Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biomimetically synthesized ZnO nanoparticles attain potent antibacterial activity against less susceptible S. aureus skin infection in experimental animals
Mohd. Ahmar Raufa,
Mohammad Owais*a,
Ravikant Rajpootb,
Faraz Ahmada,
Nazoora Khana and
Swaleha Zubair *c
*c
aInterdisciplinary Biotechnology Unit, Aligarh Muslim University, Aligarh, 202002, India. E-mail: owais_lakhnawi@yahoo.com; Fax: +91-571-2721776; Tel: +91-571-2720388
bICGEB, Aruna Asif Ali Marg, New Delhi, 110067, India
cDepartment of Computer Science, Aligarh Muslim University, Aligarh, 202002, India. E-mail: swalehazubair@yahoo.com
First published on 21st July 2017
Abstract
In the present study, we explored the antibacterial potential of ZnO–nanoparticles that were biomimetically synthesized employing conditioned culture supernatant of Staphylococcus aureus (S. aureus). The in situ synthesized ZnO–NPs were characterized employing various biophysical techniques including UV-vis spectroscopy, electron microscopy, DLS and XRD analysis etc. Electron microscopic studies revealed the disintegration of bacterial cell wall upon interaction with synthesized ZnO–NPs. Various employed tests provided evidence that synthesized ZnO–NPs effectively inhibited less susceptible Gram-positive and Gram-negative bacterial isolates. Moreover, the synthesized ZnO–NPs inhibited the biofilm synthesis of tested microbes in vitro. Finally, the biomimetically synthesized ZnO–NPs successfully treated the S. aureus mediated experimental skin infection in BALB/c mice as well.
Introduction
Synthesis of nanoparticles employing biological entities offers a great deal of interest. In general, various physical as well as chemical processes have been widely employed for synthesis of metal based nanoparticles. Of late, various biological entities have also been used for fabrication of nanoparticles. The biomimetically synthesized nanoparticles possess unusual optical,1 chemical,2 photoelectrochemical3 and electronic4 properties. Nature also relies on various biochemical processes for fabrication of metal particles with nano-sized dimensions. The biomimetic synthesis relies on various ongoing biochemical processes related to living beings. The biomimetic approach of synthesis would benefit from the development of clean, nontoxic and environmentally acceptable “green chemistry” procedures involving organisms ranging from bacteria to fungi and even plants.5–9 Both unicellular and multicellular organisms can induce synthesis of NPs in intra as well as extracellular niche.10–12 Synthesis of nanoparticles using environmentally benign materials offer various associated benefits that include least toxicity issues, eco-friendliness, and rapport for pharmaceutical and other biomedical applications. Earlier strategies used in the synthesis of metal nanoparticles involved several physical and chemical procedures.13 Chemical methods generally lead to the adsorption of toxic chemicals on the surface of formed NPs that may adversely affect their utility in medical applications.14 While chemical synthesis methods pose several potential hazards, including carcinogenicity, genotoxicity, and cytotoxicity etc., in contrast, biomimetic methods offer a more safer approach to NP synthesis.Zinc oxide (ZnO) based nanoparticles (ZnO–NPs) represent an important class of commercially viable product with unique magnetic, catalytic, ultraviolet-protective, semiconducting, antimicrobial and anticancer properties.15,16 ZnO–NPs have characteristic photocatalytic and photo-oxidizing ability as well.17 ZnO–NPs have wide-range applications in cosmetic products including sunburn lotions.18 ZnO is recognized as a ‘GRAS’ (generally recognized as safe) substance by the Food and Drug Administration (FDA 21CFR182.8991), USA. ZnO–NPs significantly inhibit the growth of a wide range of both Gram positive and Gram negative bacteria under normally visible lighting conditions.19–21 The antibacterial activity of zinc oxide nanoparticles can be attributed to their ability to generate increased levels of reactive oxygen species (ROS), mostly, singlet oxygen, hydroxyl radicals, and H2O2 that can efficiently kill microbial cells.22,23 The main advantages of using inorganic oxides NPs compared to organic chemical based antimicrobial agents lies in their mode of killing microbes, overall stability, tangibility, and long shelf life etc.
In the present study, we explored the biological synthesis of zinc oxide nanoparticles by co-incubation of zinc acetate ion with conditioned culture supernatant (bacteria free) of S. aureus. The in situ synthesized nanoparticles were characterized employing various spectroscopic techniques. We evaluated antibacterial/anti-biofilm effects of in situ synthesized ZnO–NPs against some common bacteria. Finally, we explored the antibacterial effect of ZnO–NPs against Staphylococcus aureus-induced skin infection in a mouse model.
Methods
Materials
Culture media Brain Heart Infusion (BHI), Luria Broth (LB) and Agar were acquired from Hi-media (India). Zinc acetate powder was purchased from Sisco Research laboratories Private Limited (India). The bacterial cultures of E. coli (ATCC 25922), P. aeruginosa (ATCC 27853) and S. aureus (ATCC 25923) were procured from Microbial Type Culture Collection (MTCC) Chandigarh, India and the clinical bacterial isolates, such as Klebsiella pneumonia, S. epidermidis, S. aureus (ATCC29213), methicillin-resistant S. aureus (MRSA1, MRSA2) and methicillin-resistant E. coli (MREC1 and MREC2) were obtained from the Department of Microbiology, Jawaharlal Nehru Medical College, AMU, Aligarh, India. The standard strains and isolates were sub-cultured in Luria Broth (LB), Mueller–Hinton (MH) and Brain Heart Infusion (BHI) broths. The cultures were stored at −20 °C in 20% glycerol, for long term preservation. All experiments were performed with the freshly grown cultures. All other chemicals and solvents used were of analytical grade and acquired locally.Biomimetic synthesis of ZnO–NPs
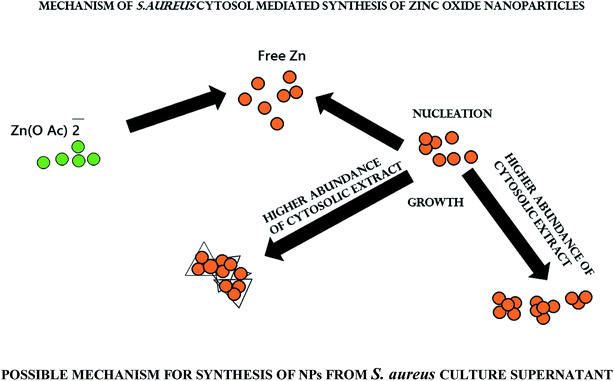
S. aureus (ATCC 29213) strain was freshly cultured on LB broth. The log-phase bacterial culture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes to remove the pellet. The culture supernatant (conditioned culture medium) was separated and subsequently used for the synthesis of ZnO–NPs.24,25 Briefly, the conditioned culture supernatant was mixed with a filter-sterilized zinc acetate solution (1 mM final concentration) to induce synthesis of zinc nanoparticles. Briefly, the culture supernatant was incubated with 1 mM Zn(O2CCH3)2 in an orbital shaker at 250 rpm and 37 °C to execute biomimetic synthesis of ZnO–NPs. The synthesis of nanoparticles was tracked by visual inspection for a change in the colour of the culture medium. After stipulated time period the mixture was centrifuged at 3500g for 10 minutes to remove any medium components, and nanoparticles were collected by centrifugation at 25
000g for 10 minutes to remove the pellet. The culture supernatant (conditioned culture medium) was separated and subsequently used for the synthesis of ZnO–NPs.24,25 Briefly, the conditioned culture supernatant was mixed with a filter-sterilized zinc acetate solution (1 mM final concentration) to induce synthesis of zinc nanoparticles. Briefly, the culture supernatant was incubated with 1 mM Zn(O2CCH3)2 in an orbital shaker at 250 rpm and 37 °C to execute biomimetic synthesis of ZnO–NPs. The synthesis of nanoparticles was tracked by visual inspection for a change in the colour of the culture medium. After stipulated time period the mixture was centrifuged at 3500g for 10 minutes to remove any medium components, and nanoparticles were collected by centrifugation at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 minutes. The as-synthesized NPs obtained were washed several times by centrifugation and dispersed in water to get rid of the residual (unused) zinc ions and traces of medium components. Finally, the obtained nanoparticles were lyophilized and stored at 0 °C till further experiments.
000g for 30 minutes. The as-synthesized NPs obtained were washed several times by centrifugation and dispersed in water to get rid of the residual (unused) zinc ions and traces of medium components. Finally, the obtained nanoparticles were lyophilized and stored at 0 °C till further experiments.
Characterization of ZnO–NPs employing UV-visible spectroscopy
Ultraviolet-visible (UV) spectra of zinc oxide NPs (ZnO–NPs) synthesized by S. aureus culture supernatant was recorded on a double beam spectrophotometer (Shimadzu) operated at a resolution of 1 nm.26Surface and size analysis of in situ ZnO–NPs employing transmission and scanning electron microscopy
Nanoparticles synthesized by using bacterial culture supernatant were characterized by TEM. Samples were prepared by placing a drop of reaction product over gold coated negative grid, allowing the solution to evaporate. TEM was performed on JEOL model electron microscope. The microscope was operated at an accelerating voltage of 1000 kV. For surface morphology as well as the size of ZnO–NPs scanning electron microscopy SEM was performed. (JSM67500F, JEOL model).X-ray diffraction analysis
XRD analysis of as-synthesized ZnO–NPs was performed in the 2θ range of 20–80° (Rigaku Miniflex II) with Cu Kα radiations (λ = 1.5406 Å) operating at a voltage of 30 kV and current of 15 mA. All the diffraction patterns were recorded as step-scans. To run a step-scan, lyophilized ZnO–NPs were mounted followed by fixing the tube voltage and current, and feeding the following parameters: starting 2-theta angle, step-size (typically 0.005 degrees), count time per step (typically 0.05–1 second) and ending 2-theta angle.Determination of particle size
DLS (Dynamic Light Scattering) measurement was employed to determine the average size and size dispersal of the synthesized ZnO–NPs using a Nanosizer 90ZS (Malvern Instruments, UK).The intensity of scattered light was sensed at 90° to the incident beam. The lyophilized powder was resuspended in aqueous buffer. Various size determining measurements were performed on the solution that was passed through 0.22 μm filter (Millipore). The data analysis was performed in default mode. The measured size was presented as the average value of 20 runs, with triplicate measurements within each run.Determination of Minimum Inhibitory Concentration (MIC)
Minimum Inhibitory Concentration (MIC) is the lowest concentration of an antimicrobial agent that may prevent the visible growth of microorganisms after stipulated time frame. MIC is considered as a significant parameter in diagnostic laboratories to ascertain sensitivity of a microorganism against a particular antimicrobial agent. The MIC value of in situ synthesized ZnO–NPs was determined by microdilution method against several ATCC strains and several clinical isolates viz. Staphylococcus aureus ATCC 25923, MRSA1, MRSA2, Enterococcus faecalis CCM 4224, Pseudomonas aeruginosa, Staphylococcus epidermidis (methicillin-susceptible), Klebsiella pneumonia and Escherichia coli ATCC 25922, MREC 1 and MREC 2. The values were estimated on the basis of viability test performed in 96-well microdilution plates according to the previously developed protocols.27Antibacterial potential of in situ ZnO–NPs as determined by agar diffusion assay
The bacterial culture was grown overnight in LB medium and centrifuged at 5000g for 5 minutes. The pellet obtained was washed with sterile PBS of 7.4 pH and again resuspended in LB medium. An aliquot (100 μl) of the suspended culture was spread evenly on LB plate with a sterile glass spreader and incubated at 37 °C. Subsequently the wells were bored using gel borer. After 1 h incubation period, the plate was exposed to increasing concentration of ZnO–NPs from 10 mg ml−1 stock solution. Zone of inhibition was determined by measuring the span of bacterial clearance after 24 h.28,29 The experimental procedure was performed under sterile conditions using level-2 bio-safety hoods. The experiments were performed in triplicate and the average of the three was calculated and compared with the reference drug vancomycin as a control.Bacterial susceptibility to antibiotics and ZnO NPs as determined by the disc-diffusion method
Following Clinical and Laboratory Standards Institute (CLSI, 2000) recommendations a disc-diffusion method was utilized to assess the antibacterial potential of ZnO–NPs as well as test antibiotics against various bacterial isolates. The methodology used is similar to agar disc diffusion protocol as described earlier with few modifications involving sterile discs that were placed on the plates using sterile forceps instead of boring of in the agar plate. Also, for determining synergistic effects, each standard antibiotic disc was impregnated with 10 μl of metallic NPs (100 ppm). Plates were labelled carefully and incubated at 37 °C for 24 hours to check the activity. Antibacterial activity was expressed as the diameter of the zone of inhibition, measured in millimetres. The assays were implemented in triplicate.30CFU assessment to evaluate bacterial susceptibility to in situ synthesized nanoparticles
The overnight grown culture of various bacterial isolates were sub-distributed into four culture tubes (adjusted density to 106–107 cells per ml). Further, 100 μl aliquot of ZnO–NPs solution from stock solution of 10 mg ml−1 or vancomycin solution (100 μg ml−1), negative control (without culture and without formulation) and positive control (culture + 100 μl PBS) was dispensed to corresponding tubes and allowed to incubate for further 4 hours at 37 °C. Thereafter, 100 μL suspension from each of the treated and control group tubes was plated in duplicate up to two different dilutions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) on to the TSB agar plates and incubated further at 37 °C. After 24 h of incubation at 37 °C, resultant colony forming units (CFU) at different dilutions were counted, averaged and expressed as log
10) on to the TSB agar plates and incubated further at 37 °C. After 24 h of incubation at 37 °C, resultant colony forming units (CFU) at different dilutions were counted, averaged and expressed as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 CFU ml−1 and the counts from two independent experiments were averaged.31
10 CFU ml−1 and the counts from two independent experiments were averaged.31
XTT biofilm assay
The XTT based biofilm assay was performed to assess antibiofilm activity of in situ synthesized NPs.32,33 Briefly, post mature biofilms formation, the wells of the plate were carefully rinsed with sterile PBS to remove non adherent cells. The mature biofilms were treated with increasing concentration of ZnO–NPs and placed at 37 °C for 48 h. After stipulated incubation period, 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) solution in PBS, was added at a final concentration of 250 mg ml−1. The obtained solution was filter sterilized using a 0.22 mm pore-size filter and stored at −80 °C until required. Menadione solution (0.4 mM) was prepared and filtered immediately just before the commencement of each assay. Adherent cells were washed with PBS (200 μl), followed by addition of XTT, and 2 μl of menadione to each well. The solution was transferred to a new plate after incubation in the dark for 4 h at 37 °C and the colorimetric change in the solution was assessed using a microtitre plate reader (BIORAD Microplate reader at 490 nm). Experiments were performed in triplicate. The data are expressed as means ± SD.Hydrophobicity index of nano-particle treated bacterial biofilm
The overnight grown bacterial cells were resuspended in LB medium and the optical density was adjusted to 1.0 ± 0.01 at a wavelength of 595 nm. 1 ml toluene was added to the cells suspension in a test tube and the tube was vortexed. The biphasic mixture of two phases was allowed to settle for 30 min and the optical density of the aqueous phase was measured. Hydrophobicity index (HI) of microbial cells was determined by using the following equation| HI% = [(Ai − Af)/Ai] × 100 |
Bacteria-nanoparticle interaction as revealed by electron microscopy
Both E. coli and S. aureus bacteria were exposed to ZnO–NPs for 60 minutes, at 37 °C, with constant agitation (at 250 rpm). The cell suspension was washed 5 times in modified-LB medium and 3 times in PBS to remove unbound or loosely associated nanoparticles. The cells (approximately 108 CFU) that interacted with ZnO–NPs at different time interval, were prepared and imaged using SEM. Additionally, the bacteria were imaged by Transmission Electron Microscopy (TEM) as well. Briefly, the bacteria, (also approximately 109 CFU) were fixed with 1% glutaraldehyde in PBS and subsequently exposed to 1% osmium tetroxide in water, for 24 hours each. The sample was transferred onto a 0.22 μm pore size filter (Millipore) and substituted with acetone, and subsequently with liquid CO2 in a critical point drying apparatus (SPI, USA). Filter paper sections were metalized with gold, by sputter coating, and imaged with a JEOL JSM-6390LV.35Confocal microscopy to determine incurred cell death in ZnO–NPs treated bacterial cells
Confocal laser scanning microscope was used to study effect of ZnO–NPs on growing bacterial cells. S. aureus and E. coli cultures were labelled with propidium iodide (PI) dye as well as syto-9 dye.36,37 PI was used to detect of dead cells as PI specifically stains only dead bacteria and gives red fluorescence whereas syto-9 stains only live cells and gives green fluorescence. The ZnO–NPs at their MIC concentrations were added to the overnight grown bacterial cultures and were incubated at 37 °C for 4 h. After the stipulated incubation period, cells were harvested by centrifugation and stained with dyes for 30 min under dark conditions. The cells were centrifuged at 2500g for 10 min and the pellet obtained was resuspended in 1 ml of sterile PBS buffer. Cells that were not treated with ZnO–NPs were taken as a control of both bacterial strains. Finally, 10 μl of the above prepared samples were then placed on a glass slide and mounted with the cover slip then cells were examined under a confocal scanning microscope.Intracellular ROS production by ZnO–NPs
Intracellular ROS generated in bacterial cells upon treatment with in situ synthesized ZnO–NPs were measured employing fluorescent probe 2,7-dichlorofluorescein diacetate (DCFH-DA).38 The DCFH-DA passively diffuses through the cell membrane and once internalized it gets de-acetylated by esterases to form a non-fluorescent 2,7-dichlorofluorescein (DCFH). The DCFH reacts with generated ROS to produce the fluorescent product 2,7-dichlorofluorescein (DCF),39 which gets trapped within the cell making it fluorescent. Briefly, the freshly cultured bacteria (106 CFU ml−1) were washed three times with fresh medium. DCFH-DA was mixed with the culture and incubated in shaking incubator for 30 min at 37 °C. The cells were pelleted down by centrifugation and washed to remove the unbound DCFH and then treated and control cells were visualized under a fluorescence microscope (Zeiss model, USA) for validation of ZnO–NPs induced ROS generation keeping into account the fact that the fluorescence intensity is proportional to the quantity of ROS produced.Potential of ZnO–NPs against skin infection
Six to eight week old male BALB/c mice were used in the current study. Animals (6 mice in each group) were anesthetized by intraperitoneal (IP) injection of a ketamine–xylazine cocktail31 and subsequently shaved on the dorsal surface. Mouse skin was rasped with sterile scalpel blades until reddened area appeared. The surface of each wound was inoculated with 50 μl of the culture suspension corresponding to 108 CFU of S. aureus. The infection was allowed to establish for 72 h. Following experimental groups were analyzed:(1) Group 1: healthy mice group with normal saline only.
(2) Group 2: mice infected with S. aureus with no further treatment.
(3) Group 3: mice exposed to S. aureus followed by treatment with ZnO–NPs (1 g per kg body weight).
(4) Group 4: mice exposed to S. aureus followed by treatment with ZnO–NPs (0.5 g per kg body weight).
Prior to application of the nanoparticles solution, each mouse was fitted with an Elizabethan collar (Braintree Scientific, Braintree, MA) to prevent removal and ingestion of applied formulation, following the protocol of luke Mortenson et al. 2014. Ten days post infection, mice were anesthetized and sacrificed. Infected skin samples were homogenized using a tissue homogenizer (Silverson Machines, East Longmeadow, MA, USA) and the residual S. aureus burden was determined by plating serially diluted homogenized samples on LB plates. For histopathological studies, skin tissues were sliced from mice belonging to various groups and fixed in 10% formaldehyde solution, dehydrated in ascending grades of ethyl alcohol, cleared in xylol and mounted in molten paraplast at 58–62 °C. The finely cut thin sections were then stained with hematoxylin and eosin stain and evaluated for any morphological changes under an Olympus BX40 microscope (PA, USA) for the comparative study.
Cytotoxicity assays
The percentage hemolysis was determined by the following equation:40
| % Haemolysis = [{AbsT − AbsC}/{Abs(100%) − AbsC}] × 100 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well were seeded in 96 well plate and allowed to adhere overnight. Next morning, the medium was aspirated and cells were incubated at 37 °C for 24 h with different concentration and for 4 h with MTT dye (5 mg per ml in PBS). The reaction mixture was aspirated and the resulting formazan crystals were dissolved by adding 200 μl of DMSO. After 10 min, absorbance was read at 490 nm in a Genetix580 microplate reader (USA).41 Un-treated sets served as control which was performed simultaneously under identical conditions.
000 cells per well were seeded in 96 well plate and allowed to adhere overnight. Next morning, the medium was aspirated and cells were incubated at 37 °C for 24 h with different concentration and for 4 h with MTT dye (5 mg per ml in PBS). The reaction mixture was aspirated and the resulting formazan crystals were dissolved by adding 200 μl of DMSO. After 10 min, absorbance was read at 490 nm in a Genetix580 microplate reader (USA).41 Un-treated sets served as control which was performed simultaneously under identical conditions.Results and discussion
S. aureus conditioned media facilitates ZnO–NPs synthesis
Various reducing agents produced during a biochemical reaction may accentuate synthesis of metal based nano-particles. Biomolecules such as proteins, carbohydrates, and enzymes play a crucial role in bio-reduction processes.42,43 It has been observed that NADH and NADH-dependent nitrate reductase enzymes play an important role in the biosynthesis of metal nanoparticles.44,45 Keeping this fact into consideration we explored the potential of conditioned S. aureus culture medium to facilitate ZnO–NPs synthesis.Characterization of biomimetically synthesized ZnO–NPs
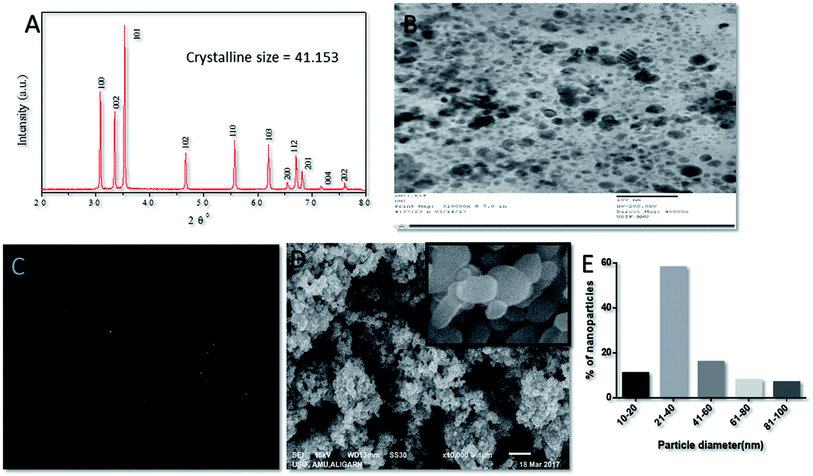
The crystal structure of in situ-synthesized ZnO–NPs was characterized by XRD analysis with Cu Kα radiation (λ = 0.15418 nm). The obtained data suggested the presence of well resolved 11 XRD peaks that were prominent at 2θ viz. 31.01°, 34.21°, 35.64°,47.10°, 56.02°, 62.15°, 65.68°, 67.51°, 69.01°, 72.08° and 76.24° which correspond to the crystal planes [100], [002], [101], [102], [110], [103], [200], [112], [201], [004] and [202] of crystalline wurtzite structure (Zincite, JCPDS5-0664) respectively (Fig. 2A). The XRD pattern of ZnO–NPs attested high quality of in situ synthesized NPs with no impurity. The average crystallite size (D) of ZnO–NPs was calculated following the Debye–Scherrer formulaD = 0.9λ/B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, θ, |
The nanostructure of synthesized ZnO–NPs was ascertained by electron microscopy as well as DLS. The scanning electron micrograph (SEM) indicates the formation of secondary ZnO–NPs. The TEM image clearly depicted that irregular shape of in situ synthesized ZnO NPs (Fig. 2B and C) that are grouped in assemblage with a size range of 10–50 nm. The size range determined by electron microscopic studies is in accordance with the data obtained in XRD and DLS results.46
SEM analysis of ZnO–NPs suggested that all NPs were stable, well dispersed and smooth. The low level of agglomeration if there was any could be because of the preparation methodology that involves deposition of particles on a copper grid followed by drying, promotes agglomeration of ZnO–NPs. TEM image offers direct observation of in situ synthesized NPs and suggest that the size of the ZnO nanoparticles was in the order of 10–50 nm and the particles appeared to be acicular in shape. This is in accordance with results obtained by the XRD.
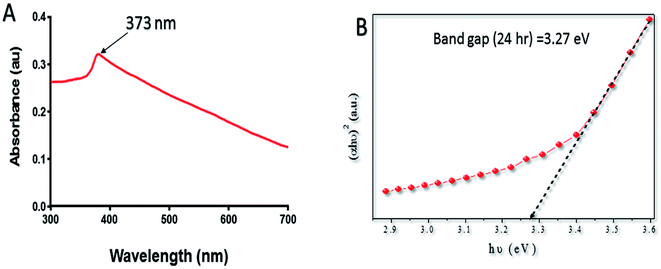
Band gap (Eg) is used to study the electronic structure of ZnO–NPs. It is the energy gap between the valence band (Ev) and the conduction band (Ec), proposes high density of states.47 The high value of the Eg is possibly because of the quantum captivity effect of the NPs. The broadening effect might be associated with the influence of various factors such as physical parameters (size and pH), carrier concentrations and the presence of oxygen vacancies, which may result in the Burstein–Moss shift.48,49 The antibacterial activity of ZnO–NPs has been linked to their ability to induce ROS species. The generation of a specific type of ROS like ˙OH, peroxide, or O˙2− by the metallic oxide NPs is associated with the electronic structures as well as the redox potentials (EH) of different ROS generation reactions. The oxidative stress offered by ZnO–NPs is considered to be the main mechanism responsible for antimicrobial activity.50
The lyophilized powder of ZnO–NPs (10 μg ml−1) was dispersed in acetate buffer by ultra-sonication and was used for performing the UV-vis measurement (Fig. 1A). The spectrum shows a characteristic absorption peak of ZnO–NPs at wavelength around 373 nm which can be attributed to the intrinsic band-gap absorption of ZnO–NPs because of the electron transitions from the valence band to the conduction band (O2–Zn3d) (Fig. 1B).51 The absorption spectrum of ZnO–NPs also suggested the narrow nano size particle distribution.
Anti-bacterial activity of in situ synthesized ZnO–NPs
The antimicrobial action of in situ synthesized ZnO–NPs was evaluated against various clinical isolates. Standard antibiotic vancomycin was taken as a control. The MIC value for the synthesized nanoparticles varied from microorganism to microorganism as shown in Table 1.| Strains | Zinc oxide NPs |
|---|---|
| a MIC observed by 96-well plate microdilution method. | |
| S. aureus ATCC 25923 | 32 |
| MRSA 1 | 256 |
| MRSA 2 | 128 |
| Staphylococcus epidermidis | 128 |
| Listeria monocytogenes | 64 |
| Klebsiella pneumonia | 256 |
| Enterococcus faecalis | 256 |
| Escherichia coli ATCC 25922 | 32 |
| MREC 1 | 64 |
| MREC 2 | 128 |
| P. aeruginosa | 32 |
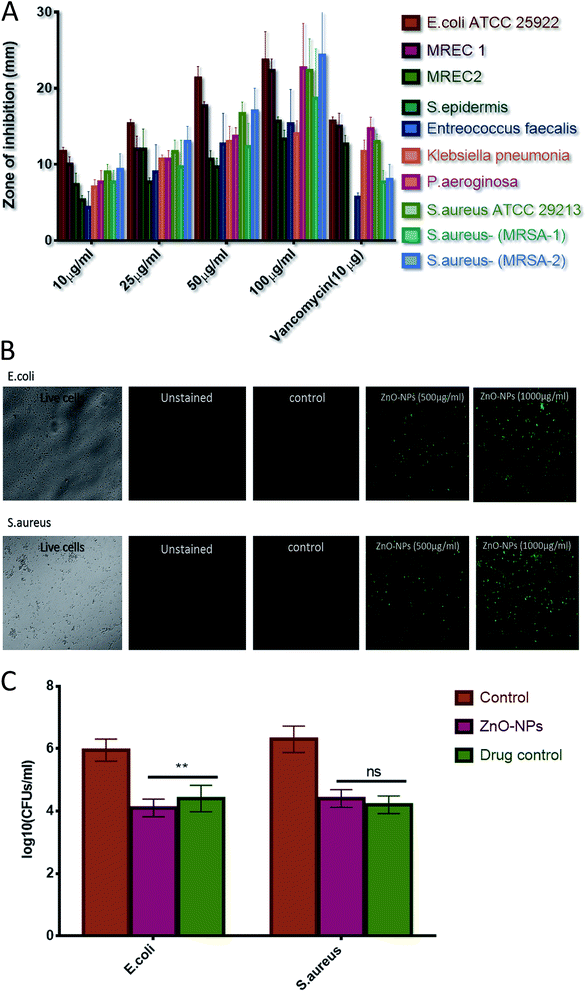
Agar diffusion assay was performed to assess the bacterial inhibition by ZnO–NPs (Fig. 3A). The zone of inhibition assay further confirms that nanoparticles possess effective bactericidal activity against the tested bacteria. The bactericidal effect of nanoparticles has been correlated to the decomposition of bacterial outer membranes by reactive oxygen species (ROS), primarily hydroxyl radicals (OH), which leads to phospholipid peroxidation and ultimately killing of bacteria.52,53 It was proposed that nano species that can physically adhere to a cell can kill the bacteria if they come in contact with the cell.54 The present study is in confirmation with earlier reports and suggest that synthesized ZnO–NPs had bactericidal effect against E. coli, P. aeroginosa, and S. aureus but showed little activity against Enterococcus faecalis. The ZnO–NPs were also successful in killing MRSAs. The internalization of zinc oxide nanoparticles in bacteria induces the production of ROS and this can affect DNA as well as total cellular machinery of bacteria.55 ROS production upon treatment of ZnO–NPs has been assessed using the fluorescence dye, DCFHDA (Fig. 3B). ZnO–NPs mediated generation of reactive oxygen species (ROS) causes alteration and decrementation of cellular proteins, DNA, and lipids etc., which lead to cell death. The generated reactive oxygen species, (such as hydrogen peroxide and superoxide anion) oxidizes DCFH-DA that leads to a formation of a highly fluorescent derivative. As shown in Fig. 3B, a significant increase in DCF fluorescence was observed after the treatment of ZnO–NPs in cells.
The combination of in situ synthesized ZnO–NPs with different antibiotics was also investigated for their antimicrobial potentials against Gram-positive (S. aureus) and Gram-negative bacteria (E. coli) using the disc diffusion method. The diameter of the zone of inhibition (mm) around the different antibiotic disks with and without ZnO–NPs against test strains was evaluated. Intriguingly, the combination of ZnO–NPs with the antibiotics erythromycin, ampicillin, and vancomycin resulted in enhanced efficacy against both Gram-positive and Gram-negative strains; however, they exhibited distinct differences in their susceptibility to anti-microbial effect. Amongst various antibiotics evaluated, the highest additive effects was observed with vancomycin followed by ampicillin and erythromycin. The differences in fold increase among these antibiotics may be due to differences in their mechanism of action (Table 2).
| Antibiotic (μg/disk) | Antibiotic only | Antibiotic + ZnO–NPs | Fold increase (%) |
|---|---|---|---|
| S. aureus | |||
| Erythromycin (10) | 14 | 18 | 28 |
| Ampicillin (10) | 12 | 16 | 33 |
| Vancomycin (10) | 9 | 13 | 44 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| E. coli | |||
| Erythromycin (10) | 16 | 19 | 18 |
| Ampicillin (10) | 13 | 17 | 30 |
| Vancomycin (10) | 11 | 15 | 38 |
Anti-bacterial susceptibility assay
A known volume of overnight grown culture of E. coli and S. aurues were distributed into four different culture tubes (adjusted density to 106–107 cells per ml). Further, a fixed volume of ZnO–NPs solution (100 μl from stock solution of 10 mg ml−1), vancomycin solution (100 μg ml−1), negative control (no bacterial culture and without formulation) and positive control (bacterial culture + 100 μl PBS) were added to each of corresponding tubes and allowed to react for further 4 hours at 37 °C. The results of CFU plating and counting assay for both the S. aureus (Gram +ve) and E. coli (Gram −ve) are shown in both graphical as well tabular forms (Table 3) (Fig. 3C). There was significant decrease (p value < 0.01 (**)) in bacterial load of E. coli in the group treated with nanoparticles when compared with vancomycin-treated group, however no significant difference (p value > 0.05 (ns)) in bacterial activity among ZnO nanoparticles and vancomycin treated group against S. aureus. In addition, it is worth mentioning that significant reduction in CFUs was evident in the case of both ZnO–NPs as well as vancomycin when compared to positive control group.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU mL−1 in tabular form
CFU mL−1 in tabular form
| Pathogens | Groups | log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU mL−1 CFU mL−1 |
|---|---|---|
| E. coli | Positive control | 6.1735 |
| ZnO–NPs | 4.0225 | |
| Drug control | 4.3145 | |
| S. aureus | Positive control | 6.2565 |
| ZnO–NPs | 4.2215 | |
| Drug control | 4.1715 |
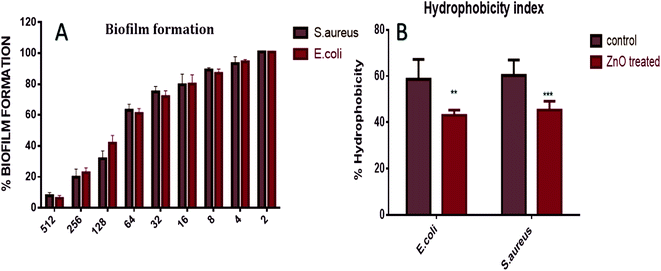
Antibiofilm & hydrophobicity index of as-synthesized ZnO–NPs
The XTT assay employed to determine anti-biofilm potential of in situ synthesized ZnO–NPs exhibited the dose dependent anti-biofilm effect. Further, the high hydrophobicity of the bacterial cell surface results in their adherence to various surfaces like mucosal epithelial cells, phagocytes etc. On treatment with ZnO–NPs, the percentage of hydrophobicity index was decreased to 52% in E. coli and 41% in S. aureus (Fig. 5). The electron microscope studies depicted well-developed biofilm formation by E. coli and S. aureus, on the infected surface while on the other hand, inhibition of biofilm formation after treatment with ZnO–NPs (Fig. 4A and B).Effect of ZnO–NPs treatment on bacterial cell as revealed by electron microscopic analysis
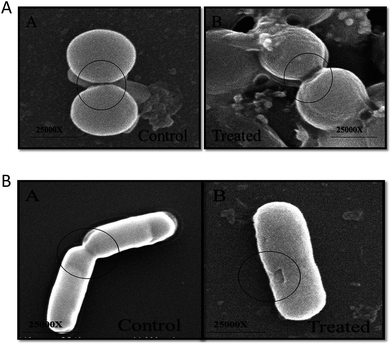
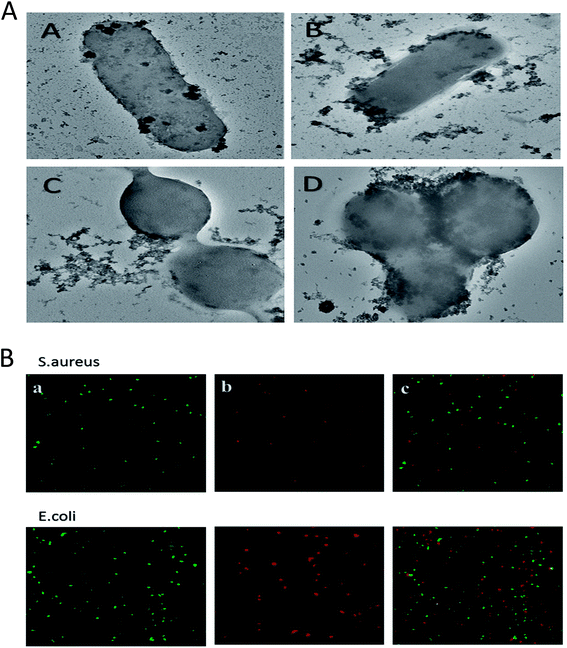
The morphological changes incurred in bacterial cells upon incubation with ZnO–NPs were deduced employing SEM. The control E. coli showed typical rod-shaped morphology. The cell was smooth with no sign of damage on the cell surface (Fig. 5B). However, in ZnO–NPs treated group, instead of normal rod-shaped cells, the bacteria with irregular fragments, appeared and the bacterial cell had swollen into a bigger size, with high agglomeration of the cells. It can be clearly seen that treatment of the bacteria with ZnO–NPs led to considerable damage to E. coli cells that eventually led to the disruption of the cell wall of bacteria. Similarly, the normal S. aureus had round, smooth morphology and the cells are almost similar in shape. Post exposure to ZnO–NPs, the cells showed abrupt morphology with damaged cell surface (Fig. 5A).ZnO–NPs adhered to the bacterial cell wall surface that eventually lead to release of their contents. The intact bacterial cell walls had enlarged soft appearance and adhered with noticeable amounts of ZnO–NPs (Fig. 6A).
Confocal scanning laser microscopy (CLSM)
CLSM was used to establish anti-microbial potential of ZnO–NPs against S. aureus and E. coli strain. PI penetrates only cells with severe membrane lesions and acquire red fluorescence whereas healthy cells with intact cell wall acquire only green fluorescence (Fig. 6B). It should be noticed that PI can only stain the cells in which the cell membrane is disrupted since it intercalates into the double-stranded nucleic acids.56 Results so obtained demonstrates that PI penetrates the bacterial and resulted in red fluorescence. The present study showed that the ZnO–NPs can significantly kill the bacterial cells.Treatment with ZnO–NPs minimizes S. aureus induced haemolytic activity
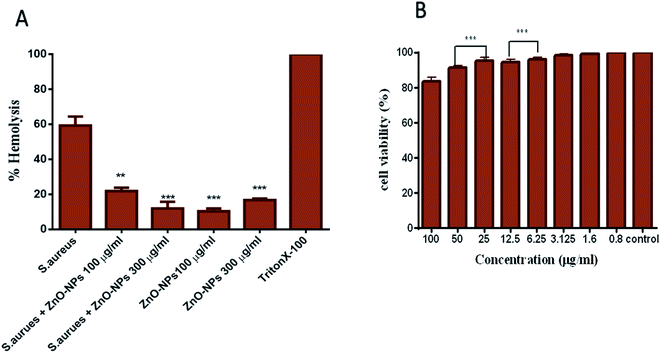
ZnO–NPs reduce hemolysis caused by Staphylococcal α-hemolysin, a major virulence factor of S. aureus which is responsible for the lysis of host cells by creating pores in phospholipid bilayers or by hydrolysing phospholipids in the bilayer of the cell. The pre-incubation of biologically synthesized ZnO–NPs inhibited the lysis caused by S. aureus. As shown in Fig. 7A, about 63% cell lysis resulted in post incubation with S. aureus, whereas in the presence of 100 and 300 μg ml−1 ZnO–NPs less than 18% and 13% (P ≤ 0.001) cell lysis was observed. Interestingly, ZnO–NPs alone caused 8% and 23% cell lysis at 100 μg ml−1 and 300 μg ml−1 concentrations, respectively.Synthesized ZnO–NPs exhibit minimal toxicity to PBMCs
The MTT assay results show that biomimetically synthesized ZnO–NPs did not show cytotoxic effects on PBMCs even at the dose quite higher to their MIC value (∼100 μg ml−1). However, exposure of cells to increasing doses of ZnO–NPs resulted in decreased cell viability. The results suggest the appropriateness of ZnO–NPs as a stable and nontoxic entity that has potent antibacterial activity and does not impart toxicity to the host cells (Fig. 7C).Antibacterial potential of ZnO–NPs against S. aureus skin infection
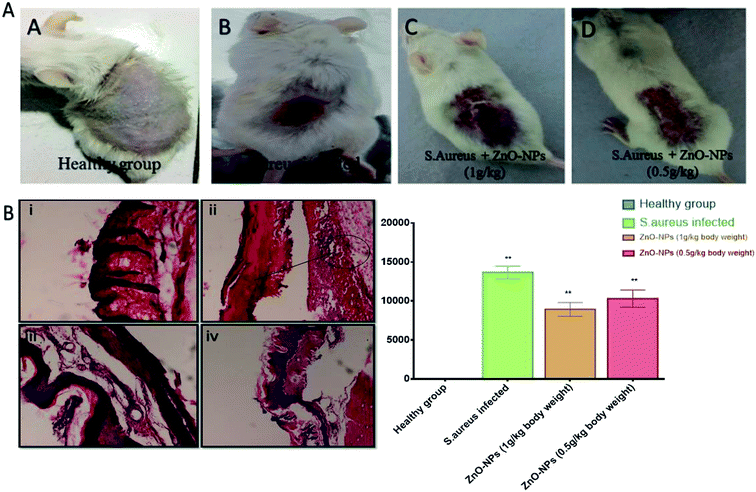
Once the efficacy of ZnO–NPs against S. aureus was established in vitro, we moved on to evaluate the efficacy of ZnO–NPs in treatment of skin infections in mice models. The cutaneous bacterial infections are characterized by reddening of the skin followed by disruption Fig. 8A. The bacterial load in the skin was assessed by allowing the enumeration of bacteria present in the given specimen by culturing in solid agar medium as shown in Fig. 8B ZnO–NPs exhibited almost 40% reduction in bacterial burden when compared to untreated control (P < 0.005) indicating the capability of in situ synthesized ZnO–NPs in treating skin bacterial infection.Histopathological analysis
We examined the skin architecture, bacterial burden and the incurred inflammatory changes in infected mice upon their treatment with ZnO–NPs. Lesions on the skin were deduced on day 10 post-infection employing hematoxylin and eosin (HE) staining. The control healthy group shows the histology of skin of a healthy animal with a normal intact epidermal layer, whereas S. aureus infected skin showed thinning of the epidermal layer with disruption along with the presence of a large number of inflammatory cells. However in the case of ZnO–NPs treated an animal, healing of epidermal layer with less damage was observed and the skin had a tendency to acquire normal architecture.Conclusion
In the present study, we demonstrated the potential of S. aureus conditioned culture medium as an agent to induce synthesis of ZnO–NPs. The employed biomimetic method of metal oxide nano-particles synthesis is simple, rapid, and eco-friendly. The structural and compositional characterization of the in situ synthesized ZnO–NPs was elucidated employing UV-vis spectroscopy, electron microscopy, and XRD analysis.The in situ synthesized ZnO nanoparticles possess antibacterial properties against range of Gram positive and negative bacteria. The synthesized ZnO–NPs were also successful in inhibiting MRSA and MRSE that are found to be resistant to a wide range of broad-spectrum antibiotics. The antimicrobial mechanisms of particles may differ from species to species of bacteria and size of the nanoparticles. The strongest indication of the susceptibility of MRSA and MRSE to ZnO–NPs may be attributed to their cell wall plasmolysis or the separation of cytoplasm from their cell wall. The damaging effect of ZnO–NPs on bacterial surface was visualized by the electron microscopic studies. In addition to that, the in situ synthesized NPs have the potential to inhibit the S. aureus-mediated lysis of RBCs. Conclusively, our data suggests that the synthesized nanoparticles have significant antibacterial properties against various pathogens with negligible toxicity and biocompatible in nature as shown in in vitro studies (RBC lysis and MTT assay results). Interestingly, in situ synthesized ZnO–NPs demonstrated strong potential to inhibit bacterial infection under in vivo conditions and eliminate S. aureus mediated skin infection in BALB/c mice.
Disclosure
The authors report no conflict of interest in this work.Acknowledgements
We are thankful to Co-ordinator, IBU, for providing required facilities to carry on this study. MAR is thankful to UGC, Govt of India, for financial assistance in the form of fellowship. The financial support from DST, Govt. of India (Ref. No. IDP/MED/2013/2014 [General]) is also duly acknowledged.References
- N. Aziz, M. Faraz, R. Pandey, M. Shakir, T. Fatma, A. Varma, I. Barman and R. Prasad, Langmuir, 2015, 31, 11605–11612 CrossRef CAS PubMed.
- L. Berti and G. A. Burley, Nat. Nanotechnol., 2008, 3, 81–87 CrossRef CAS PubMed.
- P. O. Saboe, E. Conte, M. Farell, G. C. Bazan and M. Kumar, Energy Environ. Sci., 2017, 10, 14–42 CAS.
- Z. Li, Y. Ding, S. Li, Y. Jiang, Z. Liu and J. Ge, Nanoscale, 2016, 8, 17440–17445 RSC.
- S. S. Shankar, A. Rai, A. Ahmad and M. Sastry, J. Colloid Interface Sci., 2004, 275, 496–502 CrossRef CAS PubMed.
- K. B. Narayanan and N. Sakthivel, Adv. Colloid Interface Sci., 2010, 156, 1–13 CrossRef CAS PubMed.
- K. N. Thakkar, S. S. Mhatre and R. Y. Parikh, Nanomedicine: Nanotechnology, Biology and Medicine, 2010, 6, 257–262 CrossRef CAS PubMed.
- S. K. Dorcheh and K. Vahabi, Fungal Metab., 2017, pp. 395–414 Search PubMed.
- J. Y. Song and B. S. Kim, Bioprocess Biosyst. Eng., 2009, 32, 79 CrossRef CAS PubMed.
- C. G. Kumar and S. K. Mamidyala, Colloids Surf., B, 2011, 84, 462–466 CrossRef CAS PubMed.
- M. Saravanan and A. Nanda, Colloids Surf., B, 2010, 77, 214–218 CrossRef CAS PubMed.
- S. Senapati, A. Syed, S. Moeez, A. Kumar and A. Ahmad, Mater. Lett., 2012, 79, 116–118 CrossRef CAS.
- S. Iravani, H. Korbekandi, S. V. Mirmohammadi and B. Zolfaghari, Results Pharma Sci., 2014, 9, 385–406 CAS.
- W. H. De Jong and P. J. A. Borm, Int. J. Nanomed., 2008, 3, 133–149 CrossRef CAS.
- A. Sirelkhatim, S. Mahmud, A. Seeni, N. H. M. Kaus, L. C. Ann, S. K. M. Bakhori, H. Hasan and D. Mohamad, Nano-Micro Lett., 2015, 7, 219–242 CrossRef CAS.
- M. J. Akhtar, M. Ahamed, S. Kumar, M. A. M. Khan, J. Ahmad and S. A. Alrokayan, Int. J. Nanomed., 2012, 7, 845–857 CAS.
- J. Bogdan, J. Zarzyńska and J. Pławińska-Czarnak, Nanoscale Res. Lett., 2015, 10, 309 CrossRef PubMed.
- T. G. Smijs and S. Pavel, Nanotechnol., Sci. Appl., 2011, 4, 95–112 CrossRef CAS PubMed.
- Y. Xie, Y. He, P. L. Irwin, T. Jin and X. Shi, Appl. Environ. Microbiol., 2011, 77, 2325–2331 CrossRef CAS PubMed.
- A. Sirelkhatim, S. Mahmud, A. Seeni, N. H. M. Kaus, L. C. Ann, S. K. M. Bakhori, H. Hasan and D. Mohamad, Nano-Micro Lett., 2015, 7, 219–242 CrossRef CAS.
- N. Jones, B. Ray, K. T. Ranjit and A. C. Manna, FEMS Microbiol. Lett., 2008, 279, 71–76 CrossRef CAS PubMed.
- G. Applerot, A. Lipovsky, R. Dror, N. Perkas, Y. Nitzan, R. Lubart and A. Gedanken, Adv. Funct. Mater., 2009, 19, 842–852 CrossRef CAS.
- K. R. Raghupathi, R. T. Koodali and A. C. Manna, Langmuir, 2011, 27(7), 4020–4028, DOI:10.1021/la104825u.
- A. R. Shahverdi, A. Fakhimi, H. R. Shahverdi and S. Minaian, Nanomedicine: Nanotechnology, Biology and Medicine, 2007, 3, 168–171 CrossRef CAS PubMed.
- C. Jayaseelan, A. A. Rahuman, A. V. Kirthi, S. Marimuthu, T. Santhoshkumar, A. Bagavan, K. Gaurav, L. Karthik and K. V. B. Rao, Spectrochim. Acta, Part A, 2012, 90, 78–84 CrossRef CAS PubMed.
- S. Gunalan, R. Sivaraj and V. Rajendran, Prog. Nat. Sci.: Mater. Int., 2012, 22, 693–700 CrossRef.
- I. Wiegand, K. Hilpert and R. E. W. Hancock, Nat. Protoc., 2008, 3, 163–175 CrossRef CAS PubMed.
- M. Oves, Int. J. Nonferrous Metall., 2012, 7, 6003–6009, DOI:10.2147/IJN.S35347.
- J. S. Kim, E. Kuk, K. N. Yu, J.-H. Kim, S. J. Park, H. J. Lee, S. H. Kim, Y. K. Park, Y. H. Park and C.-Y. Hwang, Nanomedicine: Nanotechnology, Biology and Medicine, 2007, 3, 95–101 CrossRef CAS PubMed.
- J. Hudzicki, Kirby-Bauer Disk Diffusion Susceptibility Test protocol, 2009 Search PubMed.
- M. Guzman, J. Dille and S. Godet, Nanomedicine: Nanotechnology, Biology and Medicine, 2012, 8, 37–45 CrossRef CAS PubMed.
- C. G. Pierce, P. Uppuluri, A. R. Tristan, F. L. Wormley, E. Mowat, G. Ramage and J. L. Lopez-Ribot, Nat. Protoc., 2008, 3, 1494–1500 CrossRef CAS PubMed.
- E. Peeters, H. J. Nelis and T. Coenye, J. Microbiol. Methods, 2008, 72, 157–165 CrossRef CAS PubMed.
- E. V. Serebriakova, I. V. Darmov, N. P. Medvedev, S. A. Alekseev and S. I. Rybak, Mikrobiologiia, 2002, 71, 237 CAS.
- M. Hartmann, M. Berditsch, J. Hawecker, M. F. Ardakani, D. Gerthsen and A. S. Ulrich, Antimicrob. Agents Chemother., 2010, 54, 3132–3142 CrossRef CAS PubMed.
- R. Ahmad, M. Mohsin, T. Ahmad and M. Sardar, J. Hazard. Mater., 2015, 283, 171–177 CrossRef CAS PubMed.
- S. Kulshrestha, S. Khan, R. Meena, B. R. Singh and A. U. Khan, Biofouling, 2014, 30, 1281–1294 CrossRef CAS PubMed.
- S. Dwivedi, R. Wahab, F. Khan, Y. K. Mishra, J. Musarrat and A. A. Al-Khedhairy, PLoS One, 2014, 9, e111289 Search PubMed.
- K. Ali, S. Dwivedi, A. Azam, Q. Saquib, M. S. Al-Said, A. A. Alkhedhairy and J. Musarrat, J. Colloid Interface Sci., 2016, 472, 145–156 CrossRef CAS PubMed.
- M. Oves, M. S. Khan, A. Zaidi, A. S. Ahmed, F. Ahmed, E. Ahmad, A. Sherwani, M. Owais and A. Azam, PLoS One, 2013, 8, e59140 CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- K. M. Moghaddam, An Introduction to Microbial Metal Nanoparticle Preparation Method, 2010, pp. 1–7 Search PubMed.
- A. Chauhan, S. Zubair, S. Tufail, A. Sherwani, M. Sajid, S. C. Raman, A. Azam and M. Owais, Int. J. Nanomed., 2011, 6, 2305–2319 CAS.
- M. Rai, A. P. Ingle, I. R. Gupta, S. S. Birla, A. P. Yadav and K. Abd-Elsalam, Curr. Nanosci., 2013, 9, 576–587 CrossRef CAS.
- M. Shah, D. Fawcett, S. Sharma, S. K. Tripathy and G. E. J. Poinern, Materials, 2015, 8, 7278–7308 CrossRef.
- P. Gajjar, B. Pettee, D. W. Britt, W. Huang, W. P. Johnson and A. J. Anderson, J. Biol. Eng., 2009, 3, 9 CrossRef PubMed.
- M. Shoeb, B. R. Singh, J. A. Khan, W. Khan, B. N. Singh, H. B. Singh and A. H. Naqvi, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2013, 4, 35015 CrossRef.
- J. Wu, W. Walukiewicz, S. X. Li, R. Armitage, J. C. Ho, E. R. Weber, E. E. Haller, H. Lu, W. J. Schaff and A. Barcz, Appl. Phys. Lett., 2004, 84, 2805–2807 CrossRef CAS.
- A. Sarkar, S. Ghosh, S. Chaudhuri and A. K. Pal, Thin Solid Films, 1991, 204, 255–264 CrossRef CAS.
- Y. Li, W. Zhang, J. Niu and Y. Chen, ACS Nano, 2012, 6, 5164–5173 CrossRef CAS PubMed.
- Z. R. Khan, M. S. Khan, M. Zulfequar and M. S. Khan, Mater. Sci. Appl., 2011, 2, 340 CAS.
- B. Halliwell and J. M. C. Gutteridge, Free Radicals in Biology and Medicine, Oxford University Press, USA, 2015 Search PubMed.
- K. R. Raghupathi, R. P. Allaker, M. A. Vargas-Reus and G. G. Ren, Antimicrob. Polym., 2012, 327–350 Search PubMed.
- N. Beyth, Y. Houri-Haddad, A. Domb, W. Khan and R. Hazan, Evidence-based Complement. Altern. Med, 2015 DOI:10.1155/2015/246012.
- Y.-N. Chang, M. Zhang, L. Xia, J. Zhang and G. Xing, Materials, 2012, 5, 2850–2871 CrossRef CAS.
- J. S. Miller and J. M. Quarles, Cytometry, 1990, 11, 667–675 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |