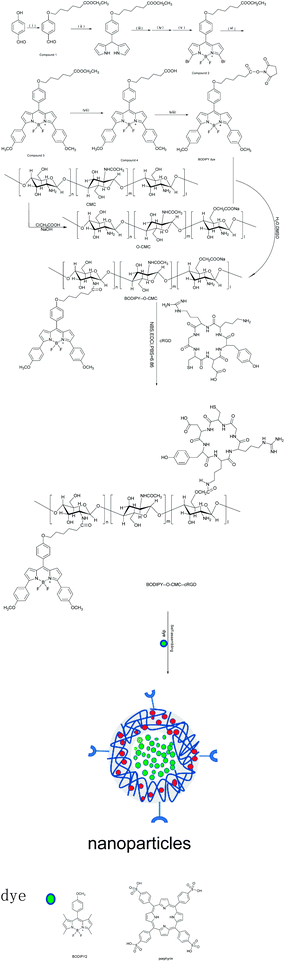
Open Access Article
Open Access ArticleA novel amphiphilic fluorescent probe BODIPY–O-CMC–cRGD as a biomarker and nanoparticle vector†
Tingting Zhua,
Ji Xionga,
Zhongbo Xuea,
Yu Sua,
Fengnan Suna,
Ran Chaicd,
Jialiang Xu a,
Yaqing Fengab and
Shuxian Meng
a,
Yaqing Fengab and
Shuxian Meng *a
*a
aSchool of Chemical Engineering and Technology, Tianjin University, No. 92, Weijin Road, Nankaiwei District, Tianjin 300072, P. R. China. E-mail: msxmail@tju.edu.cn; Fax: +86 022 27892323; Tel: +86 022 27892323
bTianjin Co-Innovation Center of Chemical Science and Engineering, Tianjin University, Tianjin 300072, P. R. China
cHebei University of Technology, P. R. China
dKey Laboratory of Hebei Province for Molecular Biophysics Institute of Biophysics, P. R. China
First published on 4th June 2018
Abstract
Fluorescent probes have been demonstrated to be promising candidates as biomarkers and biological carriers. Our study focuses on the development of a novel amphiphilic fluorescent probe with good photostability, high water solubility, excellent specificity and promising loading capability for tumor diagnosis and treatment. At first, BODIPY dye and O-carboxymethyl chitosan were prepared via a chemical reaction. Then, the prepared BODIPY dye and cRGD were bonded to O-carboxymethyl chitosan successively via an acylation reaction. Finally, we obtained the desired amphiphilic fluorescent probe: BODIPY–O-CMC–cRGD, which was based on the fluorescence resonance energy transfer (FRET) principle for selective visualization of tumors in vitro. Through a series of experiments, we found that this fluorescent probe possessed better fluorescence characteristics and tumor targeting properties. Simultaneously, by self-assembly, the amphiphilic probe encapsulated the other flexible structure of BODIPY2 and the rigid structure of porphyrin, which formed distinct nanoparticles with different particle sizes. Hence, we could observe different phagocytosis processes of the two nanoparticles in the tumor cells via the fluorescence of dyes by confocal laser scanning microscopy. Therefore, the results suggest that the fluorescent probe has advantages in tumor detection, and the constructed tumor-specific nanoparticles show high clinical potential to be utilized not only in visual and precise diagnosis but also in excellent drug delivery for tumor treatment. Henceforth, we will prepare new targeted and visualized pharmaceuticals by replacing BODIPY2 and porphyrin with antineoplastic drugs for future tumor treatment.
1 Introduction
The unique optical properties of fluorescent probes make them appealing as novel probes in biological studies1 such as in biological imaging, biological diagnostics, biological therapy and biological carriers. Especially, over the last few decades, a large variety of fluorescent probes showing photostability,2 biocompatibility3 and bioselectivity4 have been developed to further advance their application in biology and medicine. A myriad of these probes, however, suffer from vital drawbacks as they have low photostability and poor water solubility.2 Therefore, the design and development of novel fluorescent probes with good photostability, excellent water solubility and high specificity are in urgent demand.Recently, amphiphilic polymers, such as amphiphilic block copolymers3 and hydrophobically modified water-soluble polymers,5 have attracted extensive attention since their self-assembled nanoparticles in the aqueous media are suitable for trapping or grafting hydrophobic substances6 in biotechnology and medicine. Surprisingly, water-soluble polymers have played an increasingly important role in the construction of amphiphilic polymers;7 especially, the chitosan polymers have been extensively used due to their unique characteristics with higher antibacterial activity,8 more excellent biocompatibility9 and biodegradability,10 lower toxicity11 and easier modifications.12 The native chitosan is, however, insoluble under physiological pH conditions; this hinders its further application. Fortunately, investigators have successfully exploited various water-soluble chitosan derivatives.13 Among the water-soluble chitosan derivatives, O-carboxymethyl chitosan (O-CMC) is an amphiprotic ether derivative that contains both –COOH groups and –NH2 groups in the same molecule, which provide it with more favorable properties, especially with regard to water solubility.14 For the fluorophore of fluorescent probes, we have chosen BODIPY, 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene as it can provide sharp fluorescence emissions, high fluorescence quantum yields, large absorption extinction coefficients, excellent photochemical stability, narrow excitation/emission bandwidth and low sensitivity to environmental variations;15 especially, its emission wavelengths lie in the visible range.16 The core of BODIPY dye is yet hydrophobic and does not meet the requirements of water-soluble probes. Therefore, the BODIPY structure should be modified to improve its hydrophilicity. It can be conjugated to the backbone of –NH2 groups of O-CMC, and the macromolecules can exhibit preeminent solubility, stability and fluorescence in aqueous systems. However, its application is further hindered in biological and medical fields due to lack of target specificity. Many targeting groups such as active targeting groups and passive targeting groups17 have been reported several times. Therein, peptides, arginine-glycine-aspartic acid, RGD, exhibit higher binding affinity to cells of the tumour neovasculature, which express αvβ3 integrin.18 Upon grafting RGD to the –COONa group of O-CMC, we obtained absolutely water-soluble fluorescent probes targeting tumor cells.
We prepared a highly water-soluble and specific-targeting fluorescent probe: BODIPY–O-CMC–cRGD, which showed a specific fluorescence resonance energy transfer (FRET) effect on the BODIPY dye when the BODIPY donor absorbed light energy.19–21 Further, BODIPY–O-CMC–cRGD used herein as a polymeric carrier could encapsulate two different dyes by self-assembly. Hence, two nanoparticles were prepared with BODIPY2 and porphyrin. The fluorescent probe and the two dyes may be traced by their different fluorescence spectra since the fluorescence characteristics of the two dyes are different from that of the BODIPY–O-CMC–cRGD probe. In conclusion, we estimate that a novel visual diagnosis and treatment nanoparticle can be exploited if two different dyes are replaced with some antineoplastic drugs in the future.
2 Experimental
2.1 Materials
Tetrahydrofuran and toluene were distilled in vacuum before use. Dichloromethane and acetone were refluxed with calcium hydride for 12 h and distilled before use. Other solvents were of analytical grade and used without further purification. 4-Hydroxybenzaldehyde, ethyl 6-bromohexanoate, pyrrole, N-bromosuccinimide (NBS), 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ), bis(triphenylphosphine)palladium dichloride (Pd(PPh3)Cl2), N-hydroxysulfosuccinimide (NHS), 1-ethyl-3-(3-(dimethylaminopropyl)carbodiimide (EDCI), and chitosan (molecular weight 100–150 kDa, DA-80) were purchased from Aladdin (Beijing, China) and used without further purification. The cyclic Arg–Gly–Cys–Asp–Tyr–Lys (c(RGDyK)) peptide was purchased from Shanghai Dechi Biosciences Co. Ltd. (Shanghai, China). The semipermeable membrane (3500 Da cut-off) used for dialysis was purchased from Shanghai Yuanye Biotechnology Co. Ltd (Shanghai, China). Deionized water was used for all experiments. The U87-VIII cells were obtained from ATCC (the American Type Culture Collection, Manassas, VA, USA).2.2 Synthesis of BODIPY dye
The BODIPY dye was synthesized via the route shown in Scheme 1. A mixture of 4-hybroxybenzaldehyde derivative and 2.2 equivalent of ethyl 6-bromohexanoate was refluxed in dry acetone for 12 h in the presence of potassium carbonate.22 We obtained compound 1 after purification. Compound 2 was obtained in a sequence of steps by nucleophilic reaction, brominated reaction and oxidation reaction under strict water-free and oxygen-free conditions.23,24 Compound 3 and compound 4 were obtained respectively by the classical Suzuki reaction25 and traditional alkaline hydrolysis.26 Finally, the expected BODIPY dye was obtained by amidation.272.3 Synthesis of O-CMC
O-CMC was prepared as reported previously.28 Herein, 2 g chitosan was first suspended in a sodium hydroxide solution (10%, w/w, 10 mL) at 60 °C for 1 h. Then, 6 g monochloroacetic acid dissolved in 4 mL isopropanol was added to the abovementioned solution over 1 h and reacted for 4 h at the same temperature continuously. To this, 40 mL ethyl alcohol (70%) was added to stop the reaction. The precipitate was filtered, washed three times with ethanol (70%) and once with anhydrous alcohol, and then dried under vacuum for 6 h at 60 °C.2.4 Synthesis of BODIPY–O-CMC and BODIPY–O-CMC–cRGD
In a flame flask, 10 mg O-CMC was dissolved in 10 mL deionized water, followed by the addition of some equivalent of BODIPY to DMSO.29 The reaction was performed at 35 °C for 12 h. The mixture was dialyzed for 48 h to remove the excess reactants. The final product BODIPY–O-CMC obtained was lyophilized. For BODIPY–O-CMC–cRGD synthesis, 20 mg BODIPY–O-CMC, 3 mg EDCI and 2 mg NHS were dissolved in 20 mL PBS, pH = 6.86. The reaction was conducted at room temperature for 12 h, and then, 6 mg cRGD peptide was added.30 After 24 h, the resultant solution was dialyzed against water (3500 Da cut-off) for 3 days, followed by lyophilization to produce the desired fluorescent probe: BODIPY–O-CMC–cRGD.2.5 Preparation of the dye-loaded nanoparticles BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin by the thin-film dispersion method
The preparation of dye-loaded polymer nanoparticles had already been described in the literature.31–33 BODIPY2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34 was synthesized by our research group. For this, 5 mg BODIPY2 was first dissolved in 5 mL anhydrous methanol in a 25 mL round-bottomed flask. Then, the solution was added to an appropriate amount of BODIPY–O-CMC–cRGD, and the suspension was kept in an ultrasonic water bath for 3 h; some mixed membranes remained at the bottom of the flask after removing methanol by evaporation in a water bath at 45 °C. The membranes was dissolved in 4 mL deionized water and then mixed with ultrasonic probes for 5 min (200 W, work 2 s, halt 2 s). Then, the crude nanoparticle solution was passed through a filter (0.45 μm) to remove unreacted BODIPY2. The filtrate thus obtained contained the nanoparticles, which were abbreviated as BODIPY–O-CMC–cRGD/BODIPY2. Porphyrin35-loaded fluorescent probe nanoparticles, BODIPY–O-CMC–cRGD/porphyrin, were prepared using the same method.
34 was synthesized by our research group. For this, 5 mg BODIPY2 was first dissolved in 5 mL anhydrous methanol in a 25 mL round-bottomed flask. Then, the solution was added to an appropriate amount of BODIPY–O-CMC–cRGD, and the suspension was kept in an ultrasonic water bath for 3 h; some mixed membranes remained at the bottom of the flask after removing methanol by evaporation in a water bath at 45 °C. The membranes was dissolved in 4 mL deionized water and then mixed with ultrasonic probes for 5 min (200 W, work 2 s, halt 2 s). Then, the crude nanoparticle solution was passed through a filter (0.45 μm) to remove unreacted BODIPY2. The filtrate thus obtained contained the nanoparticles, which were abbreviated as BODIPY–O-CMC–cRGD/BODIPY2. Porphyrin35-loaded fluorescent probe nanoparticles, BODIPY–O-CMC–cRGD/porphyrin, were prepared using the same method.
2.6 Confocal laser scanning microscopic imaging of BODIPY–O-CMC, BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin
The imaging of BODIPY–O-CMC, BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin in tumour cells was performed using a confocal laser scanning microscope. Briefly, U87-VIII cells were seeded at a density of 1 × 105 cells per well into a six-well plate containing DMEM and incubated for 24 h under an atmosphere of 5% CO2. Then, these cell culture media were replaced with 200 μL of the sample solution (30 μg mL−1). The cells containing BODIPY–O-CMC and BODIPY–O-CMC–cRGD were incubated for 4 h, but the cells containing BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin were incubated for 4 h, 8 h, and 12 h. The cells were rinsed with PBS three times, fixed with 200 mL of 4% paraformaldehyde in PBS for 10 min at room temperature and further washed with PBS three times. The tumour cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI) for the cell nucleus. The fluorescence images of the stained U87-VIII cells were obtained by CLSM using an argon laser. The maximum excitation wavelengths for BODIPY dye, BODIPY 2, and porphyrin were 561, 488, and 594 nm, respectively, and the corresponding emission wavelengths for the three samples were 615, 514, and 670 nm. The maximum excitation and emission wavelengths for DAPI were 360 and 488 nm, respectively.2.7 Cytotoxicity of BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin
To examine the toxicity of BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin in living cells, U87-VIII cells were seeded in 96-well plates with a density of 105 cells in every well and incubated in DMEM with samples for 48 hours. After incubation, the medium was removed, and the cells were washed with PBS three times. The medium of the plates was replaced by MTT followed by incubation at 37 °C under an atmosphere of 5% CO2 for 4 h. The medium was removed followed by the addition of 150 mL of DMSO. The absorbance wavelength was set at 560 nm to measure the results using a microplate reader.3 Results and discussion
3.1 Analysis of the FTIR spectrum and 1H-NMR spectrum
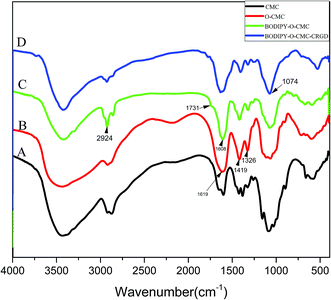
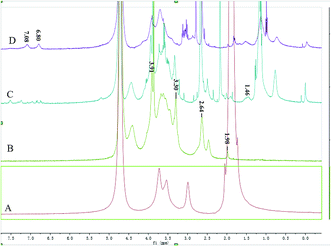
Fig. 1 shows the FTIR spectra of O-CMC, BODIPY–O-CMC and BODIPY–O-CMC–cRGD. In addition, the 1H NMR spectra of O-CMC, BODIPY–O-CMC and BODIPY–O-CMC–cRGD are depicted in Fig. 2. By considering these characteristics, we confirmed the synthesis of O-CMC, BODIPY–O-CMC and BODIPY–O-CMC–cRGD. O-CMC is an amphiprotic ether- and water-soluble derivative of chitosan, containing –CH2COO− groups and –NH2 groups in the same molecule. Fig. 1(B) shows the FTIR spectra of O-CMC. The band assignment of O-CMC was as follows (cm−1): 3467 (O–H stretch overlapped with N–H stretch), 2925 (C–H stretch), 1619 (C–O stretch of carboxyl methyl group overlapped with amide N–H bend), 1326 (C–N stretch), and 1160–1031 (bridge O stretch overlapped with C–O stretch). Compared with the peaks of commercial chitosan, the bands at 1160–1031 cm−1 changed dramatically; this was attributed to the C–O–C group of the carboxymethyl segment of –CH2–O–CH2COOH. Fig. 2(B) shows the 1H-NMR spectra of O-CMC. The proton assignment of O-CMC was as follows (ppm): 1.98 (CH3, acetamido group of chitosan), 2.64 (CH, carbon 2 of glucosamine ring), 3.30 (CH, carbon 2 of glucosamine ring with the substituted amino group), 3.4–3.8 (CH, carbon 3, 4 and 6 of glucosamine ring), 3.91 (CH2, carboxymethyl group), and 4.2–4.5 (H, carbon 1 and 5 of glucosamine ring). In comparison with the case of chitosan, there was a new peak at 3.91 ppm, which was attributed to the protons of 6-substituted carboxymethyl (–O–CH2COO−) group. Hence, the results of both FTIR and 1H-NMR spectra confirmed the successful synthesis of O-CMC and indicated that the carboxymethyl substitution occurred mainly at the C6 position.Compared with that of O-CMC, the BODIPY–O-CMC spectra in Fig. 1(C) show the disappearance of the –NH2 associated band at 1619 cm−1 of N–H bending in the primary amine and the appearance of the –NHassociated band at 1608 cm−1 of N–H bending in the second amine. The results indicated that the –NH2 groups of O-CMC were partly converted into –NH groups. There was a strong absorption band at 2924 cm−1 mainly ascribed to the –CH2 bending peak of BODIPY and a slight absorption band at 1731 cm−1 ascribed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bending peak of the amide group (O
O bending peak of the amide group (O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N–H) of BODIPY–O-CMC. In Fig. 2(C), the coupling of BODIPY to O-CMC was confirmed by the appearance of signals at 1.46 ppm, which corresponded to the CH2 protons of the –CH2–C(
C–N–H) of BODIPY–O-CMC. In Fig. 2(C), the coupling of BODIPY to O-CMC was confirmed by the appearance of signals at 1.46 ppm, which corresponded to the CH2 protons of the –CH2–C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–N(–H)– segment. The relevant signal of BODIPY was much weaker than the broad and strong proton signals of O-CMC residues. The FTIR spectrum and 1H-NMR spectrum of BODIPY–O-CMC illustrated that the carboxyl group of BODIPY was successfully coupled to the –NH2 groups of O-CMC. In addition, for more accurate evaluation, BODIPY–O-CMC was assessed by UV spectroscopy and fluorescence spectroscopy, as shown in Fig. 4(C and D).
O)–N(–H)– segment. The relevant signal of BODIPY was much weaker than the broad and strong proton signals of O-CMC residues. The FTIR spectrum and 1H-NMR spectrum of BODIPY–O-CMC illustrated that the carboxyl group of BODIPY was successfully coupled to the –NH2 groups of O-CMC. In addition, for more accurate evaluation, BODIPY–O-CMC was assessed by UV spectroscopy and fluorescence spectroscopy, as shown in Fig. 4(C and D).
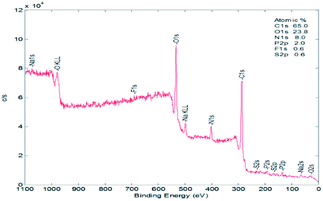
For BODIPY–O-CMC–cRGD, Fig. 1(D) shows that the peak at 1074 cm−1 enhances significantly due to the –C–O stretching and bending vibration of cRGD; Fig. 2 (D) shows the peculiar signals at 7.09 and 6.80 ppm, which are attributed to the aromatic protons of cRGD. The results indicated that cRGD was attached to BODIPY–O-CMC. In addition, this conclusion was proven by confocal experiments. The degrees of substitution by BODIPY dye and cRGD groups, calculated from the content of elements B, F, S and C of the XPS (in Fig. 3), were 2.75% and 3.26%, respectively.
3.2 Analysis of UV-vis absorption and fluorescence spectra
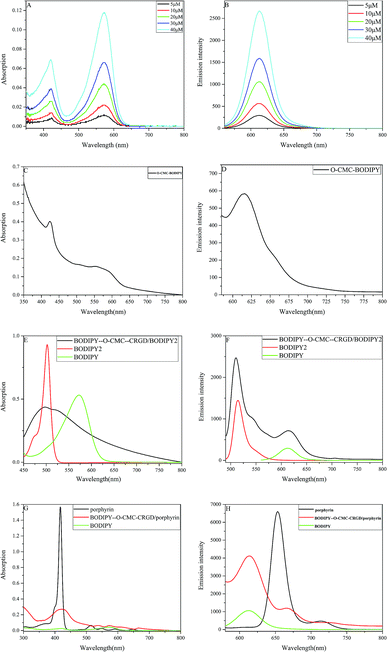
The BODIPY dye was synthesized as a highly fluorescent diaryl derivative carrying aryl groups at the 3- and 5-positions of the core skeleton. Herein, 4-hydroxybenzaldehyde reacted with ethyl 6-bromohexanoate to afford a long-chain fatty acid ester, which was easily conjugated with the –NH2 group of O-CMC. After the Suzuki reaction of compound 2 and arylboronic acid, compound 3 was found to have a longer wavelength when compared with the precursor intermediates. We investigated the absorption and emission spectra of the ultimate BODIPY dye in CH2Cl2 at different concentrations. As shown in Fig. 4(A), the absorption spectra of the BODIPY dye contained narrow spectral bands with two absorption peaks in the visible region. The intense band at 560 nm was attributed to the central skeleton of BODIPY and the pronounced shoulder on the high-energy side of the main band resulted from the five-membered ring containing nitrogen. Fig. 4(B) shows that the BODIPY derivative in CH2Cl2 has an emission spectrum with a maximum at 615 nm. These results indicated that the BODIPY dye had a Stokes shift of 55 nm. Fig. 4(C and D) show the UV and fluorescence spectra of BODIPY–O-CMC for more accurate conjugated evaluation. The spectra shown in Fig. 4(A–D) had signals of BODIPY.Dye-loaded polymeric nanoparticles were prepared by self-assembly with the fluorescent probe BODIPY–O-CMC–cRGD and two other dyes of different structures, namely BODIPY2 and porphyrin. BODIPY–O-CMC–cRGD had strong amphiphilic property owing to the presence of both hydrophobic BODIPY grafted onto moieties and hydrophilic cyclic peptide grafted onto the carboxyl group of O-CMC. In this study, two water-insoluble dyes were encapsulated into the fluorescent probe by hydrophobic interactions. Obviously, the two nanoparticles had good solubility in water, as observed from the Fig. 5, and were characterized by UV and fluorescence spectra. In an aqueous solution, as depicted in the UV-Vis absorption spectra shown in Fig. 4(E), the BODIPY–O-CMC–cRGD/BODIPY2 nanoparticles exhibited a prominent broad peak at 475–540 nm probably due to the overlap UV-Vis absorption of BODIPY dye and BODIPY2 of the consequences of colloidal solution. We would continue to verify the mechanism in our future work. Correspondingly, in the fluorescence spectra in Fig. 4(F), the BODIPY–O-CMC–cRGD/BODIPY2 nanoparticles manifested two optimal emission bands at 514 nm of BODIPY2 and 615 nm of BODIPY dye when excited at 480 nm. Analogously, the absorption spectra and fluorescence spectra of BODIPY dye and porphyrin of BODIPY–O-CMC–cRGD/porphyrin nanoparticles similar to that of pure BODIPY dye and porphyrin could be discriminated distinctly in Fig. 4(G and H), respectively. The results indicated that the two dyes were successfully loaded into the BODIPY–O-CMC–cRGD nanoparticles.
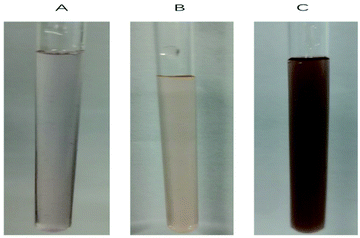 | ||
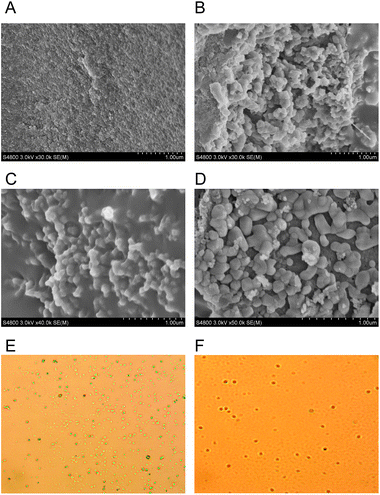
| Fig. 5 Deionized water solution of (A) BODIPY–O-CMC–cRGD, (B) BODIPY–O-CMC–cRGD/BODIPY2 and (C) BODIPY–O-CMC–cRGD/porphyrin. | ||
3.3 Analysis of SEM and optical microscopy images
The surface morphologies of BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin were examined by optical microscopy and SEM, as shown in Fig. 6. The optical microscopy image showed that the self-assembled particles were almost spherical in shape. In SEM images, the two nanoparticles presented irregular shapes. The size of BODIPY–O-CMC–cRGD/porphyrin nanoparticles was bigger than that of BODIPY–O-CMC–cRGD/BODIPY2; this was because the structure of porphyrin was rigid, whereas the structure of BODIPY2 was flexible. The SEM images coincided with those of the optical microscope.3.4 Analysis of particle size
The particle sizes and zeta potential of BODIPY–O-CMC, BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin were measured. From Table 1, it can be noted that they have unimodal particle size distribution, and particle sizes increase because the hydrophobic dye has been inserted into BODIPY–O-CMC. The sizes of porphyrin were larger than those of BODIPY2, and the particle size provided evidence supporting the results of SEM.| Sample | Diameter (nm) | Zeta potential (mv) |
|---|---|---|
| BODIPY–O-CMC | 312 ± 0.415 | −21.5 ± 9.11 |
| BODIPY–O-CMC–cRGD | 416 ± 0.460 | −19.4 ± 4.56 |
| BODIPY–O-CMC–cRGD/BODIPY2 | 656 ± 0.659 | −19.7 ± 5.76 |
| BODIPY–O-CMC–cRGD/porphyrin | 1182 ± 0.739 | −24.1 ± 3.73 |
3.5 Cytotoxicity assay
Fluorescent materials as biological probes require low toxicity. Thus, the cytotoxicity of BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin was measured by the MTT assay in U87-VIII cells. The viability of U87-VIII cells was determined at different concentrations of samples (from 0.1 to 100 μg mL−1). As shown in Fig. 7, the administration of BODIPY–O-CMC–cRGD at low concentrations did not affect the cellular viability when compared with the case of control cells. This result indicated that the cytotoxicity of BODIPY–O-CMC–cRGD towards the U87-VIII cells was very low. Therefore, BODIPY–O-CMC–cRGD can be applied to the living cell system as a fluorescent probe for visualized diagnoses and drug delivery in the future. Over 75% of the cells were viably indicating the low toxicity of BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin towards U87-VIII cells.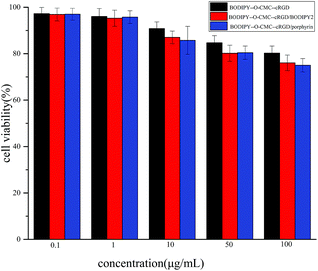 | ||
| Fig. 7 Cytotoxicity of BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin was measured after incubation with U87-VIII cells for 48 h by MTT assays. | ||
3.6 Confocal microscopic study of BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin
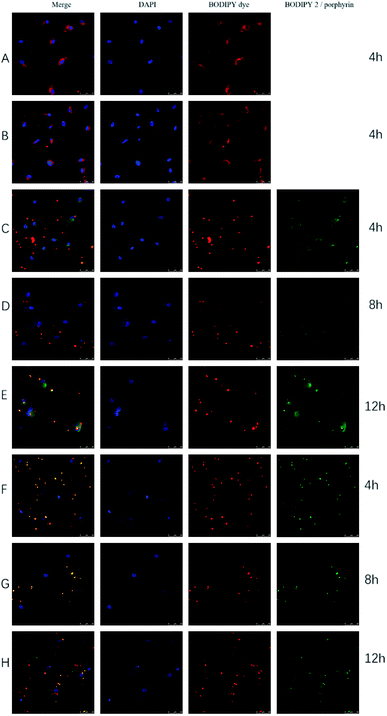
The intracellular localization of BODIPY–O-CMC, BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin nanoparticles that were taken up by the cells via the endocytic pathway was visually traced by confocal laser scanning microscopy (CLSM). Fig. 8 showed that all the fluorescent materials exhibited strong and stable fluorescence signals via CLSM. Although the specificity of cRGD had already been reported,18 the images presented in Fig. 8(A and B) did not reveal apparent specificity. The reason could be that BODIPY–O-CMC and BODIPY–O-CMC–cRGD possessed similar particle sizes and electronegativities, whose effects were more favorable to cell endocytosis than to cell affinity of cRGD. Fig. 8(C–E) showed that the nanoparticles of BODIPY–O-CMC–cRGD/BODIPY2 were endocytosed into the cells. These results demonstrated that the fluorescent probe BODIPY–O-CMC–cRGD was an efficiently targeted delivery vector for tumor cells, and it could take pure BODIPY derivatives into tumour cells. By further comprehensive comparison of Fig. 8(C, D, and E), all released processes were distinctly observed. Fig. 8(C) showed that only few nanoparticles were taken up by the tumor cells after 4 h of incubation, and BODIPY2 still remained encapsulated in BODIPY–O-CMC–cRGD. Fig. 8(D) showed that more nanoparticles were endocytosed into tumor cells, but part of BODIPY2 was released from the nanoparticles after 8 h of incubation. Moreover, BODIPY2 remained in the cells, and BODIPY–O-CMC–cRGD was far away from tumor cells. Fig. 8(E) showed that many nanoparticles had entered the cells, and more BODIPY2 was discharged from the nanoparticles. Simultaneously, BODIPY2 accumulated in the cells. Hence, from Fig. 7(C–E), it could be observed that the BODIPY–O-CMC–cRGD/BODIPY2 nanoparticles possessed prominent biocompatibility and were released in a slowly dynamic process from the tumor cells. The nanoparticles were endocytosed into tumor cells, and the fluorescence of BODIPY dye, which was the internal segment of BODIPY–O-CMC–cRGD, and BODIPY2, which was encapsulated in BODIPY–O-CMC–cRGD, changed from the state of overlapping to separation after a period of time. This indicated that BODIPY2 diffused into tumor cells. Fig. 8(F–H) showed that there were no BODIPY–O-CMC–cRGD/porphyrin nanoparticles inside the cell after 4 h, 8 h, and 12 h of incubation. It turns out that the structure of porphyrin was rigid, leading to an inflexible framework of BODIPY–O-CMC–cRGD/porphyrin nanoparticles, which was against the tumor cell endocytosis, whereas the structure of BODIPY2 was flexible. Based on all the abovementioned results, we could depict the prospects that a novel fluorescent probe, BODIPY–O-CMC–cRGD, loaded with flexible structures of antineoplastic drugs replacing dyes would be a promising candidate as a visualized and targeted delivery vehicle facilitating more accurate tumor diagnosis and therapy in clinical applications. We predict that this study will revolutionize the conventional medical practices in the coming decades.4 Conclusions
In this study, we have successfully prepared a fluorescent probe, BODIPY–O-CMC–cRGD, based on the FRET principle as a tumor cell tracer and an excellent delivery vehicle. The BODIPY dye-conjugated O-CMC had good photostability, and the BODIPY dye's fluorescence emission wavelength basically remained unchanged. The results of UV absorption spectra, fluorescence emission spectra, FTIR spectra, 1H-NMR spectra and SEM illustrated that BODIPY–O-CMC–cRGD, BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin were prepared successfully. The cytotoxicity assay made clear that BODIPY–O-CMC–cRGD had low toxicity. Confocal laser scanning microscopic analysis proved our successful fabrication of the two nanoparticles BODIPY–O-CMC–cRGD/BODIPY2 and BODIPY–O-CMC–cRGD/porphyrin with target specificity and dye release for tumor cell imaging. It can be envisioned that the proposed fluorescent probe BODIPY–O-CMC–cRGD may be used in targeted imaging and diagnosis. Furthermore, we consider that it has potential as a drug delivery vehicle, and we may prepare a novel drug integrated with visualized diagnoses and therapies in the future by replacing dyes with some antineoplastic drugs. In other words, we preconceive that the therapeutic agents will foster the development of theranostic nanoparticles with integrated targeted probe diagnosis and visualized drug therapy.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported financially by the National Natural Science Foundation of China (No. 21676187, No. 21476162), China International Science and Technology Project (No. 2012DFG41980, 2016YFE0114900), and National Key R&D Program of China (21761132007).References
- H. Xu and K. S. Suslick, Adv. Mater., 2010, 22, 1078–1082 CrossRef PubMed.
- D. R. Larson, W. R. Zipfel, R. M. Williams, S. W. Clark, M. P. Bruchez, F. W. Wise and W. W. Webb, Science, 2003, 300, 1434–1436 CrossRef PubMed.
- N. Boens, V. Leen and W. Dehaen, Chem. Soc. Rev., 2012, 41, 1130–1172 RSC.
- S. L. Niu, G. Ulrich, R. Ziessel, A. Kiss, P.-Y. Renard and A. Romieu, Org. Lett., 2009, 11, 2049–2052 CrossRef PubMed.
- L. Shang, S. Dong and G. U. Nienhaus, Nano Today, 2011, 6, 401–418 CrossRef.
- K. Letchford and H. Burt, Eur. J. Pharm. Biopharm., 2007, 65, 259–269 CrossRef PubMed.
- M.-H. Hsiao, M. Larsson, A. Larsson, H. Evenbratt, Y.-Y. Chen, Y.-Y. Chen and D.-M. Liu, J. Controlled Release, 2012, 161, 942–948 CrossRef PubMed.
- Y.-S. Wang, L.-R. Liu, Q. Jiang and Q.-Q. Zhang, Eur. Polym. J., 2007, 43, 43–51 CrossRef.
- A. R. Katritzky, J. Cusido and T. Narindoshvili, Bioconjugate Chem., 2008, 19, 1471–1475 CrossRef PubMed.
- G. Cai and H. Jiang, J. Mater. Sci.: Mater. Med., 2009, 20, 1315–1320 CrossRef PubMed.
- T. Wang, J. Zhang, P. Li and M. Shen, Chin. J. Biochem. Pharm., 2002, 23, 21–23 Search PubMed.
- B. Singh, Y. J. Choi, I. K. Park, T. Akaike and C. S. Cho, J. Nanosci. Nanotechnol., 2014, 14, 564–576 CrossRef PubMed.
- L. G. Engibaryan, A. I. Chernukhina, G. A. Gabrielyan and L. S. Gal'braikh, Fibre Chem., 2005, 37, 285–288 CrossRef.
- T. Baran, A. Mentes and H. Arslan, Int. J. Biol. Macromol., 2015, 72, 94–103 CrossRef PubMed.
- N. Boens, V. Leen and W. Dehaen, Chem. Soc. Rev., 2012, 41, 8212 RSC.
- X.-D. Jiang, X. Liu, T. Fang, C. Sun and L. Xiao, Tetrahedron Lett., 2018, 59, 546–549 CrossRef.
- X. Huang, X. Peng, Y. Wang, Y. Wang, D. M. Shin, M. A. El-Sayed and S. Nie, ACS Nano, 2010, 4, 5887–5896 CrossRef PubMed.
- C. R. de Oliveira, R. D. Marqueti, M. R. Cominetti, E. S. V. Douat, J. U. Ribeiro, C. L. S. Pontes, A. Borghi-Silva and H. S. Selistre-de-Araujo, Acta Cir Bras., 2015, 30, 134–142 CrossRef PubMed.
- J. Chen, Y. Li, K. Lv, W. Zhong, H. Wang, Z. Wu, P. Yi and J. Jiang, Sens. Actuators, B, 2016, 224, 298–306 CrossRef.
- H. Wang, P. Zhang, Y. Hong, B. Zhao, P. Yi and J. Chen, Polym. Chem., 2017, 8, 5795–5802 RSC.
- P. Zhang, H. Wang, Y. Hong, M. Yu, R. Zeng, Y. Long and J. Chen, Biosens. Bioelectron., 2018, 99, 318–324 CrossRef PubMed.
- K. Yamada, T. Toyota, K. Takakura, M. Ishimaru and T. Sugawara, New J. Chem., 2001, 25, 667–669 RSC.
- G. Ulrich, R. Ziessel and A. Harriman, Angew. Chem., Int. Ed., 2008, 47, 1184–1201 CrossRef PubMed.
- R. W. Wagner and J. S. Lindsey, Pure Appl. Chem., 1998, 70, AR1 CrossRef.
- V. Lakshmi and M. Ravikanth, J. Org. Chem., 2011, 76, 8466–8471 CrossRef PubMed.
- J. M. Khurana, S. Chauhan and G. Bansal, Monatsh. Chem., 2004, 135, 83–87 CrossRef.
- S. Poty, P. Desogere, C. Goze, F. Boschetti, T. D'Huys, D. Schols, C. Cawthorne, S. J. Archibald, H. R. Maecke and F. Denat, Dalton Trans., 2015, 44, 5004–5016 RSC.
- X.-G. Chen and H.-J. Park, Carbohydr. Polym., 2003, 53, 355–359 CrossRef.
- H. Lee, W. Akers, K. Bhushan, S. Bloch, G. Sudlow, R. Tang and S. Achilefu, Bioconjugate Chem., 2011, 22, 777–784 CrossRef PubMed.
- X. Guan, X. Hu, S. Liu, X. Sun and X. Gai, RSC Adv., 2016, 6, 1151–1157 RSC.
- Z. Cao, X. Wang, X. Cheng, J. Wang and R. Tang, Int. J. Polym. Mater. Polym. Biomater., 2017, 66, 495–506 CrossRef.
- D. Gourevich, B. Gerold, F. Arditti, D. Xu, D. Liu, A. Volovick, L. Wang, Y. Medan, J. Gnaim, P. Prentice, S. Cochran and A. Melzer, in Nano-Biotechnology for Biomedical and Diagnostic Research, ed. E. Zahavy, A. Ordentlich, S. Yitzhaki and A. Shafferman, 2012, vol. 733, pp. 135–144 Search PubMed.
- H. Guo, D. Zhang, C. Li, L. Jia, G. Liu, L. Hao, D. Zheng, J. Shen, T. Li, Y. Guo and Q. Zhang, Int. J. Pharm., 2013, 458, 31–38 CrossRef PubMed.
- K. Wang, Y. Xiao, Y. Wang, Y. Feng, C. Chen, J. Zhang, Q. Zhang, S. Meng, Z. Wang and H. Yang, Sci. Rep., 2016, 6, 23061 CrossRef PubMed.
- Z. Zeng, B. Zhang, C. Li, X. Peng, X. Liu, S. Meng and Y. Feng, Dyes Pigm., 2014, 100, 278–285 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra02125b |
| This journal is © The Royal Society of Chemistry 2018 |