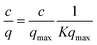Open Access Article
Open Access ArticleIon-specificity in protein binding and recovery for the responsive hydrophobic poly(vinylcaprolactam) ligand
Zizhao Liua,
S. Ranil Wickramasinghea and
Xianghong Qian *b
*b
aDepartment of Chemical Engineering, University of Arkansas, Fayetteville, AR 72701, USA
bDepartment of Biomedical Engineering, University of Arkansas, Fayetteville, AR 72701, USA. E-mail: xqian@uark.edu; Tel: +1-479-575-8401
First published on 21st July 2017
Abstract
The conformational switch between the hydrophobic state and hydrophilic state of thermo-responsive poly(vinylcaprolactam) (PVCL) has great potential for protein binding and elution applications during protein purification as a hydrophobic interaction chromatography ligand. The lower critical solution temperature (LCST) of PVCL is strongly salt type and salt concentration dependent. Here PVCL polymer chains were grafted on regenerated cellulose membranes using controllable atom-transfer radical polymerization (ATRP) for protein binding/elution studies. Protein binding and recovery demonstrates strong salt type and salt concentration dependency. The effects of salt ions on the static and dynamic binding interactions were elucidated and correlated well with the conformational and hydrophobicity changes of the responsive ligand.
1. Introduction
The past decade has seen the rapid development of upstream technology for biopharmaceuticals. As a consequence, the concentration of recombinant protein products has markedly increased from milligrams per liter (mg L−1) to grams per liter (g L−1).1 At the same time, the regulatory agencies demand high purity of products, which greatly heightens the need for the dramatic improvement of efficiency in the downstream processing and purification.1,2 The bottleneck has shifted from bio-processing to downstream purification of products. To date, estimated cost for downstream processing can go as high as 50 to 80 percent of the total manufacturing cost.2 Therefore, one of the main objectives associated with the downstream processing is cost reduction while maintaining the high quality of the products.Hydrophobic interaction chromatography (HIC) plays an important role in downstream processing. Proteins of interest bind to hydrophobic ligands at high salt concentration buffer which promotes hydrophobic interaction and elute at low salt concentration buffer which does not favor protein–ligand binding. N-Alkyl (C1–C8) and aryl (phenyl) are the two most common types of ligands for HIC. The factors that affect the performances of these conventional HIC ligands have been studied under different buffer conditions in particular, salt type and salt concentration.3,4 Theoretical models based on phenomenological solvophobic theory5 or preferential interactions6 were developed to quantitatively explain and predict the salt concentration effects on the chromatographic behaviors. However, ion specificity remains to be a theoretical challenge in understanding the interactions between protein and ligand. Moreover, protein denaturation and subsequent aggregation using these conventional hydrophobic ligand is a major concern. Currently significant efforts have been dedicated to overcome the limited capacity and poor recovery of the conventional HIC membrane chromatography. A new class of salt- and temperature-responsive ligands that can switch hydrophobic and hydrophilic conformation under different buffer conditions has been explored for HIC applications using buffers containing (NH4)2SO4 or NaCl only.7–10 As the degree of hydration and dehydration of these temperature/salt ion responsive ligands is highly salt ion and salt concentration specific, the effects of salt ion type and salt ion concentration on protein-responsive ligand binding are investigated. Here we focus on poly(vinylcaprolactam) (PVCL) which exhibits higher binding capacity than other responsive ligands.10 Poly(N-isopropylacrylamide) (PNIPAM) has also been investigated as a thermo-responsive ligand for HIC applications.8 The main advantage of PVCL lies in its low toxicity and biocompatibility. As the amide bond of the monomer is located on its seven-member ring, its hydrolysis in the polymer does not generate low molecular weight species. On the other hand, hydrolysis of PNIPAM produces low molecular weight amines, which are toxic to biological systems. Another advantage of PVCL as well as PNIPAM is that its LCST (32–50 °C) is close to the room temperature (25 °C), at which downstream processing typically occurs. It means that only low concentrations of salt are needed in order to reduce its LCST to below the room temperature. Low salt is preferred as high salt could denature the protein.
Thermo-responsive polymers such as PVCL exhibit a lower critical solution temperature (LCST), above which the polymers adopt a collapsed hydrophobic conformation. However, at a temperature below their LCSTs, these polymers adopt an extended coil-like hydrophilic conformation. The presence of the salt tends to reduce the transition temperature. The reduction of LCST is found to be strongly dependent on the salt type and salt concentration.11 The higher the concentration is, the larger the reduction will be. It is also ion-specific with cations and anions each following a different order depending on the charge and size of the ion. Our earlier studies11–14 investigating the specific interactions of ions with the thermo-responsive PNIPAM show that the cations interact directly with the amide oxygen. The anion–polymer interaction is mediated by the cations even though larger size anions could potentially form hydrophobic interaction with the isopropyl group. For PNIPAM, the LCST transition only depends slightly on the molecular weight of the polymer chain. However, the LCST of PVCL depends strongly on the polymer molecular weight and concentration.15 For very dilute PVCL solutions, the LCST of PVCL varies from about 32 °C for long polymer chains (molecular weight (MW) ≥ 275![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) to over 50 °C for short chains (MW ∼ 9000). This hydrophilic-to-hydrophobic LCST transition can be switched by increasing the temperature and/or salt concentration. Since the binding and elution of the proteins are based on the hydrophobicity change of the thermo-responsive ligands, relative high recovery of the proteins is expected. Our earlier studies10 show that binding capacity and recovery can be further optimized by designing comb-like ligands.
000) to over 50 °C for short chains (MW ∼ 9000). This hydrophilic-to-hydrophobic LCST transition can be switched by increasing the temperature and/or salt concentration. Since the binding and elution of the proteins are based on the hydrophobicity change of the thermo-responsive ligands, relative high recovery of the proteins is expected. Our earlier studies10 show that binding capacity and recovery can be further optimized by designing comb-like ligands.
Salt ion type and salt ion concentration appear to have a strong effect on the conventional hydrophobic based protein–ligand interactions.4,5,16–18 It has been shown that the effects of different ions on protein–ligand binding correlate with the Hofmeister series.4,16,19 Hofmeister series refers to the different ability of different ions to denature protein.20 Cations and anions have their respective orders.21 The direct Hofmeister series for the anions follows the order PO43− > SO42− > CH3COO− > Cl− > Br− > NO3− > I− > ClO4− > SCN−. For the cations, the order follows NH4+ > K+ > Na+ > Li+ > Mg2+ > Ca2+. Ions on the left side generally have greater ability to decrease the solubility of hydrocarbon (salting-out), promote hydrophobic interaction as well as stabilize proteins. However, inverse Hofmeister effects have also been observed.22 It was thought that the ion order depends on both the surface hydrophobicity and surface polarity. So far, the exact nature of ion specificity on physical and biological phenomena remains unexplained and under considerable debate. It is not sure whether it is caused by the changes in the bulk water structure in different salt solutions or by the direct ion–protein interactions.23
Earlier studies have also shown that the LCST transition of thermo-responsive polymers is strongly ion specific.11,24 Mikheeva et al. found two thermal cooperative transition of PVCL hydrogel at 31.5 °C and 37.6 °C and showed that NaCl solution lowers LCST while SDS increases LCST.25 Zavgorodnya et al. studied Hofmeister anion effect on PVCL hydrogels.26 Khokhlov et al. investigated the complexation effect of poly(VCL-co-methacrylic acid).27 It was found that insoluble inter-macromolecular complex formed under acidic condition. Our previous theoretical studies11,13 on the effects of salt ions on LCST of PNIPAM demonstrate that the stronger hydration of ions tends to decrease the LCST whereas the direct cation–amide oxygen binding tends to increase the LCST. The strength of the cation–amide oxygen interaction is dictated by the competition between the electrostatic and the hydration forces. For the singly charged alkali cations (Li+, Na+, K+ and Rb+), electrostatic interactions dominate. The larger the cation is, the weaker the binding interaction with the amide oxygen. For the doubly charged cations (Mg2+ and Ca2+), the strong hydration of these divalent ions overcomes the electrostatic attraction between the ion and amide oxygen leading to the very weak binding between the cations and the amide oxygen. Moreover, our simulation results show that the LCST transition dynamics and the degree of hydration/dehydration are ion specific. Experimentally, responsive HIC membrane chromatography using PNIPAM and its copolymers as ligands shows relatively low capacity and recovery.8,28 The PVCL ligands grafted on the regenerated cellulose membranes using atom-transfer radical polymerization exhibit both higher capacity and higher recovery than the PNIPAM ligands.7,9,10 Our previous results show effective binding of BSA and IgG4 in the high salt buffer (1.8 M (NH4)2SO4) solution. High recovery (over 97%) of BSA was also obtained at an appropriate ligand density. In addition, our results show that binding capacity tends to increase with the increase of the polymer chain density as well as the salt concentration at binding.7 The architecture of the ligand affects both the binding capacity and recovery.10 Here the effects of salt ion type and salt ion concentration on the binding capacity and recovery of BSA and IgG4 proteins were investigated using PVCL as a responsive ligand. The effects of salt ion type and concentration on the LCST transition of the solution phase PVCL were investigated using turbidity test initially. This will be followed by the investigation on the dynamic/static binding capacity and recovery of the responsive membranes grafted with PVCL ligands at different salt conditions. Common salts used for industrial bioprocessing with hydrophobic interaction chromatography including NaCl, (NH4)2SO4, Na2SO4 and K2SO4 were investigated. In addition, ZnSO4 and Al2(SO4)3 representing divalent and trivalent cations were also studied. Other divalent sulfate salts such as CaSO4, MgSO4 and CuSO4 all exhibit rather low solubility and are not suitable for the current study.
2. Materials
N-Vinylcaprolactam (98%), 2-bromo-2-methylpropionyl bromide (BIB, 98%), 4-(dimethylamino)pyridine (DMAP, ≥99%), copper(I) chloride (≥99.995%), copper(II) chloride (≥99.995%) and N,N,N′,N′′,N′′-pentamethyldiethylenetriamine (PMDETA, 99%) were purchased from Sigma Aldrich (St. Louis, MO). Triethylamine (TEA, ≥99%) and aluminum sulfate (≥97%) were obtained from Alfa Aesar (Ward Hill, MA). Methanol (99.8%), acetonitrile (99.8%) and zinc sulfate heptahydrate (≥99.5%) were obtained from EMD Chemicals (Billerica, MA). Boric anhydride was purchased from Avantor Performance Materials (Center Valley, PA). Anhydrous acetonitrile was obtained by distilling acetonitrile with boric anhydride. Sodium chloride (≥99.5%), ammonium sulfate (≥99.0%) and sodium sulfate (≥99.0%) were bought from Macron™ Fine Chemicals (Center Valley, PA). Regenerated cellulose membranes (0.45 μm pore size, RC55, 47 mm diameter) were purchased from Whatman Ltd. (Pittsburgh, PA). Bovine serum albumin (BSA) (>99%, pI 4.7, 66 kDa) was obtained from Avantor Performance Materials (Center Valley, PA). Purified human IgG4 monoclonal antibody (pI 7.1, 146 kDa) was provided by Eli Lilly (Indianapolis, IN).3. Experiments
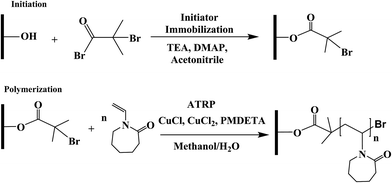
3.1 Membrane surface modification
Membranes were modified through surface-initiated ATRP as reported by our earlier studies.7,29–31 Basically, regenerated cellulose (RC) membranes were first immobilized with ATRP initiator 2-bromoisobutyryl bromide (80 mM) in acetonitrile for 3 hours. Then the monomer, copper(I) chloride, copper(II) chloride, ligand N,N,N′,N′′,N′′-pentamethyldiethylenetriamine (PMDETA), and solvent, methanol/water mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) were mixed together and degassed with argon for 15–20 min. The ratio among monomer
1, v/v) were mixed together and degassed with argon for 15–20 min. The ratio among monomer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CuCl
CuCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CuCl2
CuCl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PMDETA is 200
PMDETA is 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2
0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Prior to polymerization reaction, flasks containing initiator immobilized membranes were de-oxygenated by vacuum and argon back-filling process three times. At last, the polymerization solution was transferred to the flask by a syringe. After 4 h ATRP, membranes were first rinsed with methanol and water mixture (1
2. Prior to polymerization reaction, flasks containing initiator immobilized membranes were de-oxygenated by vacuum and argon back-filling process three times. At last, the polymerization solution was transferred to the flask by a syringe. After 4 h ATRP, membranes were first rinsed with methanol and water mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) three times and then washed with DI water overnight. The schematic of the modification procedure is shown in Fig. 1.
1, v/v) three times and then washed with DI water overnight. The schematic of the modification procedure is shown in Fig. 1.
3.2 Characterization
3.3 Protein binding experiments for HIC membranes
| Membrane volume = π × Dmembrane2/4 × d |
4. Results and discussion
4.1 Salt effects on solution phase PVCL polymers
In order to study the effects of salt ion type and salt ion concentration on the LCST of PVCL solutions, PVCL polymers are synthesized using free radical polymerization based on the protocol reported.32 The hydrodynamic diameters (Dh) of synthesized PVCL in aqueous solution at different temperatures were determined using dynamic light scattering (DLS) with a DelsaNano HC particle analyzer instrument (Beckman Coulter, Miami, FL) at a fixed scattering angle of 165°. Results were processed by DelsaNano program (v.3.7) with CONTIN algorithm. Similar to the Dh results observed before, the increase in the Dh is likely caused by the aggregation of PVCL when the temperature increased above LCST when it became hydrophobic. The LCST of the PVCL synthesized here is around 37 °C as shown by the DLS results in Fig. 2. Previously reported PVCL synthesized by free radical polymerization has a LCST ranging from 30–50 °C, depending on the molecular weight.33 The polydispersity seems to fluctuate as temperature increases. However, it decreases slightly above the LCST when the polymer is collapsed.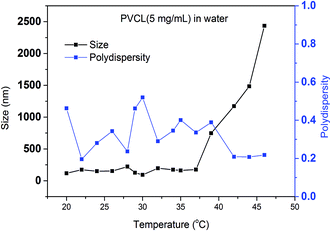 | ||
| Fig. 2 Size measurement of synthesized PVCL under different temperatures by dynamic light scattering and calculated polydispersity. | ||
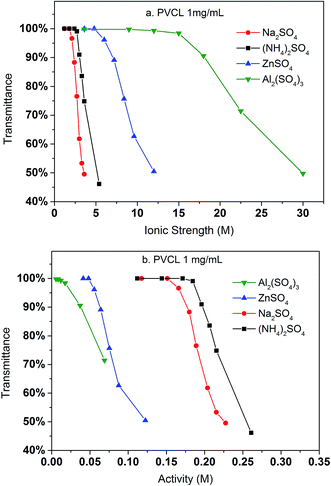
Turbidity measurements were conducted to further investigate the effects of salt on the change of LCST for the synthesized PVCL in salt solutions. Similar to PNIPAM, earlier studies show that the presence of KCl decreases the LCST of PVCL.34 Here, systematic studies were carried out to investigate the effects of salt type on the reduction of LCST for the synthesized PVCL. Different sulfate salts were used for the investigation as sulfate ions demonstrate a strong effect on the reduction of LCST for PNIPAM.35 Turbidity (transmittance) of PVCL solution was measured in monovalent (Na+, NH4+), divalent (Zn2+) and trivalent (Al3+) sulfate salt solutions at room temperature of around 23 ± 1 °C. The impact of cations on the turbidity thus the change of LCST was plotted as a function of the ionic strength (Fig. 3a) and the activity (Fig. 3b) for the four salt solutions. Our results show that the impact of ions on the reduction of PVCL LCST follows: Na+ > NH4+ > Zn2+ > Al3+ based on nominal ionic strength or Al3+ > Zn2+ > Na+ > NH4+ based on the ionic activity. Clearly the results based on activity should be more meaningful than the ones based on nominal ionic strength since higher valence ions have a stronger tendency to associate with each other even though they have a higher degree of solvation. Moreover, it was observed earlier that Al3+ ion largely exists as Al(OH)4− in aqueous solution forming a polymeric network.36,37 Based on the turbidity as a function of ionic activity as shown in Fig. 3b, a reverse Hofmeister series is observed. This agrees with earlier studies on the effects of cations on the LCST of PNIPAM.38 It is worthwhile to point out that PVCL/PNIPAM and proteins are quite different as proteins generally contain multiple charged residues whereas PVCL/PNIPAM do not have net charges. Our earlier studies11–13 show that monovalent cations bind directly with the amide oxygen on PNIPAM whereas anions interact indirectly with the hydrophobic residues on the polymer. The direct cation–amide oxygen interaction strength is modulated by the competition between the favorable electrostatic interaction and the unfavorable dehydration force. Our earlier studies further show that divalent ions actually do not bind directly or only bind weakly with the amide oxygen. Clearly the unfavorable dehydration force is dominant for the higher valence ions as they typically have significant hydration free energy. The presence of these high valent ions leads to a relative high surface tension of the solution due to the strong solvation free energies of these ions. The strong surface tension stabilizes the hydrophobic conformation of the polymer leading to an enhanced reduction in LCST. The NH4+ and Na+ ions, on the other hand, have relatively small hydration free energies. NH4+ ion has a similar radius to K+. The hydration free energy is more negative for Na+ than for NH4+. As a result, the increase in surface tension in Na2SO4 salt solution tends to be slightly higher than in the same concentration of (NH4)2SO4 salt solution as shown in Fig. 4.
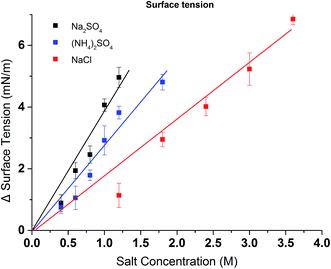 | ||
| Fig. 4 Excess surface tension of (NH4)2SO4, Na2SO4 and NaCl at various concentrations (water surface tension is 73.54 ± 0.16 mN m−1). All data were averaged by five measurements. | ||
It should be pointed out that the concentration of PVCL for the DLS study is five times that of the turbidity test. It appears that DLS requires a high concentration of the polymer as it measures the size change of the particles. Here it is only possible when the PVCL chains collapse onto each other to form aggregates at the transition temperature. However, for turbidity test, as light transmittance was measured, the conformational change of PVCL from coil-like to collapsed globular structure can be detected. This could even occur when the polymer chain collapses onto itself at the relative low PVCL concentrations.
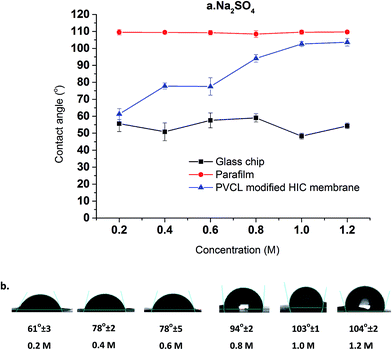
Compared to a non-responsive ligand where the hydrophobicity does not change with salt ion concentration and salt ion type, the hydrophobicity of the thermo-responsive ligand will be different at different solution conditions. This is expected to have a strong impact on the binding and elution of proteins to/from these ligands. As a result, the change in hydrophobicity for the grafted PVCL polymers on membrane substrates in different salt solutions were investigated by contact angle measurement and correlated with the reduction in LCST from the turbidity test. The hydrophobicity change was investigated by measuring the static contact angles of various concentrations of salt solution drops on membrane surface. Here only Na2SO4 salt solutions were investigated as Na2SO4 salt demonstrates a stronger effect on the capacity for protein binding compared to conventionally used (NH4)2SO4 salt in biotechnology industry. Glass chip and parafilm were also tested under the same conditions for comparison purposes. As shown in Fig. 5, the contact angle of the membrane surface increases from 60° to 100° when the Na2SO4 salt concentration increases from 0.2 to 1.2 M indicating that higher salt concentration results in an increase in hydrophobicity. Besides grafted PVCL ligands, surface morphology will also affect the hydrophobicity after surface modification. The morphologies of modified membranes under salt solutions as well as at temperatures above its LCST up to 45 °C were imaged using atomic force microscopy (AFM). No obvious change in roughness and morphology has been observed at 500 nm and 1 μm scales (results not shown). Therefore, the increase in the hydrophobicity of PVCL grafted membrane is likely to due to the conformational changes occurring at the molecular level. Additionally, a sudden increase in the hydrophobicity of the membrane surface was observed when the salt concentration reaches around 0.6 M. This corresponds exactly to the earlier PVCL ligand turbidity results which show that at least 0.6 M Na2SO4 is needed in order to reduce the LCST from 37 °C to room temperature. By comparison, no significant contact angle change was observed for the glass chip and parafilm under the same salt conditions. This indicates that higher surface tension from higher salt concentration does not increase the hydrophobicity of the substrate surface. Enhanced binding between the protein and membrane substrate grafted with non-responsive hydrophobic ligand at higher salt concentrations is likely due to the stronger hydrophobic force resulting from the higher salt concentration.
4.2 Salt effects on protein binding studies
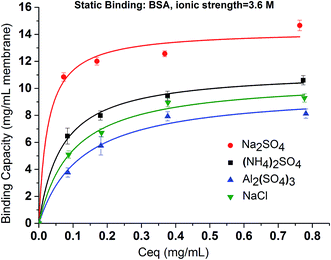
In order to elucidate the binding mechanism and binding energetics using PVCL ligands, BSA static binding isotherms were determined in 3.6 M ionic strength of (NH4)2SO4, Na2SO4, Al2(SO4)3 and NaCl solutions. Fig. 6 shows BSA isotherms fitted with Langmuir model based on equationwhere q and qmax represent the binding capacity and maximum binding capacity, respectively. K is the equilibrium constant and c is the protein concentration at equilibrium. The linear regression equations and parameters are shown in Table 1. The trend for static binding follows Na2SO4 > (NH4)2SO4 > NaCl > Al2(SO4)3 in term of capacity, binding constant K and binding free energy ΔG at the same ionic strength of 3.6 M. These binding results agree with the turbidity measurement of the solution phase LCST transition under different sulfate salt solutions. As discussed earlier, divalent sulfate salt solutions have a stronger effect on the transition temperature than the monovalent chloride salt solutions. This is due likely to the higher surface tension induced by sulfate ions than by the chloride ions at the same ionic strength as measured experimentally and shown in Fig. 4. The reason that the binding capacity is lower in Al2(SO4)3 solution at the same ionic strength as other salt solutions is due to the significantly reduced activity of the Al3+ ions resulting from its hydrolysis and subsequent polymerization.36,37
| Na2SO4 | (NH4)2SO4 | NaCl | Al2(SO4)3 | |
|---|---|---|---|---|
| Fitting equation | c/q = 0.064c + 0.002 | c/q = 0.080c + 0.005 | c/q = 0.095c + 0.007 | c/q = 0.105c + 0.011 |
| Fitting coefficient | R2 = 0.994 | R2 = 0.996 | R2 = 0.997 | R2 = 0.992 |
| qmax (mg mL−1) | 15.63 | 12.20 | 10.53 | 9.52 |
| Ionic strength (M) | 3.6 | 3.6 | 3.6 | 3.6 |
| Activity (M) | 0.23 | 0.22 | 2.80 | 0.006 |
| ΔG (kJ mol−1) | −45.5 | −44.9 | −44.5 | −44.3 |
| K (107 × M−1) | 9.45 | 7.56 | 6.36 | 5.76 |
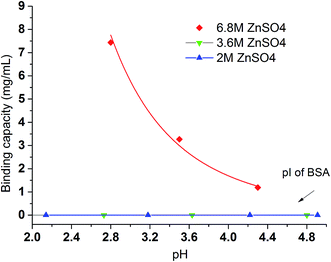
In contrast to Na2SO4 solution, there is no detectable binding between the PVCL ligand and BSA in 3.6 M ionic strength of ZnSO4 solution. This is probably due to the significantly reduced activity coefficient in the case of Zn2+ ion than the monovalent Na+ ion and that the solution at 3.6 M ionic strength does not reduce the LCST to room temperature as the binding experiments were performed. This can be seen from Fig. 3b that the LCST transition occurs only when the ionic strength of the ZnSO4 solution is larger than 5 M. As seen from Fig. 7, binding was observed when the ionic strength increases to 6.8 M. However, binding in 3.6 M ionic strength of Al2(SO4)3 salt solution has already been observed as shown in Fig. 6 despite the fact that the LCST transition occurs at much higher ionic strength in Al2(SO4)3 salt solution as shown in Fig. 3a. In order to reconcile the discrepancies observed in these two salt solutions, the nature of binding and the interaction of salt cations with BSA have to be taken into account. Besides being a thermo- and ionic strength responsive ligand, PVCL is a mild hydrophobic ligand that can bind proteins at higher salt conditions similar to conventional hydrophobic ligand. Thus under 3.6 M ionic strength Al2(SO4)3 solution when the PVCL ligand has not gone through the hydrophilic-to-hydrophobic transition yet, relative low binding interaction has already occurred. This accounts for the observed binding shown in Fig. 6. Similarly, in the case of ZnSO4, some binding at 3.6 M ionic strength should also be expected even though the ligand remains to be in a hydrophilic state. However, no binding at this condition was observed. This is probably due to the relative strong specific Zn2+–protein interaction which leads to a reduction of the hydrophobicity for the BSA protein. On the other hand, due to the strong hydrolysis of the Al3+ ion to form Al(OH)4−,37 no direct cation interaction with the protein is present leading to conventional hydrophobic binding between the protein and ligand. A strong pH dependence for BSA binding is observed when the ionic strength of ZnSO4 reaches 6.8 M as shown in Fig. 7. This is likely due to the fact that Zn2+ also has a tendency to form Zn(OH)42− in aqueous solution depending on the pH of the solution. As the pH decreases, the fraction of Zn2+ over Zn(OH)42− becomes higher. The strong Zn2+–protein interaction at high ionic strength of 6.8 M could also lead to the denaturation of the protein resulting in a higher BSA binding capacity observed. On the other hand, Zn(OH)42− ion dominated at higher pH probably does not form direct binding interaction with the protein and is less likely to denature the protein.
Dynamic binding experiments were performed using BSA and IgG4 as model proteins in a bind and elute mode. Binding conditions were varied with respect to the salt type for BSA and salt concentration for both BSA and IgG4 whereas the protein feed concentration and elution conditions were kept the same. Chromatograms of protein loading (100% breakthrough), membrane washing and membrane eluting steps were shown in Fig. 8. It can be seen that, for IgG4, the higher the salt concentration is, the longer the time it takes for a breakthrough to start. This indicates that more IgG4 are bound to the membranes at higher salt concentrations with corresponding higher binding capacities. However, a different salt concentration effect was observed for the breakthrough curve of BSA. Comparing the dynamic binding and breakthrough curves under the highest and lowest Na2SO4 concentrations, the longest time delay for the start of BSA breakthrough is only 0.5 min whereas the corresponding time delay is about 9 min for IgG4. This indicates that capacity for IgG binding is higher and that the effect of salt concentration on IgG4 binding is stronger than the corresponding BSA. It also took significantly longer time for IgG4 to reach 100% breakthrough than BSA due to the stronger binding interaction between the IgG4 and the substrate and the subsequent higher binding capacity. The delayed elution indicates that IgG4 is more hydrophobic than BSA resulting in a higher binding capacity for the IgG4 protein as shown in our earlier work.7,9 Our results appear to indicate that salt concentration affects BSA binding kinetics after breakthrough occurred whereas salt concentration affects IgG4 mainly on the time for the breakthrough to start. The change in slope in the BSA breakthrough curves suggests that probably different kinetics exist for BSA binding at different salt concentrations. Earlier results39 show that BSA adsorption onto the hydrophobic ligand at relatively low salt concentrations is a two-stage process involving adsorption and the subsequently spreading. It is known that BSA is relatively soft with adiabatic compressibility of 2.05 × 10−10 m2 N−1.40 The presence of salt ions and their concentrations will affect BSA conformations upon adsorption when the kinetics is relatively slow and binding interaction is relatively weak. IgG has a lower adiabatic compressibility around 6 × 10−11 m2 N−1, which means it is more rigid than BSA.41,42 In the case of stronger binding interactions, more rigid proteins and faster kinetics as shown in the IgG4 chromatogram, the two-step adsorption and spreading process is less apparent thus similar breakthrough slopes are observed.
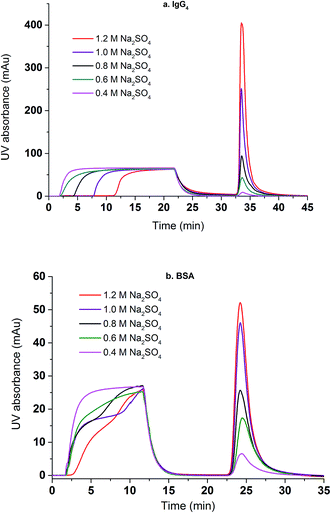 | ||
| Fig. 8 The effects of Na2SO4 concentration on the breakthrough and elution curves for IgG4 (a) and BSA (b). | ||
Fig. 9 compares the dynamic binding capacities in mg mL−1 and percentage recoveries for IgG4 and BSA when the binding ionic strength of Na2SO4 varies from 1.2 to 3.6 M. For both proteins, the binding capacity increases with the increase of the ionic strength but with different behaviors. Overall, the binding capacity for IgG4 is higher than for BSA indicating that IgG4 has a stronger hydrophobic interaction with PVCL as discussed earlier.7,9 In addition, the binding capacity for IgG4 improves more rapidly as the salt ionic strength increases. On the contrary, the increase in BSA binding capacity becomes less obvious when the ionic strength reaches beyond 3 M. These results are consistent with the observed breakthrough curves for these two proteins. As IgG is more hydrophobic, the stronger interaction between the protein and ligand leads to fast adsorption kinetics. As the salt concentration increases, the more hydrophobic proteins tend to have a stronger attractive hydrophobic force with the ligand leading to rapid increase in the binding capacities for these more hydrophobic proteins. On the other hand, the increase in the ionic strength of the Na2SO4 solution will lead to the unfolding and denaturation of the more flexible BSA protein. As was discussed earlier, Na+ ion has a stronger impact on protein denaturation than the NH4+ ion. The unfolded or denatured protein may prefer to aggregate rather than adsorb onto the membrane substrate depending on the magnitudes of the relative forces involved. As a result, the increase of the dynamic binding capacity slows down at higher ionic strength of the salt solution. If the capacity is measured by the number of proteins adsorbed instead by the mass, the capacities for the two proteins are likely to be more close to each other at 3.6 M salt condition as the MW of IgG4 is almost 3 times larger than that of BSA. Since the adsorption of the proteins follows Langmuir isotherm, maximum capacity is reached when there is a complete monolayer coverage. The reason that similar molar adsorption capacity is observed is probably due to the soft nature of the BSA protein and its tendency to spread upon adsorption.
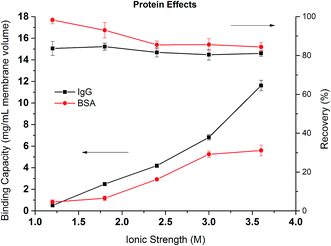 | ||
| Fig. 9 Dynamic binding capacities and recoveries of IgG4 and BSA as a function of the ionic strength of Na2SO4. | ||
Comparing the recoveries for the two proteins, it can be seen that the recovery of BSA is very high at lower ionic strength reaching over 90%. However, the recovery decreases to around 85% when the ionic strength increases to more than 2 M. It is known that hydrophobic force increases as the salt concentration increases leading to the stronger binding of the protein to the membrane substrate. The stronger hydrophobic interaction leads to larger deformation or spreading of the protein after adsorption resulting in irreversible binding and the reduction in recovery. The recovery of IgG4 remains more or less the same at around 80% but is always lower than the corresponding BSA. As mentioned earlier, the stronger hydrophobic force in the IgG interaction with PVCL ligand leads to irreversible binding and subsequent reduced recovery.
In order to investigate the effects of different salt and their concentrations on the dynamic binding capacity and recovery, BSA binding and recovery tests were conducted in NaCl, (NH4)2SO4, Na2SO4 salt solutions with ionic strength varying from 1 to 5.5 M. Due to the low solubility of K2SO4 in water, only two conditions were tested. As shown in Fig. 10, except in Na2SO4 solution at ionic strength above 2 M, BSA recovery remains high (>90%) in (NH4)2SO4 and NaCl solutions during the range of ionic strengths and for low ionic strength Na2SO4 solutions. This is consistent with our previous discussion that NaCl and (NH4)2SO4 are weaker denaturants which lead to more reversible binding even at high ionic strengths. As expected, the binding capacity is strongly salt condition dependent. The higher the ionic strength, the higher the binding capacity due to the stronger hydrophobic interactions induced. The divalent (NH4)2SO4 and Na2SO4 solutions have larger binding capacities than the corresponding NaCl solution. The binding capacity follows the order: Na2SO4 > (NH4)2SO4 > NaCl. The order is in agreement with our turbidity results for PVCL as well as static binding experiments. However, protein binding is more complex as both ligand and protein are affected by the ionic strength and the specific salt ions present. Nevertheless, PVCL responsive ligand for HIC application is clearly dominated by the ligand conformational transition at mild salt conditions.
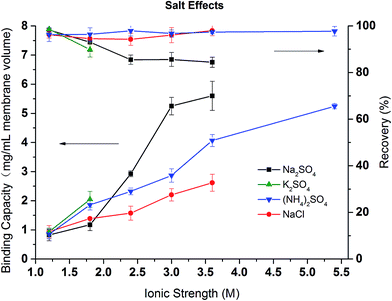 | ||
| Fig. 10 The effects of salt and ionic strength on the dynamic binding capacity and recovery of BSA on PVCL modified membrane substrate. | ||
5. Conclusions
The specificity of salt ions on the static and dynamic binding capacity and recovery was investigated for protein binding to the thermo- and salt responsive PVCL modified membrane substrates. Turbidity results show the effects of salt cations on the reduction of LCST for PVCL follows the order with Na+ > NH4+ > Zn2+ > Al3+ at the same ionic strength and with Al3+ > Zn2+ > Na+ > NH4+ at the same activity. The contact angle of PVCL grafted membrane surface increases with Na2SO4 concentration indicating the corresponding increase in the hydrophobicity. The increase in hydrophobicity correlates well with the hydrophilic-to-hydrophobic transition of the PVCL ligand and the binding capacity increase for BSA binding to the PVCL ligand. Static and dynamic binding studies of BSA and IgG show that the capacity follows the order Na+ > NH4+ > Al3+ > Zn2+ at the same ionic strength. The recovery is almost independent of the salt concentration for (NH4)2SO4 and NaCl. However, for Na2SO4, the recovery decreases when Na2SO4 concentration increases above 0.8 M due to the irreversible binding and aggregation of the protein. In addition, our results show that the binding capacity of IgG is higher than BSA due to the higher hydrophobicity of IgG protein.Acknowledgements
Partial financial support from Arkansas Bioscience Institute (ABI) is gratefully acknowledged.References
- S. M. Cramer and M. A. Holstein, Curr. Opin. Chem. Eng., 2011, 1, 27–37 CrossRef CAS.
- G. Guiochon and L. A. Beaver, J. Chromatogr. A, 2011, 1218, 8836–8858 CrossRef CAS PubMed.
- D. Nagrath, F. Xia and S. M. Cramer, J. Chromatogr. A, 2011, 1218, 1219–1226 CrossRef CAS PubMed.
- B. K. Nfor, N. N. Hylkema, K. R. Wiedhaup, P. D. E. M. Verhaert, L. A. M. van der Wielen and M. Ottens, J. Chromatogr. A, 2011, 1218, 8958–8973 CrossRef CAS PubMed.
- W. R. Melander, D. Corradini and C. Horváth, J. Chromatogr. A, 1984, 317, 67–85 CrossRef CAS PubMed.
- M. Record and C. F. Anderson, Biophys. J., 1995, 68, 786–794 CrossRef CAS PubMed.
- H. H. Himstedt, X. Qian, J. R. Weaver and S. R. Wickramasinghe, J. Membr. Sci., 2013, 447, 335–344 CrossRef CAS.
- Q. Wu, R. Wang, X. Chen and R. Ghosh, J. Membr. Sci., 2014, 471, 56–64 CrossRef CAS.
- A. Vu, X. Qian and S. R. Wickramasinghe, Sep. Sci. Technol., 2017, 52, 287–298 CrossRef CAS.
- Z. Liu, S. R. Wickramasinghe and X. Qian, RSC Adv., 2017, 7, 27823–27832 RSC.
- H. Du, R. Wickramasinghe and X. Qian, J. Phys. Chem. B, 2010, 114, 16594–16604 CrossRef CAS PubMed.
- H. Du and X. Qian, in Responsive Membranes and Materials, John Wiley & Sons, Ltd., 2012, pp. 229–242, DOI:10.1002/9781118389553.ch10.
- H. Du, S. R. Wickramasinghe and X. Qian, J. Phys. Chem. B, 2013, 117, 5090–5101 CrossRef CAS PubMed.
- H. Du and X. Qian, J. Polym. Sci., Part B: Polym. Phys., 2011, 49, 1112–1122 CrossRef CAS.
- F. Meeussen, E. Nies, H. Berghmans, S. Verbrugghe, E. Goethals and F. Du Prez, Polymer, 2000, 41, 8597–8602 CrossRef CAS.
- W. Melander and C. Horváth, Arch. Biochem. Biophys., 1977, 183, 200–215 CrossRef CAS PubMed.
- T. W. Perkins, D. S. Mak, T. W. Root and E. N. Lightfoot, J. Chromatogr. A, 1997, 766, 1–14 CrossRef CAS.
- E. Müller and A. Faude, J. Chromatogr. A, 2008, 1177, 215–225 CrossRef PubMed.
- P. Lo Nostro and B. W. Ninham, Chem. Rev., 2012, 112, 2286–2322 CrossRef CAS PubMed.
- F. Hofmeister, Archiv für experimentelle Pathologie und Pharmakologie, 1888, 24, 247–260 Search PubMed.
- N. Schwierz, D. Horinek and R. R. Netz, Langmuir, 2013, 29, 2602–2614 CrossRef CAS PubMed.
- N. Schwierz, D. Horinek and R. R. Netz, Langmuir, 2010, 26, 7370–7379 CrossRef CAS PubMed.
- H. Du, Z. Liu, R. Jennings and X. Qian, Sep. Sci. Technol., 2017, 52, 320–331 CrossRef CAS.
- L. Patra, A. Vidyasagar and R. Toomey, Soft Matter, 2011, 7, 6061–6067 RSC.
- L. M. Mikheeva, N. V. Grinberg, A. Y. Mashkevich, V. Y. Grinberg, L. T. M. Thanh, E. E. Makhaeva and A. R. Khokhlov, Macromolecules, 1997, 30, 2693–2699 CrossRef CAS.
- O. Zavgorodnya, V. Kozlovskaya, X. Liang, N. Kothalawala, S. A. Catledge, A. Dass and E. Kharlampieva, Mater. Res. Express, 2014, 1, 035039 CrossRef.
- I. M. Okhapkin, I. R. Nasimova, E. E. Makhaeva and A. R. Khokhlov, Macromolecules, 2003, 36, 8130–8138 CrossRef CAS.
- Z. Liu, S. R. Wickramasinghe and X. Qian, Sep. Sci. Technol., 2017, 52, 299–319 CrossRef CAS.
- Z. Liu, H. Du, S. R. Wickramasinghe and X. Qian, Langmuir, 2014, 30, 10651–10660 CrossRef CAS PubMed.
- H. H. Himstedt, Q. Yang, L. P. Dasi, X. Qian, S. R. Wickramasinghe and M. Ulbricht, Langmuir, 2011, 27, 5574–5581 CrossRef CAS PubMed.
- X. Qian, J. Lei and S. R. Wickramasinghe, RSC Adv., 2013, 3, 24280–24287 RSC.
- A. Laukkanen, L. Valtola, F. M. Winnik and H. Tenhu, Macromolecules, 2004, 37, 2268–2274 CrossRef CAS.
- F. Meeussen, E. Nies, H. Berghmans, S. Verbrugghe, E. Goethals and F. Du Prez, Polymer, 2000, 41, 8597–8602 CrossRef CAS.
- Y. Maeda, T. Nakamura and I. Ikeda, Macromolecules, 2001, 35, 217–222 CrossRef.
- Y. Zhang, S. Furyk, D. E. Bergbreiter and P. S. Cremer, J. Am. Chem. Soc., 2005, 127, 14505–14510 CrossRef CAS PubMed.
- R. J. Moolenaar, J. C. Evans and L. D. McKeever, J. Phys. Chem., 1970, 74, 3629–3636 CrossRef CAS.
- A. L. Petrou, Coord. Chem. Rev., 2002, 228, 153–162 CrossRef CAS.
- H. Fu, X. Hong, A. Wan, J. D. Batteas and D. E. Bergbreiter, ACS Appl. Mater. Interfaces, 2010, 2, 452–458 CAS.
- E. Haimer, A. Tscheliessnig, R. Hahn and A. Jungbauer, J. Chromatogr. A, 2007, 1139, 84–94 CrossRef CAS PubMed.
- M. J. W. Povey, J. D. Moore, J. Braybrook, H. Simons, R. Belchamber, M. Raganathan and V. Pinfield, Food Hydrocolloids, 2011, 25, 1233–1241 CrossRef CAS.
- B. Gavish, E. Gratton and C. J. Hardy, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 750–754 CrossRef CAS.
- S. V. Thakkar, S. B. Joshi, M. E. Jones, H. A. Sathish, S. M. Bishop, D. B. Volkin and C. R. Middaugh, J. Pharm. Sci., 2012, 101, 3062–3077 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |