Open Access Article
Open Access ArticleRapid production of Pd nanoparticle by a marine electrochemically active bacterium Shewanella sp. CNZ-1 and its catalytic performance on 4-nitrophenol reduction
Haikun Zhang and
Xiaoke Hu *
*
Yantai Institute of Costal Zone Research, Chinese Academy of Sciences, Yantai 264000, China. E-mail: xkhucas@163.com; Fax: +86 535 2109127; Tel: +86 535 2109127
First published on 23rd August 2017
Abstract
Microbial recovery of Pd through Pd(II) reduction is emerging as a clean alternative to traditional physical and chemical reclaiming treatments. Shewanella species are proven to be capable of excellent metal reduction properties and have thus drawn great attention for Pd(II) reduction. So far, however, only Shewanella oneidensis MR-1 has been explored for Pd nanoparticles (Pd-NPs) production. In this study, we report a marine electrochemically active bacterium Shewanella sp. CNZ-1 with the ability for Pd-NPs generation in the presence of sodium lactate as an electron donor. SEM-EDX, TEM, FTIR, XRD and XPS analyses were performed to analyze the Pd(II) reduction process and products. The results showed that Shewanella sp. CNZ-1 was more efficient than Shewanella oneidensis MR-1 in reducing Pd(II) under current conditions. In addition, Pd(II) reduction by CNZ-1 was inhibited by low pH value (3–6) and high concentration of NaCl (3–9%), whereas Pd(II) adsorption by CNZ-1 was unaffected by these factors. Further study showed that hydrogenase and some amides were involved in Pd(II) reduction by CNZ-1. Besides, Pd-NPs produced by CNZ-1 cells (bioCNZ-1-Pd-NPs) seemed more effective than those produced by MR-1 cells (bioMR-1-Pd-NPs) in mediating 4-nitrophenol (4-NP) reduction by NaBH4. For the bioCNZ-1-Pd-NPs supplemented system, the k1 (zero-order rate constant) values were 2.49-, 2.76- and 1.91-fold of the bioMR-1-Pd-NPs supplemented system when the concentrations of NPs were 33 mg L−1, 83 mg L−1 and 167 mg L−1, respectively. The Pd recovery process by CNZ-1 is simple, efficient and environmentally friendly, and this biotechnology has potential practical application in treating mining wastewater.
1. Introduction
Palladium (Pd) is a member of the precious platinum-group metals and possesses many unusual physicochemical and electronic properties. Based on these characteristics, Pd nanoparticles (Pd-NPs) have thus been widely used in biomedicine, chemicals, catalysts, sensors and biosensors.1–16 For example, Pd-NPs can be used as catalysts for a range of reactions, including alkene hydrogenation,1,2 Heck and Suzuki reactions,5,6 Cr(VI) reduction,5,7 xenobiotics reduction8–11 and in fuel cells.12 In addition, Pd-NPs can be used in sensors and biosensors owing to their good electro-catalytic activity, and previous studies have showed their capability for the detection of O2,13 hydrazine,14 formaldehyde,15 and hydrogen peroxide.16 With the wide use of Pd-NPs, Pd certainly will be in short supply in the near future. For the sustainable use of the Pd resource, it is necessary to develop economical, environmentally friendly and efficient methods for Pd-recovery to meet the increasing demand of Pd.Microbial recovery of Pd through Pd(II) reduction is emerging as a clean alternative to traditional physical and chemical reclaiming treatments. The advantages of this method are as follows: (i) the biomass can grow in large amounts rapidly and cheaply; (ii) this method is sensitive enough to recover Pd at ppm concentrations, which is often below the economic threshold of traditional recovery methods.17 According to previous studies, Pd-NPs that produced by microorganisms performed comparably to or better than the commercial catalyst during their applications in catalyzing hydrogenation,2 the environmental pollutants (Cr(VI),7 halogenated aromatics,9,10 flame-retardants11 and so on) reduction, and the production of electricity in fuel cell.12 So far, many bacteria including Shewanella,10,18,19 Desulfovibrio,7,9,20 Cupriavidus,21,22 Pseudomonas,21,22 Paracoccus21,22 and Escherichia17,22 genus etc. were proven to be capable of producing Pd-NPs inside or outside the cell. Among them, members of genus Shewanella are famous members of electrochemically active bacteria (EAB) that capable of using various metals/metalloids as terminal electron acceptors for their anaerobic respiration.23 It's reported that members of genus Shewanella are widely used to synthesize various metal NPs with great potentials for practical applications.22–24 Therefore, studies on production process and mechanism of Pd-NPs through genus Shewanella are attracting more and more attentions.10,18,19 However, to the best of our knowledge, only Shewanella oneidensis MR-1 was reported for Pd recovery18,19,22 and more Shewanella species need to be explored due to their potential practical application in producing Pd-NPs.
Shewanella species are thought to be of marine origin.23 Marine ecosystems are an excellent resource of metal tolerant microorganisms.24 In recent years, marine bacteria are being explored as the potential bio-factories of metal tolerant microorganisms for synthesizing metallic NPs.25 In our previous studies, we isolated a marine electrochemically active bacterium Shewanella sp. CNZ-1 from the sediment of Bohai straits (China). Strain CNZ-1 was proven to be capable of extracellular reducing various N-substituted aromatic compounds due to its good electrochemically activity.26 Accordingly, in this study, we further (i) investigated the ability of Shewanella sp. CNZ-1 for Pd(II) reduction under various factors; (ii) compared the biosynthesis process of Pd-NPs and their catalytic performance on 4-nitrophenol (4-NP) reduction between Shewanella sp. CNZ-1 and Shewanella oneidensis MR-1 (type strain of Shewanella genus); (iii) preliminarily investigated the mechanism of Pd-NPs production through Shewanella sp. CNZ-1.
2. Materials and methods
2.1 Chemicals
Na2PdCl4 was used to prepare Pd(II) solution and it was purchased from Shanghai Macklin Biochemical Co., Ltd (China). 4-NP was purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan). All other reagents used in this study were of the highest analytical grade.2.2 Bacterial strains and growth conditions
Shewanella sp. CNZ-1 (GenBank accession number KX384589, CNZ-1), an electrochemically active bacterium, was isolated from marine sediment by Zhang et al. (Bohai straits, N 38° 30.29′, E 121° 14.10′, China).26 Shewanella oneidensis MR-1 (MR-1), the type stain of Shewanella species, was obtained from Pro. Haichun Gao's lab (Zhejiang University, China). Luria-Bertani (LB) broth contains 5 g L−1 yeast extract, 10 g L−1 peptone, and 10 g L−1 NaCl. Mineral salt medium (MSM) contains 2 g L−1 sodium lactate, 1.0 g L−1 NH4Cl, 0.8 g L−1 Na2HPO4, 0.2 g L−1 KH2PO4, 0.2 g L−1 MgCl2, 0.1 g L−1 CaCl2·2H2O, 20 g L−1 NaCl (pH 7.2).262.3 Preparation of bio-Pd-NPs (under various conditions)
After strain CNZ-1/MR-1 was cultured overnight in 100 mL LB broth in a rotary incubator shaker (150 rpm, 30 °C), the CNZ-1/MR-1 cells were harvested by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 5 min) and washed twice with a sterile phosphate buffer solution (20 mM, pH 7.0). Then, the cell pellets were re-suspended with MSM for the following studies. The experimental systems utilised 135 mL serum bottles containing 100 mL deoxygenated sterile MSM and 0.28 mmol L−1 Pd(II). The CNZ-1/MR-1 cells were added into the systems at a final concentration of 0.5 g L−1. After 10 min of N2 exposure and 24 h of incubation, bio-Pd-NPs were collected by centrifugation (10
000 rpm, 5 min) and washed twice with a sterile phosphate buffer solution (20 mM, pH 7.0). Then, the cell pellets were re-suspended with MSM for the following studies. The experimental systems utilised 135 mL serum bottles containing 100 mL deoxygenated sterile MSM and 0.28 mmol L−1 Pd(II). The CNZ-1/MR-1 cells were added into the systems at a final concentration of 0.5 g L−1. After 10 min of N2 exposure and 24 h of incubation, bio-Pd-NPs were collected by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 5 min), washed with deionised water and finally dried in an oven at 60 °C, respectively. Meanwhile, additional samples were taken at 10 min and 24 h with a sterile needle and a syringe for the analysis of the Pd species, respectively. In addition, various influence factors for Pd(II) reduction by CNZ-1/MR-1, including pH (3–9, MSM adjusted by 1 M HCl and NaOH), concentration of NaCl (2–9%) and initial Pd(II) concentration (0.14–1.41 mmol L−1), were systematically studied. To further investigate the mechanism of Pd(II) reduction through strain CNZ-1, Pd(II) reduction by heat-killed cells (121 °C, 20 min) and Cu(II)-treated cells (0.4 mmol L−1 Cu(II), 30 °C, 15 min) were also performed.
000 rpm, 5 min), washed with deionised water and finally dried in an oven at 60 °C, respectively. Meanwhile, additional samples were taken at 10 min and 24 h with a sterile needle and a syringe for the analysis of the Pd species, respectively. In addition, various influence factors for Pd(II) reduction by CNZ-1/MR-1, including pH (3–9, MSM adjusted by 1 M HCl and NaOH), concentration of NaCl (2–9%) and initial Pd(II) concentration (0.14–1.41 mmol L−1), were systematically studied. To further investigate the mechanism of Pd(II) reduction through strain CNZ-1, Pd(II) reduction by heat-killed cells (121 °C, 20 min) and Cu(II)-treated cells (0.4 mmol L−1 Cu(II), 30 °C, 15 min) were also performed.
2.4 Characterisation of bio-Pd-NPs
To identify the location of the reduced product, CNZ-1 cells attached with Pd-NPs were characterized using scanning electron microscopy-energy dispersive X-ray (SEM-EDX, Hitachi S-4800, Japan), transmission electron microscopy (TEM, JEM-1400, Japan) and Fourier transform infrared spectroscopy (FTIR, Jasco FT/IR-4100, Japan). To identify the structure and valence state of Pd-NPs, the reduced products were further analysed using ultraviolet visible spectrometer (UV-Vis, PerkinElmer Lambda 365, USA), X-ray diffraction patterns (XRD, BRUKER D8 ADVANCE, Germany) and X-ray photoelectron spectroscopy (XPS, ESCALAB 250Xi, England). For SEM-EDX study, the bacterial cells associated with reduced product were filtered through micropore filtration unit, washed three times with normal saline and fixed with glutaraldehyde (3%); it was then washed with Tris–HCl buffer followed by deionized water; the sample was finally dried with ethanol in ambient conditions, mounted on an aluminum stub. For TEM analysis, the samples were prepared as described Bunge et al.21 For XRD, XPS and FTIR analyses, samples were obtained as following steps: (i) the cells were first exposed to 0.28 mmol L−1 Pd(II) for 10 min and 24 h anaerobically; (ii) then separated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min; (iii) the pellet was washed with deionised water and finally dried in an oven at 60 °C. The supernatant was used for subsequent UV-Vis analysis.
000 rpm for 5 min; (iii) the pellet was washed with deionised water and finally dried in an oven at 60 °C. The supernatant was used for subsequent UV-Vis analysis.
2.5 Enhanced 4-NP reduction in the presence of bio-Pd-NPs
The catalytic performance of bio-Pd-NPs was further evaluated. The electron donor and electron acceptor are NaBH4 and 4-NP, respectively. The total volume of the reaction mixture is 3 mL, which contains 33–167 mg L−1 bio-Pd-NPs, 1 g L−1 NaBH4, and 150 mg L−1 4-NP in ultrapure water. Control assays without bio-Pd-NPs and with CNZ-1/MR-1 powders (drying in the oven at 60 °C) only were also performed. The assay mixtures were incubated at room temperature. Each test was conducted three times and the mean was taken to eradicate any discrepancies.2.6 Analytical methods
The Pd(II) concentration was determined by inductively coupled plasma source mass spectrometer (ICP-MS). The cell and 4-NP concentrations were determined by UV-Vis spectrophotometer at their characteristic absorption peaks (600 nm and 400 nm, respectively). For analysis of reduction products, the supernatant was filtrated using 0.22 μm filter membrane and then directly injected into the mass spectrometer. The mobile phase was methanol at 1.0 mL min−1. The reduction efficiency was calculated using eqn (I), as follows:
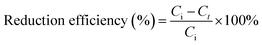 | (I) |
A zero-order model was applied to describe the kinetics of 4-NP reduction by NaBH4. The zero-order rate constant k1 (mol L−1 d−1) was determined according to the following eqn (II), where t (h), C0 (mg L−1) and Ct (mg L−1) are the reaction time, the initial and residual 4-NP at time zero and t, respectively.
| C0 − Ct = k1t | (II) |
A pseudo-first-order model was used to describe the kinetics of 4-NP reduction by NaBH4. The first-order rate constant k2 (min−1) was determined according to the following eqn (III), where t (h), C0 (mg L−1) and Ct (mg L−1) are the reaction time, the initial and residual 4-NP at time zero and t, respectively.
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C0/Ct = k2t C0/Ct = k2t
| (III) |
3. Results and discussion
3.1 Efficient Pd(II) reduction by Shewanella sp. CNZ-1
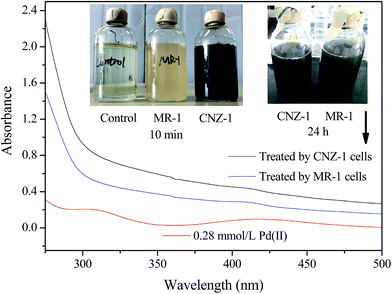
The ability of Shewanella sp. CNZ-1 for Pd(II) bioreduction was first investigated (Fig. 1). After ten minutes of nitrogen exposure, the colour of Pd(II) solution changed from pale yellow to black brown in the CNZ-1 cells treated system (SCNZ-1). In comparison, the colour of MR-1 cells treated system (SMR-1) changed from pale yellow to light brown. The reason for this difference may be that Pd(II) reduction mechanism by MR-1 and CNZ-1 are quite different and we preliminarily discussed this question below. After 24 h of cultivation, the colour of both systems was turn into dark brown, indicating the production of Pd-NPs by CNZ-1 and MR-1 cells. The black appearance provided a visible signature for the formation of bio-Pd-NPs. The UV-Vis sorption results showed that a broad absorption band extending throughout the visible near-ultraviolet region (∼300–500 nm) was appeared, indicating the formation of Pd-NPs, which was consisted with previous studies.27,283.2 Characterization of bio-Pd-NPs
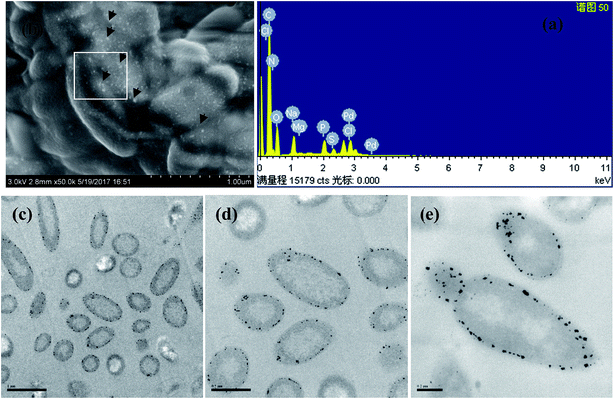
To confirm the reductive deposition of Pd(II) visually, SEM-EDX, TEM, XRD and XPS analyses were further performed. After exposed to Pd(II) solution for 24 h, a number of Pd-NPs were observed on the surface of CNZ-1 cells (Fig. 2a). EDX analysis of Pd-NPs on the cell wall (insert checked pattern in Fig. 2a) showed that Pd peak was appeared and the weight percent of Pd was ∼15.38% (Fig. 2b), indicating that the possible location of Pd-NPs was the surface of CNZ-1 cells. TEM images of thin sections of CNZ-1 cells were obtained to further clarify the deposition location of Pd-NPs (Fig. 2c–e). The result showed that Pd-NPs were located in the periplasmic space of CNZ-1 cells. As indicated by Konishi et al., the space between the outer and inner membranes is a preferable location for easy recovery of biogenic metal NPs.29 The superficial deposition of Pd-NPs by CNZ-1 is beneficial for their recovery and potential use in interfacial bio-catalysis.XRD patterns of different bio-Pd-NPs were presented in Fig. 3a, the diffraction peaks at ∼40°, 46°, 68°, 81° and 84° could be indexed to the (111), (200), (220), (311) and (222) reflections of the fcc Pd (JCPDS 46-1043) structure. It addition, other diffraction peaks of bio-Pd-NPs may attributed to the protein on the surface of CNZ-1 cells (Fig. 3b). XPS was used to further characterize the chemical state information and electronic properties of bio-Pd-NPs. Samples taken from control system that without adding Pd(II) did not show Pd 3d spectra signal (data not shown). Fig. 3b revealed that after 10 min reduction by CNZ-1, Pd(II) ions that absorbed on the surface of CNZ-1 cells were partially reduced. After 24 h of reaction between CNZ-1 cells and Pd(II) ions, all attached Pd(II) ions were reduced to Pd0 NPs. As shown in Fig. 3c, the binding energies of Pd 3d5/2 and Pd 3d3/2 had shifted due to the formation of Pd-NPs. In addition, all attached Pd(II) ions were also completely reduced in SMR-1 (XPS data not shown). It is noticeable that the peak area of Pd0 in SCNZ-1 is greater than that in SMR-1, indicating more Pd-NPs were produced in SCNZ-1 at 10 min (Fig. 3b and d). This finding was consistent with the different colour of SCNZ-1 and SMR-1 at 10 min as shown in Fig. 1.
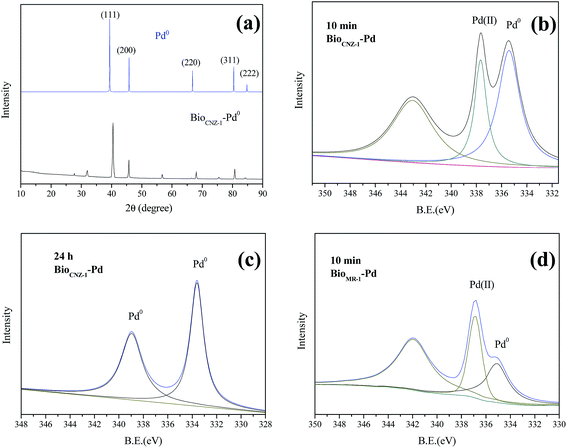 | ||
| Fig. 3 The XRD spectra of bioCNZ-1-Pd0 (a); the XPS spectra (Pd 3d) of bioCNZ-1-Pd0 at 10 min (b) and 24 h (c); the XPS spectra (Pd 3d) of bioMR-1-Pd0 at 10 min (d). | ||
3.3 Mechanism study
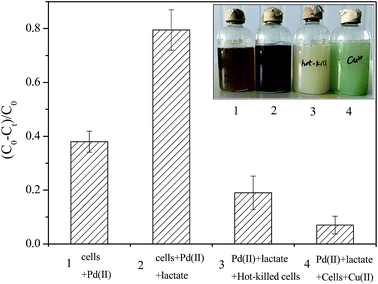
According to the calculation of Pd(II) concentration in SCNZ-1 and SMR-1, we found that ∼36% Pd(II) (0.28 mmol L−1) was reduced during in 10 min and ∼80% Pd(II) was reduced in 24 h in SCNZ-1; for SMR-1, ∼8% Pd(II) (0.28 mmol L−1) was reduced during in 10 min and ∼72% Pd(II) was reduced in 24 h. In current experiment conditions, CNZ-1 was more efficient than MR-1 in Pd(II) reduction. It seemed that the mechanism for the reductive deposition of Pd(II) by CNZ-1 and MR-1 is rather different. At present, the mechanism of microbial Pd(II) reduction was still unclear. Lloyd et al. reported that enzyme was involved in Pd(II) reduction by a sulfate-reducing bacterium.30 Ng et al. reported that [NiFe]-hydrogenase HyaB could facilitate Pd(II) reduction by MR-1 in the presence of lactate as electron donors.19 However, Rotaru et al. found that Pd-NPs were still deposited on autoclaved cells of Cupriavidus necator that had no hydrogenase activity, suggesting a hydrogenase independent formation mechanism. Rotaru et al. also believe that amine group on the cells surface was important in Pd(II) recovery and even a non-enzymatic protein could stimulate Pd(II) reduction as efficiently as bacterial cells.31In this study, the effect of hydrogenase in Pd(II) reduction through CNZ-1 was also studied and results were shown in Fig. 4. Four groups (group 1: CNZ-1 cells + Pd(II); group 2: CNZ-1 cells + Pd(II) + sodium lactate; group 3: hot-killed CNZ-1 cells + Pd(II) + sodium lactate; group 4: CNZ-1 cells + Pd(II) + sodium lactate + Cu(II)) were introduced to verify the roles of electron donor and hydrogenase during Pd(II) reduction as described by Lloyd et al.30 Results showed that the black brown precipitate can be observed obviously in the groups 1 and 2, which was not seen in groups 3 and 4. It's inferred that one or more hydrogenases were involved in the Pd(II) reduction by CNZ-1 in the presence of a lactate electron donor. Besides, since the Pd(II) reduction efficiencies by CNZ-1 and MR-1 were different under the same reaction conditions, it was speculated that the involved enzymes for Pd(II) reduction in CNZ-1 and MR-1 were different.
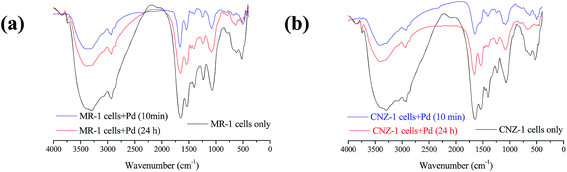
In addition, Pd(II) reduction by CNZ-1 was also observed without electron donor supplemented. A possible reason is that chemical groups of the extracellular secretion on CNZ-1 cell surface contributed to the Pd(II) reduction process.21 FTIR spectra can reveal possible physical and chemical interactions between Pd(II) and groups on the cell surface, we investigated this further with FTIR analysis and the results were shown in Fig. 5. As can be seen, some peaks decreased or disappeared after treated with CNZ-1 and MR-1 cells 10 min and 24 h. These changes are centred at 1250–1750 cm−1, attributed to N–H in plane bending and C–N stretching of amides and carboxyl groups. Previous study has reported that amine groups on surface of MR-1 cell were involved in Pd(II) reduction.31 In the FTIR spectra of CNZ-1 cells, the changes in above-mentioned feature absorption region are even more obvious than MR-1 cells. It's inferred that some cellular protein, mainly functional amine groups,31 were also involved in Pd(II) adsorption and reduction through CNZ-1. These findings imply that some structural properties of CNZ-1 cells account for the formation of Pd-NPs, which have implications for better use of these Shewanella strains.
Our current data imply that more general and structural properties of the cells account for the observed formation of Pd(0) nanoparticles among the bacterial strains investigated here. This may have implications for potential biotechnological applications, possibly also for processing increasingly toxic metals with bacteria not known for their dissimilatory metal-reducing properties.
3.4 Effects of various factors on Pd(II) reduction
Previous study showed that MR-1 could reduce Pd(II) in the presence of various electron donors including H2, formate, lactate, pyruvate and ethanol. Moreover, the most efficient electron donor for Pd(II) reduction by MR-1 was proven to be lactate.10 Thus, 2 g L−1 sodium lactate was selected for further experiment in this study. The effect of initial Pd(II) concentration on the Pd(II) bioreduction efficiency by CNZ-1 was determined after 24 h (Fig. 6a). The addition of Pd(II) at 0.14, 0.28, 0.42, 0.56, 0.84 and 1.41 mmol L−1 result in 0.135 (97.1%), 0.224 (80.0%), 0.239 (57.1%), 0.282 (50.0%), 0.295 (34.9%) and 0.282 mmol L−1 (20.0%) Pd(II) removal in the presence of 0.5 g L−1 CNZ-1 cells. The results showed that the Pd(II) removal percentage decreased as initial Pd(II) concentration increased, indicating a relationship between initial Pd(II) concentration and biomass. Similarly, Yates et al. found that Pd(II) reduction efficiency by another electrochemically active bacterium Geobacter sulfurreducens decreased from 51% to 34% as initial concentration of Pd(II) increased from 100 to 200 mg L−1, respectively.32 The effects of pH and NaCl concentration on Pd(II) reduction by CNZ-1 were illustrated in Fig. 6b. It was found that pH value and NaCl concentration were important factors for CNZ-1 to producing Pd-NPs. When the pH value was less than 6, the colour of Pd(II) solution did not change obviously. In our previous study, we found that strain CNZ-1 grew best under pH 7.3, whereas it failed to survive at pH 3.8.26 When the pH value is higher than 6, colour change could be observed easily in the reaction system due to the formation of Pd-NPs. In addition, Pd(II) reduction through CNZ-1 was inhibited by 3–9% of NaCl. Yong et al. reported that formate hydrogenlyase was sensitive to NO3− and Cl−, we thus inferred that the activity of hydrogenase related to lactate was inhibited by low pH value and high NaCl concentration.33 However, it's notable that Pd(II) adsorption by CNZ-1 could be observed even at low pH value (3–6) and high NaCl concentrations (3–9%). At pH 3.2 and 9% NaCl, Pd(II) adsorption efficiency could still reach ∼84% and ∼74%, respectively.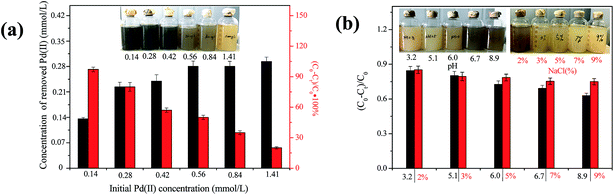 | ||
| Fig. 6 The effects of initial Pd(II) concentration (a), pH and NaCl concentration (b) on Pd(II) reduction by CNZ-1. | ||
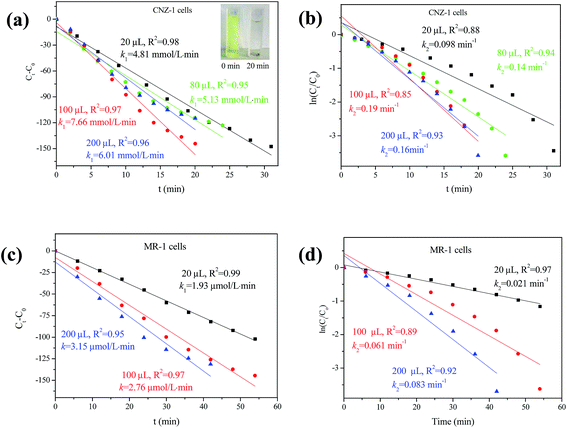
3.5 Enhanced 4-NP reduction in the presence of bio-Pd-NPs
The catalytic activity of bio-Pd-NPs was evaluated through 4-NP reduction in the presence of NaBH4. As E0 for 4-nitrophenol/4-aminophenol and H3BO3/BH4− is −0.76 and −1.33 V vs. SHE, respectively, this reaction could not proceed without additional catalyst.34 Thus, it was used to test the catalytic performance of bio-Pd-NPs in this study. Results showed that no reduction (<3%) of 4-NP occurred in control assays during 50 min whereas 4-NP was completely reduced during 20–30 min with the addition of bioCNZ-1-Pd-NPs (33–167 mg L−1). In comparison, catalytic reduction of 4-NP completely by bioMR-1-Pd-NPs (33–167 mg L−1) needed more than 40 min. The reduction products were analyzed by mass spectrum, and 4-aminophenol was proven to be the primary product (data not shown). The zero-order and pseudo-first-order model were used to describe the kinetics of 4-NP reduction mediated by bioCNZ-1-Pd-NPs (Fig. 7a and b) and bioMR-1-Pd-NPs (Fig. 7a and b), respectively. The kinetic parameters of the zero-order and pseudo-first-order equations were listed in Fig. 7. It was observed that the experimental data were well explained by the zero-order model in both reaction systems (R2 > 0.95). For the bioCNZ-1-Pd-NPs supplemented system, the k1 (zero-order rate constant) values were 2.49-, 2.76- and 1.91-fold of the bioMR-1-Pd-NPs supplemented system when the concentrations of NPs were 33 mg L−1, 83 mg L−1 and 167 mg L−1, respectively. These results indicated that bioCNZ-1-Pd-NPs have potential application in environmental pollutants control.3.6 Implications
In this study, we have demonstrated that Shewanella sp. CNZ-1, a marine electrochemically active bacterium, is capable of adsorbing Pd(II) and producing Pd-NPs in the presence of sodium lactate. Shewanella species are proven to be capable of excellent metal reduction properties and have thus drawn great attention in Pd(II) reduction.18,19,23 So far, however, only Shewanella oneidensis MR-1 was explored for Pd nanoparticles (Pd-NPs) production. In this study, we report a marine electrochemically active bacterium Shewanella sp. CNZ-1 with the ability for Pd-NPs generation in the presence of sodium lactate as electron donor. The current study was to contribute to the list of Shewanella strains with the ability for Pd-NPs production. In addition, CNZ-1 was found to be more efficient than MR-1 under current conditions (2% NaCl, pH 7.2, 2 g L−1 sodium lactate as electron donor, 0.28 mmol L−1 Pd(II) as electron acceptor). Compared to MR-1, CNZ-1 could produce Pd-NPs in a very short period of time (10 min), which is beneficial for its further practical application.The effects of various factors on Pd(II) reduction by CNZ-1 were studied and the optimal conditions for Pd recovery with highest efficiency were thereby assessed on all of basic factors. We noticed that excessive Pd(II) concentration (1.41 mmol L−1), low pH values (3–6) and high concentration of Cl− (3–9%) affected Pd(II) reduction by CNZ-1, whereas nearly no influence on Pd(II) adsorption by CNZ-1. Because the enzymatic and genomic mechanisms of Pd(II) reduction by CNZ-1 are unknown yet, further studies need to be done for guiding the application of CNZ-1. On the basis of experiment results obtained in this study, a simple two-step process including Pd(II) adsorption and Pd(II) reduction was proposed for obtaining microbial produced Pd-NPs through CNZ-1 (Fig. 8).
4. Conclusion
In this study, we demonstrated that Pd-NPs could be produced rapidly by Shewanella sp. CNZ-1 cells with sodium lactate as electron donor. It is found that CNZ-1 was more efficient than Shewanella oneidensis MR-1 for producing Pd-NPs under current conditions. Pd(II) reduction by CNZ-1 was inhibited by low pH value and high concentration of NaCl, whereas effects of the two factors on Pd(II) adsorption by CNZ-1 were not obviously. In addition, bioCNZ-1-Pd-NPs seemed more effective in mediating 4-NP reduction by NaBH4 compared to bioMR-1-Pd-NPs. The Pd recovery process by CNZ-1 is simple, efficient and environmental friendly, this biotechnology thus has potential application in treating mining wastewater.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This subject was supported by the National Natural Science foundation of China (No. 51608519) and National Natural Science foundation of Shandong Province, China (No. ZR2016EEB10). Thanks to Pro. Haichun Gao for providing Shewanella oneidensis MR-1.References
- Q. M. Kainz, R. Linhardt, R. N. Grass, G. Vilé, J. Pérez-Ramírez, W. J. Stark and O. Reiser, Palladium nanoparticles supported on magnetic carbon-coated cobalt nanobeads: highly active and recyclable catalysts for alkene hydrogenation, Adv. Funct. Mater., 2014, 24, 2020–2027 CrossRef CAS.
- N. J. Creamer, I. P. Mikheenko, P. Yong, K. Deplanche, D. Sanyahumbi, J. Wood, K. Pollman, M. Merroun, S. Selenska-Pobell and L. E. Macaskie, Novel supported Pd hydrogenation bionanocatalyst for hybrid homogeneous/heterogeneous catalysis, Catal. Today, 2007, 128, 80–87 CrossRef CAS.
- N. I. Hulkoti and T. C. Taranath, Biosynthesis of nanoparticles using microbes-a review, Colloids Surf., B, 2014, 121, 474–483 CrossRef CAS PubMed.
- J. Cui and L. Zhang, Metallurgical recovery of metals from electronic waste: a review, J. Hazard. Mater., 2008, 158, 228–256 CrossRef CAS PubMed.
- K. Deplanche, J. A. Bennett, I. P. Mikheenko, J. Omajali, A. S. Wells, R. E. Meadows, J. Wood and L. E. Macaskie, Catalytic activity of biomass-supported Pd nanoparticles: influence of the biological component in catalytic efficacy and potential application in ‘green’ synthesis of fine chemicals and pharmaceuticals, Appl. Catal., B, 2014, 147, 651–665 CrossRef CAS.
- Y. Li, Y. Dai, Z. Yang and T. Li, Controllable synthesis of palladium nanoparticles and their catalytic abilities in Heck and Suzuki reactions, Inorg. Chim. Acta, 2014, 414, 59–62 CrossRef CAS.
- A. C. Humphries, K. P. Nott, L. D. Hall and L. E. Macaskie, Continuous removal of Cr(VI) from aqueous solutions catalyzed by palladized biomass of Desulfovibrio vulgaris, Biotechnol. Lett., 2004, 26, 1529–1532 CrossRef CAS PubMed.
- A. Safavi and S. Momeni, Highly efficient degradation of azo dyes by palladium/hydroxyapatite/Fe3O4 nanocatalyst, J. Hazard. Mater., 2012, 201–202, 125–131 CrossRef CAS PubMed.
- V. S. Baxter-Plant, I. P. Mikheenko and L. E. Macaskie, Sulphate-reducing bacteria, palladium and reductive dehalogenation of chlorinated aromatic compounds, Biodegradation, 2003, 14, 83–90 CrossRef CAS PubMed.
- W. De Windt, P. Aelterman and W. Verstraete, Bioreductive deposition of palladium(0) nanoparticles on Shewanella oneidensis with catalytic activity towards reductive dechlorination of polychlorinated biphenyls, Environ. Microbiol., 2005, 7, 314–325 CrossRef CAS PubMed.
- S. Harrad, M. Robson, S. Hazrati, V. S. Baxter-Plant, K. Deplanche, M. D. Redwood and L. E. Macaskie, Dehalogenation of polychlorinated biphenyls and polybrominated diphenyl ethers using a hybrid bioinorganic catalyst, J. Environ. Monit., 2007, 9, 314–318 RSC.
- P. Yong, M. Paterson-Beedle, I. P. Mikheenko and L. E. Macaskie, From bio-mineralisation to fuel cells: biomanufacture of Pd and Pt nanocrystals for fuel cell electrode catalyst, Biotechnol. Lett., 2007, 29, 539–544 CrossRef CAS PubMed.
- K. Jukk, N. Kongi, L. Matisen, T. Kallio, K. Kontturi and K. Tammeveski, Electroreduction of oxygen on palladium nanoparticles supported on nitrogen-doped graphene nanosheets, Electrochim. Acta, 2014, 137, 206–212 CrossRef CAS.
- P. K. Rastogi, V. Ganesan and S. Krishnamoorthi, Palladium nanoparticles decorated gaur gum based hybrid material for electrocatalytic hydrazine determination, Electrochim. Acta, 2014, 125, 593–600 CrossRef CAS.
- A. Safavi, N. Maleki, F. Farjami and E. Farjami, Electrocatalytic oxidation of formaldehyde on palladium nanoparticles electrodeposited on carbon ionic liquid composite electrode, J. Electroanal. Chem., 2009, 626, 75–79 CrossRef CAS.
- Y. Liu, G. Sun, C. Jiang, X. T. Zheng, L. Zheng and C. M. Li, Highly sensitive detection of hydrogen peroxide at a carbon nanotube fiber microelectrode coated with palladium nanoparticles, Microchim. Acta, 2014, 181, 63–70 CrossRef CAS.
- K. Deplanche, I. Caldelari, I. P. Mikheenko, F. Sargent and L. E. Macaskie, Involvement of hydrogenases in the formation of highly catalytic Pd (0) nanoparticles by bioreduction of Pd (II) using Escherichia coli mutant strains, Microbiology, 2010, 156, 2630–2640 CrossRef CAS PubMed.
- Y. Tuo, G. Liu, B. Dong, H. Yu, J. Zhou, J. Wang and R. Jin, Microbial synthesis of bimetallic PdPt nanoparticles for catalytic reduction of 4-nitrophenol, Environ. Sci. Pollut. Res., 2017, 24, 5249–5258 CrossRef CAS PubMed.
- C. K. Ng, T. K. C. Tan, H. Song and B. Cao, Reductive formation of palladium nanoparticles by Shewanella oneidensis: role of outer membrane cytochromes and hydrogenases, RSC Adv., 2013, 3, 22498–22503 RSC.
- J. R. Lloyd, A. N. Mabbett, D. R. Williams and L. E. Macaskie, Metal reduction by sulphate-reducing bacteria: physiological diversity and metal specificity, Hydrometallurgy, 2001, 59, 327–337 CrossRef CAS.
- M. Bunge, L. S. Søbjerg, A. E. Rotaru, D. Gauthier, A. T. Lindhardt, G. Hause, K. Finster, P. Kingshott and R. L. Meyer, Formation of palladium (0) nanoparticles at microbial surfaces, Biotechnol. Bioeng., 2010, 107, 206–215 CrossRef CAS PubMed.
- S. De Corte, T. Hennebel, B. De Gusseme, W. Verstraete and N. Boon, Bio-palladium: from metal recovery to catalytic applications, Microb. Biotechnol., 2012, 5, 5–17 CrossRef CAS PubMed.
- D. Saffarini, K. Brockman, A. Beliaev, R. Bouhenni and S. Shirodkar, Shewanella oneidensis and extracellular electron transfer to metal oxides, Bacteria-Metal Interactions, Springer International Publishing, 2015, pp. 21–40 Search PubMed.
- P. Manivasagan, S. Y. Nam and J. Oh, Marine microorganisms as potential biofactories for synthesis of metallic nanoparticles, Crit. Rev. Microbiol., 2016, 42, 1007–1019 CrossRef CAS PubMed.
- M. Agnihotri, S. Joshi, A. R. Kumar, S. Zinjarde and S. Kulkarni, Biosynthesis of gold nanoparticles by the tropical marine yeast Yarrowia lipolytica NCIM 3589, Mater. Lett., 2009, 63, 1231–1234 CrossRef CAS.
- H. Zhang and X. Hu, Catalytic reduction of NACs by nano Fe3O4/quinone composites in the presence of a novel marine exoelectrogenic bacterium under hypersaline conditions, RSC Adv., 2017, 7, 11852–11861 RSC.
- S. Momeni and I. Nabipour, A simple green synthesis of palladium nanoparticles with Sargassum alga and their electrocatalytic activities towards hydrogen peroxide, Appl. Biochem. Biotechnol., 2015, 176, 1937–1949 CrossRef CAS PubMed.
- T. Yonezawa, K. Imamura and N. Kimizuka, Direct preparation and size control of palladium nanoparticle hydrosols by water-soluble isocyanide ligands, Langmuir, 2001, 17, 4701–4703 CrossRef CAS.
- Y. Konishi, K. Ohno, N. Saitoh, T. Nomura, S. Nagamine, H. Hishida, Y. Takahashi and T. Uruga, Bioreductive deposition of platinum nanoparticles on the bacterium Shewanella algae, J. Biotechnol., 2007, 128, 648–653 CrossRef CAS PubMed.
- J. R. Lloyd, P. Yong and L. E. Macaskie, Enzymatic recovery of elemental palladium by using sulfate-reducing bacteria, Appl. Environ. Microbiol., 1998, 64, 4607–4609 CAS.
- A. E. Rotaru, W. Jiang, K. Finster, T. Skrydstrup and R. L. Meyer, Non-enzymatic palladium recovery on microbial and synthetic surfaces, Biotechnol. Bioeng., 2012, 109, 1889–1897 CrossRef CAS PubMed.
- M. D. Yates, R. D. Cusick and B. E. Logan, Extracellular palladium nanoparticle production using Geobacter sulfurreducens, ACS Sustainable Chem. Eng., 2013, 1, 1165–1171 CrossRef CAS.
- P. Yong, N. A. Rowson, J. P. G. Farr, I. R. Harris and L. E. Macaskie, Bioreduction and biocrystallization of palladium by Desulfovibrio desulfuricans NCIMB 8307, Biotechnol. Bioeng., 2002, 80, 369–379 CrossRef CAS PubMed.
- S. K. Ghosh, M. Mandal, S. Kundu, S. Nath and T. Pal, Bimetallic Pt-Ni nanoparticles can catalyze reduction of aromatic nitro compounds by sodium borohydride in aqueous solution, Appl. Catal., A, 2004, 268, 61–66 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |