Open Access Article
Open Access ArticleProtein adsorption and desorption behavior of a pH-responsive membrane based on ethylene vinyl alcohol copolymer
Hui Ye ab,
Lilan Huangab,
Wenrui Liab,
YuZhong Zhang*ab,
Lizhi Zhaoab,
Qingping Xinab,
Shaofei Wangab,
Ligang Linab and
Xiaoli Dingab
ab,
Lilan Huangab,
Wenrui Liab,
YuZhong Zhang*ab,
Lizhi Zhaoab,
Qingping Xinab,
Shaofei Wangab,
Ligang Linab and
Xiaoli Dingab
aState Key Laboratory of Separation Membranes and Membrane Processes, Tianjin Polytechnic University, Tianjin 300387, China. E-mail: zhangyz2004cn@163.com; Fax: +86-22-83955806; Tel: +86-22-83955810
bSchool of Materials Science and Engineering, Tianjin Polytechnic University, Tianjin 300387, China
First published on 18th April 2017
Abstract
Protein adsorption and desorption behavior was investigated for a pH-responsive ethylene vinyl alcohol copolymer (EVAL) membrane with an interconnected porous structure. The transition of electrostatic behavior and conformation change of the poly(dimethylaminoethyl methacrylate) (poly(DMAEMA)) chain contributed to the pH-responsive protein adsorption and desorption. Protein adsorption was conducted under acidic and neutral conditions. Protein desorption was conducted under alkaline conditions. The protonated poly(DMAEMA) chain was positively charged and extended into the BSA solution below its pKa, providing a three-dimensional space for BSA adsorption. The maximum static protein adsorption capacity was obtained at pH 6.4. The dynamic adsorption capacities of membrane EVAL10 at 10% and 50% breakthrough were 45 and 99 mg BSA per g of membrane, respectively. The Q50% of membrane EVAL10 was equivalent to 22.6 mg BSA per mL of membrane, almost 95% of the static adsorption capacity. BSA was quickly desorbed from the membrane and 94% recovery of BSA was observed at pH 9.0 in the dynamic desorption process, due to a deprotonated and collapsed conformation of the poly(DMAEMA) chains. The dynamic adsorption capacity of the membrane did not change significantly after four sequential cycles.
1. Introduction
The conventional resin-based chromatography used for protein recovery and purification has several major limitations, e.g., high-pressure drop and long process time. Membrane chromatography, which makes use of a microporous membrane as chromatographic support material, is considered to have a lower pressure drop, higher processing speed and higher throughput process than resin-based chromatography. A predominantly convective transport mechanism of the solutes through the membrane pores to the adsorption sites is superior to the intraparticle diffusion transport mechanism of resin-based chromatography. Until now, various types of membranes have been investigated for protein purification,1,2 i.e., membrane chromatography based on ion exchange,3–6 affinity,7–9 and hydrophobic interactions.10,11Stimuli-responsive membrane can change structural and functional properties (i.e. pore-size, porosity, surface charge and hydrophilicity) in response to environment conditions such as temperature, pH, electric or magnetic field and ionic strength.12–14 Recently, there is an increased interest in the stimuli-responsive surfaces which can change their affinity towards biomolecules under external stimuli.15–17 Raja Ghosh et al.18 prepared the hydrogel-paper composite membranes for temperature-responsive protein recovery by hydrophobic interaction membrane chromatography. The hydrophobic or hydrophilic states of membranes could be regulated by the addition or removal of salts for selective antibody binding. Subsequently, Raja Ghosh et al.11 fabricated temperature-responsive membrane by grafting poly-(N-isopropyl acrylamide) (PNIPAM) onto filter paper surface. The protein binding could be controlled for temperature-based separation. However, the salt- and temperature-responsive membranes possibly led to certain problems and challenges, such as high pressure drop and deposition of salts within equipment in the salt-based mode, or protein stability problems in the temperature-based mode. The pH-responsive membranes can be potentially useful for selective bio-separation, but very few developments on pH-responsive membranes for protein purification have been reported up to now.
Poly(dimethylaminoethyl methacrylate) (poly(DMAEMA)) is a weak cationic polyelectrolyte with tertiary amine groups, which can ionize under alkaline conditions (pKa 8.5–9.5). The degree of ionization of weak polyelectrolytes can be flexibly adjusted by pH value, thus the weak polyelectrolytes seem more favorable to control electrostatic interactions between weak polyelectrolytes and feed solutes compared to strong polyelectrolytes.19 The poly(DMAEMA) has been employed for preparing ion exchange, antimicrobial and pH-responsive membrane, but studies to develop protein separation performance based on the electrostatic behavior and conformation change of poly(DMAEMA) are still limited. Moreover, the permeability and selectivity of pH-responsive membranes grafted with poly(DMAEMA) chains can be regulated at high pH, which could be potential application for separating mixtures containing alkaline components, e.g. soy protein concentrates,20,21 whey protein concentrates22,23 and traditional Chinese medicine (TCM) extracts.24,25
Ethylene vinyl alcohol copolymer (EVAL) is a promising biomedical material with excellent hydrophilicity and biocompatibility.26,27 Incorporation of hydroxy groups increases the wettability of membrane and reduces the non-selective (irreversible) adsorption between the biomolecules and the membrane surface, especially in protein separation.28,29 To our knowledge, the protein separation performance of EVAL membrane functionalized with grafted pH-responsive polymers has not yet been reported.
The pH-responsive EVAL ultrafiltration membrane has been reported in an earlier paper from our group.30,31 The pH-responsive properties of the poly(DMAEMA)-grafted EVAL membrane were observed in the pH range of 9–10, but the surface of the membrane was dense. Such dense surface had a negative impact on convective transport and led to protein rejection. In the present study, the protein adsorption and desorption behavior was investigated for a pH-responsive EVAL membrane prepared by grafting DMAEMA onto EVAL microfiltration membrane. The effective surface charge characteristic of membrane in response to pH was measured. Protein adsorption measurements (BSA as model protein) were used to evaluate the pH-responsive protein separation of the membranes. The protein adsorption and desorption were investigated in dynamic process and the effect of flow rate on dynamic adsorption was studied.
2. Experimental
2.1 Materials
EVAL (ethylene content of 44 mol%) was purchased from Kuraray (Japan). Dimethylsulfoxide (DMSO) (solvent) and 1-octanol (non-solvent additive) were purchased from Tianjin Guangfu Chemical Reagent Co. Ltd. (China). DMAEMA (98%) was purchased from J&K Chemicals (Beijing, China) and was purified under depressurized condition to remove the inhibitor before usage. BSA (molecular weight of 68 kDa) was sourced from Sigma-Aldrich. Benzophenone (BP) purchased from TCI (Shanghai) Development Co. Ltd was used as photoinitiator. The sodium dihydrogen phosphate (NaH2PO4), sodium hydrogen phosphate (Na2HPO4), hydrochloric acid (HCl) and sodium hydroxide (NaOH) were used of analytical reagent grade and obtained from Tianjin Fengchuan Chemical Reagent Co. Ltd. (China).2.2 Preparation of original EVAL membrane
The microfiltration EVAL membrane with high pore interconnectivity was used as porous support. Such original EVAL membrane was prepared by phase inversion method.32 15 wt% EVAL polymer and 20 wt% of 1-octanol were dissolved in 65 wt% DMSO at 70 °C and ultrasonicated for 30 min to eliminate bubbles. Solutions were cast on a glass plate, and immersed in a water bath at 40 °C until fully cured. After that, the membranes were taken out and washed by ethanol to remove remaining solvent and additive.2.3 Preparation of poly(DMAEMA)-grafted EVAL membrane
The poly(DMAEMA)-grafted EVAL membrane was prepared using UV-induced graft polymerization, as detailed in our previous publication.30 Firstly, the EVAL membrane was washed by ethanol for 24 h to remove the impurities and additives. Then, EVAL membrane was dried and pre-coated in 50 mL solution with 5 g L−1 BP in acetone for 2 h. Secondly, BP-adsorbed EVAL membrane was placed between two pieces of quartz plate, immersed in DMAEMA solution and irradiated under UV light. The grafted membrane was subsequently extracted with ethanol by Soxhlet extraction for 48 h to remove redundant DMAEMA monomer and poly(DMAEMA) homopolymer. The grafting degree (GD) was calculated as follows:
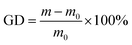 | (1) |
2.4 Morphology of membranes
The surface morphology of the membranes was examined by a field emission electron microscopy (FE-SEM, Hitachi S-4800, Japan). The accelerating voltage used was 10 kV. Samples of the membranes were attached with double-sided tape to steel stabs and sputter-coated with gold prior to FE-SEM measurements.2.5 Zeta potential measurements
The effective surface charge characteristics of the membranes were evaluated from zeta potential measurements using a SurPASS electrokinetic analyzer (Anton Parr GmbH, Austria). The membrane was placed in an adjustable gap cell and filled with 10 mM KCl solution. The gap height was set to 100 μm and the pressure was varied between 0–300 mbar. The pH was adjusted with a 0.1 M HCl solution or a 0.1 M NaOH as needed. Each membrane was measured twice at each direction. The zeta potential (ξ) was calculated using the Helmholtz–Smoluchowski equation:
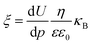 | (2) |
2.6 Static protein adsorption
Adsorption capacity (Qe) was calculated by eqn (3) as follows:
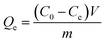 | (3) |
The effect of pH (3.6, 4.8, 6.4, 7.4, and 9.0) on the adsorption capacity was measured. The pH value was measured by using a pH meter (EL20, Mettler Toledo, Switzerland).
Langmuir isotherm model:
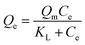 | (4) |
Freundlich isotherm model:
| Qe = KFCe1/n | (5) |
The pseudo-first-order model:
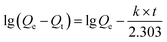 | (6) |
The pseudo-second-order model:
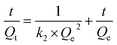 | (7) |
2.7 Dynamic adsorption and desorption
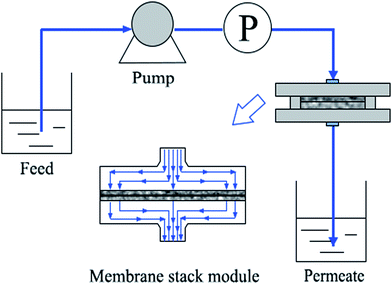
Dynamic adsorption and desorption of poly(DMAEMA)-grafted EVAL membrane was measured using a custom-made dead-end filtration cell (Fig. 1). A total internal volume of the test cell was 10 mL and an effective membrane area was 15.9 cm2. Two membranes were packed into the stack holder to form membrane stack. The feed-side pressure was maintained by a peristaltic pump (BT100-2J, Baoding Longer Precision Pump Co., Ltd.).Buffer A (10 mM PBS, pH 6.4) and buffer D (10 mM PBS, pH 9.0) were used as adsorption and desorption buffer, respectively. A typical protocol for a single run was as follows: (1) the cell was initially filled with buffer A and 60 mL buffer A was used to equilibrate membranes, (2) 0.6 mg mL−1 BSA solution was loaded through the stack holder at the flow rate of 3.8–28.3 L m−2 h−1, (3) un-adsorbed BSA was washed from membrane stack with buffer A, (4) bounded BSA was eluted with buffer D until a stable baseline was observed. After each procedure, the cell was emptied and filled the solution of the next step. The permeate solution was collected and the BSA concentration was measured at 280 nm as previously described. The dynamic binding capacities were calculated at 10% and 50% breakthrough (Q10% and Q50%).
The mass of desorbed BSA from the membrane was calculated from the area under the elution peak. Desorption efficiency was calculated as the quotient of the mass of desorbed BSA in elution to the mass of adsorbed BSA on the membrane, multiplied by 100%.
3. Results and discussion
3.1 Morphology of membranes
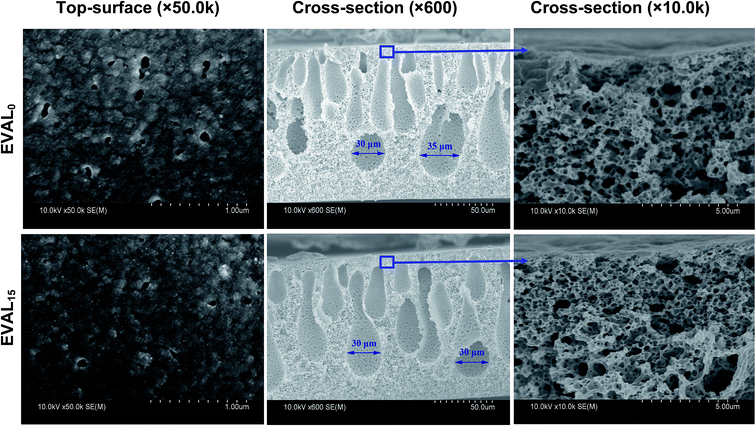
The poly(DMAEMA)-grafted EVAL membrane was prepared using UV-induced graft polymerization in our previous work. Surface composition, morphology and alkali-responsive properties (i.e., contact angle, permeability, and selectivity) of the membrane were observed. The GD of membranes was obtained by varying the grafting condition, including initial BP concentration, UV irradiation time and monomer concentration. In this work, the original EVAL membrane with micron-sized membrane pore was used for preparing poly(DMAEMA)-grafted EVAL membrane. The morphology of the membranes is presented in Fig. 2. The micron-sized membrane pores were observed on the membrane surface. The maximum pore size was approximately 0.1 μm–0.2 μm, the BSA molecule dimension was 8.0 × 8.0 × 3.0 nm,33 thus the BSA molecule could permeate through the pore channels of the EVAL membrane freely. Both of the original and the poly(DMAEMA)-grafted EVAL membrane displayed high pore interconnectivity. Such membrane structure provided an interconnected path for convective flow in the adsorption process. After grafting polymerization, the membrane pores were narrowed, but the membrane pore morphology was intact.3.2 Zeta potentials
The effect of pH on zeta potentials of the original and the poly(DMAEMA)-grafted EVAL membrane is presented in Fig. 3. The zeta potential of membrane EVAL0 was negative over the entire studied pH range, most likely due to the preferential adsorption of negative anions from the electrolyte solution.34,35 In contrast, the zeta potentials of membranes EVAL5, EVAL10 and EVAL15 significantly increased due to the presence of the poly(DMAEMA) chains on the membrane surface. The zeta potentials of the membranes were increased by decreasing pH.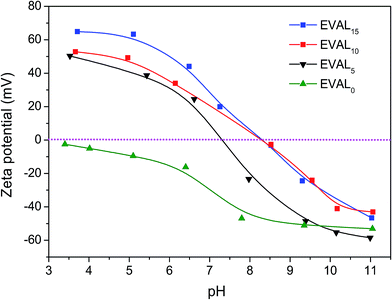 | ||
| Fig. 3 The effect of pH on zeta potentials of the original and the poly(DMAEMA)-grafted EVAL membrane. | ||
This could be ascribed to protonation of the tertiary amine of the poly(DMAEMA) chains at lower pH. The pKa value of membrane EVAL10 was higher than that of EVAL5 because the effective charge density on the membrane surface was improved by increasing GD. However, the pKa values of EVAL10 and EVAL15 were similar, probably not all poly(DMAEMA) chains of EVAL15 exposed on the membrane surface, thus the effective charge density on membrane surfaces of EVAL15 was approximate to that of EVAL10.
3.3 Static protein adsorption
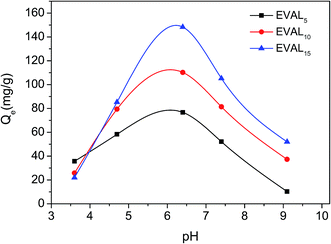 | ||
| Fig. 4 The effect of pH on static protein adsorption of poly(DMAEMA)-grafted EVAL membrane (1 mg BSA per mL, 10 mM PBS). | ||
As shown in Fig. 4, the effect of pH on adsorption capacity was significant. The maximum BSA adsorption capacity was obtained at pH 6.4. The poly(DMAEMA)-grafted EVAL membrane surface was positively charged at pH 6.4, while the BSA molecule (pI 4.7–4.9) was negatively charged. Thus, electrostatic interaction led to significant BSA adsorption. At pH 3.6, very weak BSA adsorption was observed because of charge repulsion. When pH was 9.0, the positive charge of membrane surface disappeared due to the deprotonated of grafted chains, thus the BSA adsorption capacity was low. The above results demonstrated that the BSA adsorption could be tuned by varying pH value. In static adsorption process, protein adsorption mainly depended on electrostatic behavior between grafted chains and protein. In this case, pH 6.4 was selected for BSA adsorption.
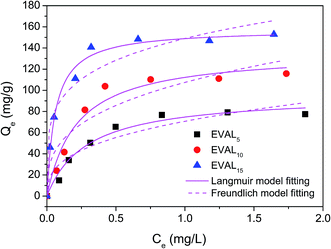 | ||
| Fig. 5 Adsorption isotherms of protein adsorption on the poly(DMAEMA)-grafted EVAL membrane (pH 6.4, 10 mM PBS). | ||
| Membrane | Langmuir model | Freundlich model | |||
|---|---|---|---|---|---|
| Qm (mg g−1) | KL (L mg−1) | R2 | KF (mg g−1)(L mg−1)1/n | R2 | |
| EVAL5 | 96.2 ± 5.9 | 0.2889 | 0.9744 | 70.69 | 0.8964 |
| EVAL10 | 136.5 ± 9.3 | 0.2164 | 0.9634 | 107.45 | 0.8711 |
| EVAL15 | 157.8 ± 4.0 | 0.0603 | 0.9889 | 148.99 | 0.9279 |
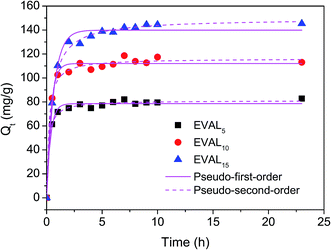 | ||
| Fig. 6 Adsorption kinetics of protein adsorption on the poly(DMAEMA)-grafted EVAL membranes (1 mg BSA per mL, pH 6.4, 10 mM PBS). | ||
| Membrane | Pseudo-first-order kinetic adsorption model | Pseudo-second-order kinetic adsorption model | ||||
|---|---|---|---|---|---|---|
| k1 (min−1) | Qe (mg g−1) | R2 | K2 (g (mg−1 min−1)) | Qe (mg g−1) | R2 | |
| EVAL5 | 2.8781 | 78.5 ± 0.8 | 0.9865 | 3.38 × 106 | 81.2 ± 0.7 | 0.9941 |
| EVAL10 | 2.6208 | 112.1 ± 1.2 | 0.9860 | 8.62 × 106 | 116.2 ± 1.3 | 0.9904 |
| EVAL15 | 1.5653 | 139.9 ± 1.5 | 0.9874 | 8.36 × 106 | 149.5 ± 1.3 | 0.9956 |
3.4 Dynamic protein absorption
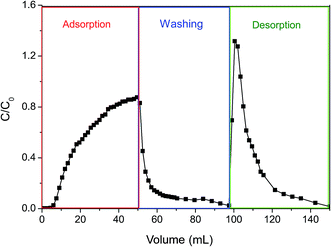 | ||
| Fig. 7 The dynamic adsorption and desorption curve of poly(DMAEMA)-grafted EVAL membrane (EVAL10, 0.6 mg BSA per mL, pH 6.4, 10 mM PBS, 3.8 L m−2 h−1). | ||
The bound protein was eluted with buffer D until a stable base-line was observed. The BSA was quickly desorbed from the membrane and 94% recovery of BSA was observed. The concentration of the eluted protein was 1.3-fold increase relative to the feed concentration. The electrostatic attraction between BSA and membrane surface disappeared and the poly(DMAEMA) chains were collapsed at pH 9.0. The high dynamic desorption efficiency at pH 9.0 depended on not only electrostatic behavior but also conformation change of pH-responsive chains. In addition, the effective pore size of the membranes increased when the poly(DMAEMA) chains were deprotonated, mass transfer resistance decreased, thus the dynamic desorption efficiency was improved.
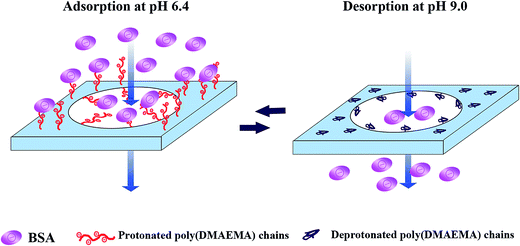
The mechanism of protein adsorption and desorption on the pH-responsive EVAL membrane is shown in Fig. 8. The poly(DMAEMA) chains, which were grafted on the membrane surface and pore wall, acted as a pH sensor and a valve which regulated the protein adsorption and desorption properties of the membrane. Be differ from ion exchange membrane, electrostatic behavior is not the only factor for protein adsorption of pH-responsive membrane. The conformation change of grafted chains also plays an important role for pH-responsive protein adsorption and desorption. In this case, the transition of electrostatic behavior and conformation change of the poly(DMAEMA) chains both contributed to the pH-responsive protein adsorption and desorption. At pH 6.4, the grafted EVAL membrane surface was positively charged and the BSA molecule was negatively charged. The significant BSA adsorption capability was obtained due to electrostatic interaction. Moreover, the protonated poly(DMAEMA) chains extended into the BSA solution in the filtration process, providing a three-dimensional space for BSA adsorption. The high dynamic desorption efficiency was obtained at pH 9.0 due to deprotonated and collapsed conformation of poly(DMAEMA) chains.
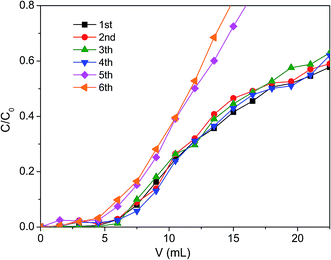 | ||
| Fig. 9 The effect of adsorption/desorption cycle numbers on dynamic adsorption of membrane EVAL10 (EVAL10, 0.6 mg BSA per mL, pH 6.4, 10 mM PBS). | ||
| Cycle number | Q10%, mg BSA per g membrane | Q50%, mg BSA per g membrane |
|---|---|---|
| 1st | 45 | 99 |
| 2nd | 45 | 99 |
| 3th | 45 | 99 |
| 4th | 45 | 99 |
| 5th | 36 | 72 |
| 6th | 36 | 72 |
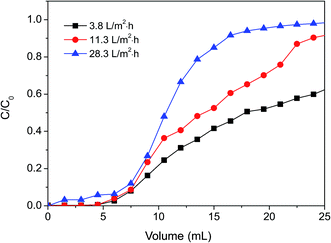 | ||
| Fig. 10 The effect of flow rate on dynamic protein adsorption of poly(DMAEMA)-grafted EVAL membrane (EVAL10, 0.6 mg BSA per mL, pH 6.4, 10 mM PBS). | ||
| Flow rate, L m−2 h−1 | Q10%, mg BSA per g membrane | Q50%, mg BSA per g membrane |
|---|---|---|
| 3.8 | 45 | 99 |
| 11.3 | 45 | 81 |
| 28.3 | 36 | 63 |
4. Conclusions
Model protein BSA adsorption and desorption of the poly(DMAEMA)-grafted EVAL membrane with the interconnected porous structure was investigated. The maximum static BSA adsorption capacity was observed at pH 6.4. Fitted on Langmuir model the monolayer adsorption of BSA molecule on the membrane surface was found. The chemical interaction between the BSA molecule and the membrane dominated the overall adsorption rate. The dynamic adsorption capacity of membrane EVAL10 at 10% and 50% breakthrough were 45 and 99 mg BSA per g membrane, respectively. The Q50% of membrane EVAL10 was equivalent to 22.6 mg BSA per mL of membrane, almost 95% of the static adsorption capacity. The BSA was quickly desorbed from the membrane and 94% recovery of BSA was observed at pH 9.0 in dynamic desorption process. The transition of electrostatic behavior and conformation change of the poly(DMAEMA) chains contributed to the pH-responsive protein adsorption and desorption. The dynamic adsorption capacity of the membrane did not change significantly after four sequential cycles. The breakthrough occurred earlier by increasing the flow rate and the dynamic capacity decreased.Acknowledgements
This research was sponsored by the National Natural Science Foundation of China (Funding No. 51373120, 51503146), Science and Technology Commission Foundation of Tianjin (No. 15JCZDJC37600), and the Research Fund for the Doctoral Program of Higher Education (Funding No. 20111201110003). Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT) of Ministry of Education of China (Grand No. IRT13084); The Science and Technology Plans of Tianjin (No. 15PTSYJC00240, 15PTSYJC00230).References
- V. Orr, L. Zhong, M. Moo-Young and C. P. Chou, Biotechnol. Adv., 2013, 31, 450–465 CrossRef CAS PubMed.
- A. Saxena, B. P. Tripathi, M. Kumar and V. K. Shahi, Adv. Colloid Interface Sci., 2009, 145, 1–22 CrossRef CAS PubMed.
- Y. Wei, J. Ma and C. Wang, J. Membr. Sci., 2013, 427, 197–206 CrossRef CAS.
- H. C. Chenette, J. R. Robinson, E. Hobley and S. M. Husson, J. Membr. Sci., 2012, 432–424, 43–52 CrossRef PubMed.
- Y. Liu, Z. Feng, Z. Shao and X. Chen, Mater. Sci. Eng., C, 2012, 32, 1669–1673 CrossRef CAS PubMed.
- M. He, C. Wang and Y. Wei, RSC Adv., 2016, 6, 6415–6422 RSC.
- K. K. R. Tetala, K. Skrzypek, M. Levisson and D. F. Stamatialis, Sep. Purif. Technol., 2013, 115, 20–26 CrossRef CAS.
- N. Anuraj, S. Bhattacharjee, J. H. Geiger, G. L. Baker and M. L. Bruening, J. Membr. Sci., 2012, 389, 117–125 CrossRef CAS PubMed.
- N. Mustafaoglu, T. Kiziltepe and B. Bilgicer, Analyst, 2016, 141, 6571–6582 RSC.
- H. H. Himstedt, X. Qian, J. R. Weaver and S. R. Wickramasinghe, J. Membr. Sci., 2013, 447, 335–344 CrossRef CAS.
- Q. Wu, R. Wang, X. Chen and R. Ghosh, J. Membr. Sci., 2014, 471, 56–64 CrossRef CAS.
- C. Zhao, S. Nie, M. Tang and S. Sun, Prog. Polym. Sci., 2011, 36, 1499–1520 CrossRef CAS.
- Z. Liu, W. Wang, R. Xie, X.-J. Ju and L.-Y. Chu, Chem. Soc. Rev., 2016, 45, 460–475 RSC.
- D. Wandera, S. R. Wickramasinghe and S. M. Husson, J. Membr. Sci., 2010, 357, 6–35 CrossRef CAS.
- M. Motornov, Y. Roiter, I. Tokarev and S. Minko, Prog. Polym. Sci., 2010, 35, 174–211 CrossRef CAS.
- Y. Stetsyshyn, K. Fornal, J. Raczkowska, J. Zemla, A. Kostruba, H. Ohar, M. Ohar, V. Donchak, K. Harhay, K. Awsiuk, J. Rysz, A. Bernasik and A. Budkowski, J. Colloid Interface Sci., 2013, 411, 247–256 CrossRef CAS PubMed.
- L. Mei, R. Xie, C. Yang, X.-J. Ju, J.-Y. Wang, Z. Zhang and L.-Y. Chu, J. Membr. Sci., 2013, 429, 313–322 CrossRef CAS.
- K. Z. Mah and R. Ghosh, J. Membr. Sci., 2010, 360, 149–154 CrossRef CAS.
- Y. Su and C. Li, J. Membr. Sci., 2007, 305, 271–278 CrossRef CAS.
- K. Nishinari, Y. Fang, S. Guo and G. O. Phillips, Food Hydrocolloids, 2014, 39, 301–318 CrossRef CAS.
- J. Yang, J. Guo, X.-Q. Yang, N.-N. Wu, J.-B. Zhang, J.-J. Hou, Y.-Y. Zhang and W.-K. Xiao, J. Food Eng., 2014, 143, 25–32 CrossRef CAS.
- L. Voswinkel and U. Kulozik, Procedia Food Sci., 2011, 1, 900–907 CrossRef CAS.
- A. Arunkumar and M. R. Etzel, J. Membr. Sci., 2015, 475, 340–348 CrossRef CAS.
- X.-X. Yang, W. Gu, L. Liang, H.-L. Yan, Y.-F. Wang, Q. Bi, T. Zhang, J. Yu and G.-X. Rao, RSC Adv., 2017, 7, 3089–3100 RSC.
- A. N. N. Murakami, R. D. d. M. C. Amboni, E. S. Prudêncio, E. R. Amante, C. B. Fritzen-Freire, B. C. B. Boaventura, I. d. B. Muñoz, C. d. S. Branco, M. Salvador and M. Maraschin, Food Chem., 2013, 141, 60–65 CrossRef CAS PubMed.
- M. E. Avramescu, W. F. C. Sager, M. H. V. Mulder and M. Wessling, J. Membr. Sci., 2002, 210, 155–173 CrossRef CAS.
- M. Avramescu, J. Membr. Sci., 2003, 216, 177–193 CrossRef CAS.
- S. M. Saufi and C. J. Fee, J. Chromatogr. A, 2011, 1218, 9003–9009 CrossRef CAS PubMed.
- S. M. Saufi and C. J. Fee, Sep. Purif. Technol., 2011, 77, 68–75 CrossRef CAS.
- H. Ye, L. Chen, A. Li, L. Huang, Y. Zhang, Y. Li and H. Li, J. Appl. Polym. Sci., 2015, 132, 41775 CrossRef.
- M. Liu, L. Zhao, S. Li, H. Ye, H. An and Y. Zhang, RSC Adv., 2016, 6, 10704–10712 RSC.
- Y. Cai, J. Li, Y. Guo, Z. Cui and Y. Zhang, Desalination, 2011, 283, 25–30 CrossRef CAS.
- D. H. Tsai, F. W. DelRio, A. M. Keene, K. M. Tyner, R. I. MacCuspie, T. J. Cho, M. R. Zachariah and V. A. Hackley, Langmuir, 2011, 27, 2464–2477 CrossRef CAS PubMed.
- M. M. Rohani and A. L. Zydney, J. Membr. Sci., 2012, 397–398, 1–8 CrossRef CAS.
- M. Kumar and M. Ulbricht, J. Membr. Sci., 2013, 448, 62–73 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |