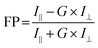Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescent silver nanoparticle based highly sensitive immunoassay for early detection of HIV infection†
Aditya Dileep Kurdekar a,
L. A. Avinash Chunduri
a,
L. A. Avinash Chunduri a,
Sai Manohar Chelli
a,
Sai Manohar Chelli b,
Mohan Kumar Haleyurgirisetty
b,
Mohan Kumar Haleyurgirisetty c,
Eswarappa Pradeep Bulagonda
c,
Eswarappa Pradeep Bulagonda d,
Jiwen Zhenge,
Indira K. Hewlettc and
Venkataramaniah Kamisetti
d,
Jiwen Zhenge,
Indira K. Hewlettc and
Venkataramaniah Kamisetti *a
*a
aLaboratories for Nanoscience and Nanotechnology Research, Department of Physics, Sri Sathya Sai Institute of Higher Learning, Prasanthi Nilayam, Puttaparthi 515134, India. E-mail: vrkamisetti@gmail.com
bDepartment of Chemistry, Sri Sathya Sai Institute of Higher Learning, Prasanthi Nilayam, Puttaparthi 515134, India
cLaboratory of Molecular Virology, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, USA
dDepartment of Biosciences, Sri Sathya Sai Institute of Higher Learning, Prasanthi Nilayam, Puttaparthi 515134, India
eDivision of Biology, Chemistry and Material Science, Office of Science and Engineering laboratories, Center for Devices and Radiological Health (CDRH), Food and Drug Administration, Silver Spring, MD 20993, USA
First published on 5th April 2017
Abstract
For the first time, we have engineered streptavidin labeled fluorescent silver nanoparticles for their application in immunosensing of biomolecules which will significantly increase sensitivity without compromising the specificity. A computational perspective for understanding the efficiency of the conjugation process is provided as a proof of concept. The fluorescence based sandwich immunoassay was demonstrated to detect HIV-1 p24 antigen in clinical specimens with improved sensitivity and specificity. The detection range of the fluorescent silver nanoparticle-based immunoassay (FSNIA) was found to be between 10 and 1000 pg mL−1 in a linear dose dependent manner. False positives were not observed with plasma samples from healthy adults (HIV−ve), hepatitis B (HBV+ve) and hepatitis C (HCV+ve). Plasma samples that were HIV−ve showed no interference with detection of HIV-1 p24 antigen. This technology can be used in resource limited settings and easily adopted for the detection of other pathogen antigens.
1 Introduction
Developing countries are witnessing a rise in infectious diseases which are the leading cause of loss of life.1,2 Although different diseases such as malaria, tuberculosis, hepatitis, pulmonary infections, to mention a few, are prevalent worldwide, the disease which has caught the attention of the globe due to its severity is Acquired Immune Deficiency Syndrome (AIDS) caused by the Human Immunodeficiency Virus (HIV). Every year nearly 2.2 million people are infected and 1.3 million die from AIDS-related illness.3 At the end of 2015, around 40 million people were living with HIV/AIDS.4 AIDS and its related illnesses can affect large part of populations if not diagnosed and treated at the earliest stage.The viral agent responsible for AIDS (Acquired Immune Deficiency Syndrome), human immunodeficiency virus (HIV), destroys cells of the immune system and progressively impairs the body's ability to defend against other infections and certain cancers.5 Antiretrovirals (ARVs) and their use in combination “cocktails” have drastically reduced mortality and morbidity, prolonged life expectancy and improved the quality of lives of infected people, although they have not been able to cure patients of HIV/AIDS.6 However, 95% of people infected with HIV live in developing countries, where the availability and access to these medicines remains unacceptably inadequate and the costs unreasonably exorbitant. Hence, rapid and accurate diagnosis of HIV is pivotal in the management and control of this global outbreak. The diagnostic tools currently available for early detection of HIV often require advanced and elaborate facilities which restrict their application in resource limited settings. Currently available diagnostic assays need further improvement for early detection and this can be achieved using nanoparticles for signal amplification coupled with sensitive and specific antibodies. Thus, the current need is rapid, sensitive and specific diagnostics at point of care which are capable of rapid identification and characterization of the disease allowing patients to get immediate treatment for effective clinical outcomes.
When it comes to detection of HIV, there are a wide variety and range of biomarkers related to HIV infection, which can even predict the progress of HIV infection to AIDS.7 Although HIV antibody tests are the most appropriate for identifying the infection, they are insufficient due to the time the antibodies take to manifest in the plasma.8 Hence, alternate efficient technologies are required for accurate diagnosis for monitoring response to therapy, and to effectively predict disease outcome. Virus isolation through viral culture, nucleic acid detection techniques to detect viral RNA,9 and tests to detect p24 antigen10 are commonly used clinical methods to identify virus or viral components in blood early, and confirm an infection. While, these methods are highly specific, each has limitations and their use is restricted to highly specific testing situation depending on the resources available. Although tests for viral RNA detection have been introduced recently, their requirements of complex instrumentation and well-trained personnel makes them unsuitable for resource limited settings.11
The p24 antigen tests stand out amongst all other tests as it is one of the earliest protein biomarkers to appear in blood after infection. The HIV-1 capsid (core) p24 is detected much earlier than antibodies during an acute HIV infection.12 A major advantage that p24 testing offers is that it is highly specific for HIV infection. Testing for p24 can be of value in: early detection of HIV infection,13 screening blood donor samples,14,15 diagnosing infection in the newborn,16,17 and monitoring antiviral therapy.18
There are many sensitive detection platforms for HIV p24 detection but user friendly point-of-care rapid test platforms are an unmet need. The most widely used test format is the enzyme-linked immunosorbent assays (ELISAs)19 which requires separate wells in the assay for each analyte to be detected and use of an enzyme which can be unstable making it less convenient. Since it is frequently necessary or helpful to detect several biomarkers simultaneously, conventional ELISAs make the process time consuming.20 Current ELISA methods are further limited by problems common to the use of enzymes, contamination of the substrate solution and the need to read the micro well shortly after completion of the enzyme/substrate reaction.21
The use of fluorescence-linked immunoassays gives advantages over ELISAs such as increased sensitivity, a larger dynamic range and greater stability of reagents.22,23 Fluorescence methods may also be multiplexed, thereby using lower volumes of reagents and decreasing the assay time.24 The ability to spectroscopically detect multiple analytes in a single well assay is not without its challenges. These include cross reactivity of antibodies,25 the need for discrete spectral bands which do not overlap too closely,26 and the possibility of interaction between the fluorophores. The use of fluorescent nanoparticles removes some of these challenges which has prompted the application of fluorescent nanoparticles as immunoassay labels.
Noble metal nanoparticles which are generally clusters of 5–100 atoms shielded by capping agents,27 are of particular interest for their exceptional properties like high tunability of their optical spectra and high surface to volume ratios allowing a higher surface area for specific bio-functionalization.28–30 In the field of protein detection, noble metal nanoparticles have played a key role in the development of highly sensitive detection platforms for rapid identification of disease biomarkers.31,32
Silver nanoparticles (AgNPs), have been extensively studied for their application in biosensing based on their photoluminescence, surface plasmon resonance, optical scattering, and labelling features.33,34 These nanoparticles are being explored for other sensing applications in composites and assembled nanostructures for their synergistic properties.35,36 AgNPs have also been exploited for the development of protocols involving metal enhanced fluorescence based detection, surface-enhanced Raman scattering (SERS) and in combination with gold to form bimetallic NPs which can either be alloys or core shell nanoparticles.37,38
Fluorescent silver nanoparticles (FSNPs) are emerging functional materials which have generated significant interest in the field of bioimaging and biosensing.39,40 These particles can interact with light as they have discreet energy levels due to quantum confinement,41 allowing intense light absorption and emission.42,43 These optical properties along with superior biocompatibility, brightness and photostability have led to their application as fluorescent biolabels.44,45 Additionally, these FSNPs can have application as sensitive probes, as their fluorescence emission is a function of their interaction with the organic scaffold.46–48
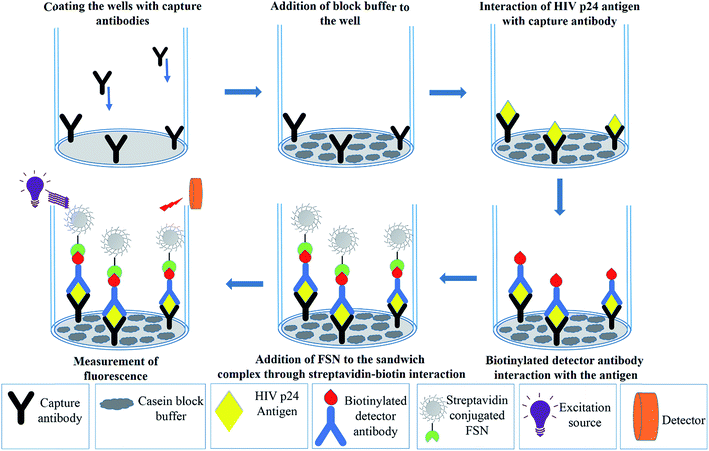
Combining the efficiency of FSNPs in biosensing with the current need for sensitive detection of biomarkers, we report the development of a versatile fluorescent silver nanoparticle immunoassay (FSNIA). The FSNPs bioconjugated to streptavidin can be used with biotin labeled antibodies or antigens for detection of target antigen or antibodies. The conjugation and functioning of FSNPs was interpreted using computational modeling. A mechanistic interpretation of the bio-conjugation was sought for immunosensing applications for which in silico studies were performed using various computational methods to explain the thermodynamics of these processes. In this regard, the energetics of individual steps was evaluated to elucidate the process of the conjugation observed. FSNIA was then employed for the detection of HIV. HIV-1 p24 was chosen as the model analyte as it is the biomarker for early detection of HIV. The results in this study can help in development of a multiplexed screening protocol based on FSNPs which could be potentially beneficial in clinical settings and in the control and prevention of the spread of AIDS.
2 Experimental
2.1 Chemicals and reagents
Glutathione (Himedia), silver nitrate (Sigma), hydrazine hydrate (Thermo Scientific), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, (Thermo Scientific, USA), N-hydroxysuccinimide (Thermo Scientific, USA), streptavidin (Scripps Lab, USA), casein block buffer (Thermo Scientific, USA), anti-HIV-1 p24 antibody (Prospec, USA), HIV-1 p24 antigen (Virogen, USA), biotinylated HIV-1 p24 detector antibody (Perkin-Elmer, USA). Phosphate buffer saline (100 mM, pH-7.2) and carbonate bicarbonate buffer (500 mM, pH-9.5) were prepared by following established procedures. Centrifugal spin columns (10, 30, 100 kDa) were purchased from GE Health Care, UK. Double distilled water was used for performing all the experiments. Plasma samples from patients and healthy individuals were obtained from the Sri Sathya Sai Institute of Higher Medical Sciences, Prasanthigram, India, (study id-SSSIHL/IEC/PSN/BS/2012/01). The details of the samples were coded and tested in a blinded manner. Samples were confirmed to be positive or negative using fourth generation Microlisa HIV Ag + Ab Elisa kit (J. Mitra & Co. Pvt. Ltd.).2.2 Synthesis of fluorescent silver nanoparticles
Fluorescent silver nanoparticles (FSNPs) were prepared following the process of chemical reduction using hydrazine hydrate as the reducing agent.49 5 mL aqueous solution of 2 mM glutathione was mixed (with vigorous stirring) with 5 mL aqueous solution of 1 mM silver nitrate till a milky white acidic solution was obtained. The acidic gel was brought to neutral pH by adding 400 μL of sodium hydroxide (1 M) until a clear solution was formed. Following the formation of a clear solution, the rate of stirring was reduced and after 5 minutes, 500 μL of hydrazine hydrate was added. The solution was now stirred for 10 hours till it turned pale yellow. The resulting solution was filtered to remove any large particulates using a nitrocellulose membrane. The filtered solution was lyophilized and stored at 4 °C for further characterization and immunoassay experiments.2.3 Characterization of the FSNPs
The morphology and size of the FSNP dispersion was characterized using a Jeol 1400 TEM at 80 kV and LIBRA 200 High Resolution Transmission Electron Microscope operated at 200 kV, respectively. The FSNPs were drop casted on the copper grid and vacuum dried before imaging. Total 514 particles were analyzed using Image J to determine the size distribution of particles. The optical characterization experiments and measurements were carried out using Shimadzu 2450 UV-Vis spectrophotometer and Spectramax M5 microplate reader. To obtain the absorbance and fluorescence spectra, the samples were accordingly diluted. FTIR spectra were obtained using Thermo Scientific Nicolet 10 FT-IR spectrometer on the potassium bromide (KBr) pellet.2.4 Bioconjugation of FSNPs with streptavidin
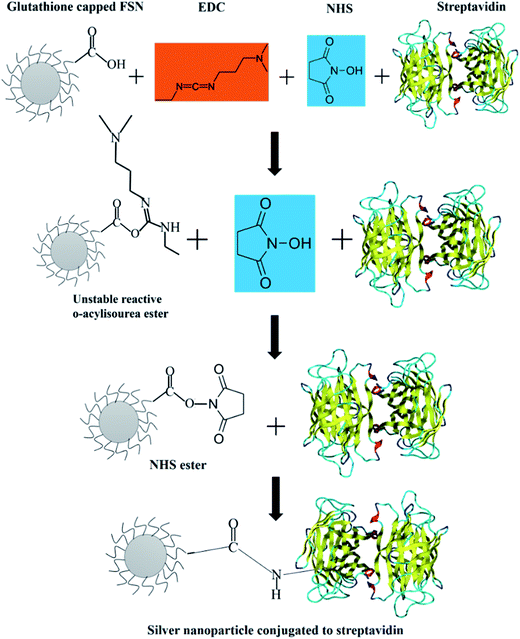
The bioconjugation of the above purified FSNPs was carried out using the EDC–NHS coupling protocol.50 10 mg of nanoparticles were dispersed in 10 mM phosphate buffer (pH = 7.4) and were washed in centrifugal spin columns. Nanoparticles were dispersed in Phosphate Buffered Saline (PBS) and the solution was sonicated with a tip sonicator for 30 s. The activation of carboxyl groups was carried out by mixing the FSNPs with 10 mmol L−1 of EDC and 10 mmol L−1 NHS in PBS buffer for 30 min. These activated particles were washed once with wash buffer. To this solution, 50 μL solution of 1 mg mL−1 streptavidin dissolved in carbonate bicarbonate buffer (pH = 9.0) was added. The above prepared solution was incubated at room temperature for 3 h following which the streptavidin conjugated nanoparticles were washed 5 times with the wash buffer.In order to gain a deeper mechanistic understanding of the bio-conjugation for immunosensing applications, in silico studies were performed using various computational methods to rationalize the thermodynamics of these processes. In this regard, the energetics of the individual steps was evaluated to elucidate the process of the conjugation observed.
2.5 Fluorescence polarization
The fluorescence polarization experiments were carried out in order to ascertain whether the process of bioconjugation has produced conjugated nanoparticles. The difference in the fluorescence polarization value is the indicator of size of the bioconjugated molecule, which increases after conjugation.53 The excitation and emission wavelengths were chosen as 401 nm and 716 nm respectively. The fluorescence polarization value was calculated from the formula given by eqn (1),
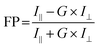 | (1) |
The I∥ and I⊥ were measured in the fluorescence polarization mode by Spectramax M5 plate reader and substituted in the above formula to obtain the FP ratio. Conjugation is confirmed by the higher magnitude of fluorescence polarization for the conjugated FSNPs compared to the unconjugated FSNPs.
2.6 Fluorescent silver nanoparticle immunoassay (FSNIA) for detection of HIV-1 p24 antigen
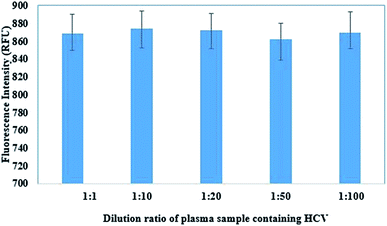
In addition to interference of a protein, studies were also carried out to see the interference effect of coexisting viruses. For our study, hepatitis C virus (HCV) was chosen as an interfering virus as it is a common co-infection in HIV patients. To study the effect of the presence of HCV as an interfering agent in the immunoassay, the HCV positive plasma sample was diluted 10, 20, 50 and 100 times. 500 pg mL−1 of p24 antigen was spiked in all the solution including the undiluted plasma sample. The immunoassay was carried out on the spiked samples.
2.7 HIV detection in blood plasma samples
 | (2) |
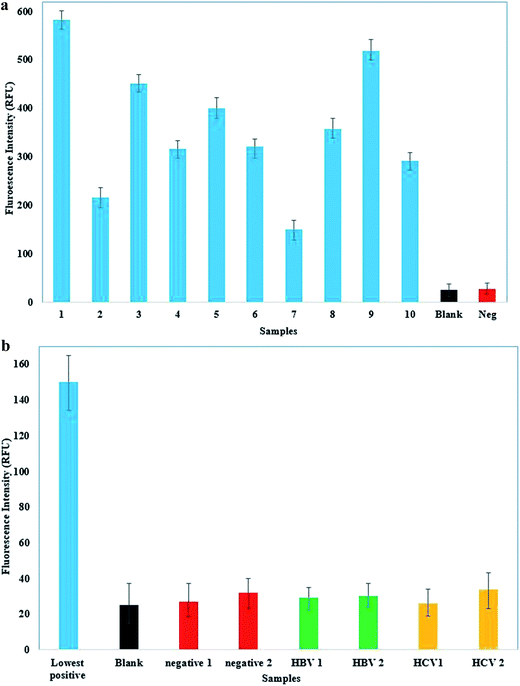
It is necessary to test the performance of the assay with clinical samples in order to evaluate its applicability to clinical studies. To validate the assay, FSNIA was evaluated using 50 HIV positive and 50 HIV negative clinical samples. These samples were prepared using the procedure described before. The cross reactivity was also evaluated with 10 HBV positive/HIV negative and 10 HCV positive/HIV negative plasma samples.
3 Results and discussion
3.1 Morphological characterization of fluorescent silver nanoparticles
The synthesis of FSNPs is discussed in Section S1† in ESM. The size and morphology of the silver nanoparticles was confirmed by TEM imaging. The TEM images as seen in Fig. 1 show the particle size to be 15.4 ± 3.7 nm of spherical morphology. The HRTEM confirmed the interplanar spacing to be 0.237 nm which is in accordance with the literature.54 The nanoparticles were found to be polycrystalline which also is a reported observation.55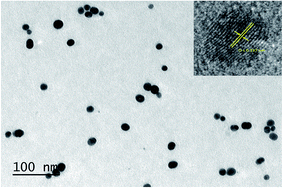 | ||
| Fig. 1 TEM image of FSNPs which indicates the size of the FSNPs to be around 15.4 ± 3.7 nm. The inset shows the HRTEM image of FSNPs which indicate a d spacing of 0.237 nm. | ||
3.2 Optical spectral characteristics of fluorescent silver nanoparticles
UV-Vis spectroscopy and photoluminescence spectroscopy studies were used to study the optical properties of the FSNPs. As illustrated in Fig. 2, the absorption spectra revealed a broad peak in the range of 370–450 nm with the peak maximum at 401 nm. On being subjected to UV light (365 nm), a red emission was observed which is visible to naked eyes. A broad emission centered at 716 nm was observed in the emission spectrum with an excitation wavelength peak at 360 nm. This emission can be attributed to hybrid states which are formed due to the interaction of glutathione and silver at their interface.56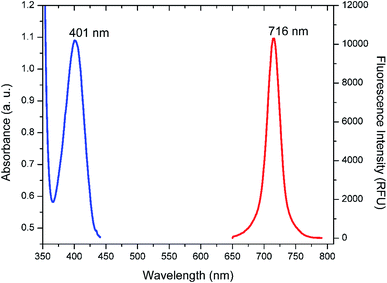 | ||
| Fig. 2 UV-Vis absorbance and fluorescence emission spectra of FSNPs. The blue peak is the absorbance spectra while the red one is fluorescence spectra of FSNPs. | ||
In order to understand the surface functionalization on the FSNPs, FTIR studies were carried out (Fig. 3a). The most important feature of the FTIR spectra obtained was the missing peak at 2510 cm−1 which is the characteristic absorption peak for S–H indicating that the S–H group is not freely available in the FSNPs. This suggests that glutathione caps the silver nanoparticles through the thiol from cysteine.57 The stretching vibration band which is the characteristic peak of carboxylic group was found to have shifted slightly to a lower wavenumber of 1602 cm−1 which can be attributed to weak interaction between the negatively charged COO− and electropositive silver core as depicted in Fig. 3b.58 The peak at 1356 cm−1 is an indicator of deformation and vibration features of C–N linkage which is predominant in this tripeptide. Meanwhile, the 1093 cm−1 peak is due to the vibration band of C–O. The peaks at 2974 cm−1 and 1464 cm−1 can be ascribed to the stretching and bending vibrations of aliphatic C–H group in glutathione.59 The 1010–880 and 850–740 bands are the out of plane vibration bands of C–H and N–H group which are typically formed by interaction of the silver nanoparticle with GSH.60
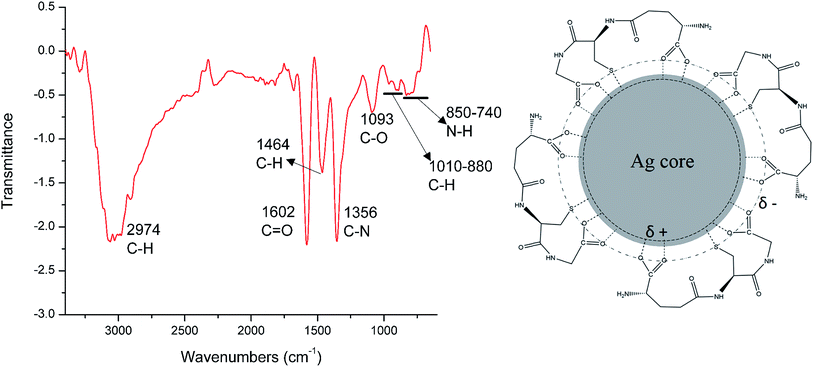 | ||
| Fig. 3 (a) FTIR spectrum of FSNPs (b) interaction between the silver core and glutathione molecules. | ||
3.3 Bioconjugation of fluorescent silver nanoparticles to streptavidin
Streptavidin conjugation to FSNPs was performed to make them specific for biomolecular recognition of biotin labeled antibodies. Since it is difficult to directly conjugate a protein to a nanoparticle with high efficiency, an intermediary coupling protocol was chosen. Now, the most well established and versatile methods that exist for conjugating or crosslinking a moiety to carboxylic acid are based on carbodiimide compounds. Since the medium is aqueous, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) was chosen. N-hydroxysuccinimide (NHS) is an essential part of EDC based coupling protocols, whose purpose is to enhance the coupling efficiency and create amine-reactive intermediates which are stable. EDC helps in coupling of NHS to carboxyls forming an NHS ester which is more stable than the O-acylisourea intermediate.61 It also permits bioconjugation to primary amines at physiological pH. The coupling reaction was carried out in PBS whose pH is 7.4, which is compatible with the bioconjugation chemistry.62A schematic representation of the conjugation protocol is presented in Scheme 2. The first step of this reaction is the interaction between the glutathione from the FSNPS with EDC. It results in the formation of an unstable o-acylisourea intermediate. This intermediate reacts with NHS and forms an amine reactive NHS ester which is considerably more stable than the o-acylisourea intermediate. The final addition to this reaction mixture is the protein, streptavidin. The NH2 from the amino acids in streptavidin reacts with the amine reactive NHS ester and results in formation of an amide bond between carboxylic group of glutathione and amine group from streptavidin.
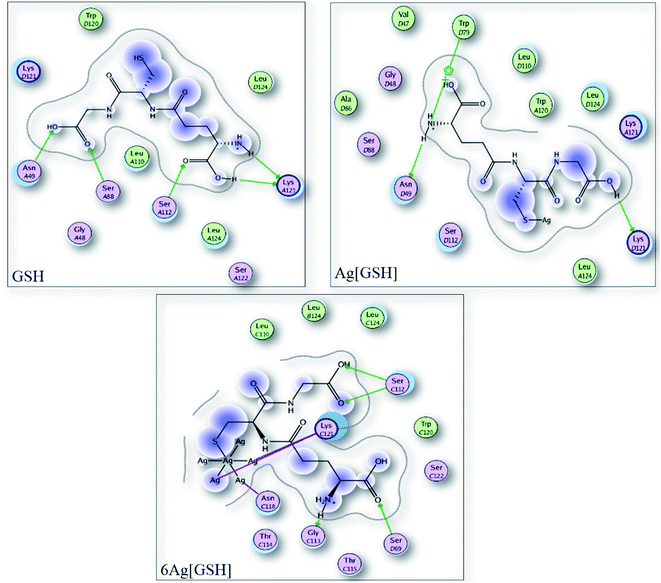
| Molecule | Hex 8.0.0 score |
|---|---|
| GSH | −262.1 |
| Ag[GSH] | −260 |
| 6 Ag[GSH] | −324.42 |
| 31 Ag[GSH] | −380.03 |
| Binding moieties | HEX score |
|---|---|
| GSH–streptavidin–biotin | −224.5 |
| Ag[GSH]–streptavidin–biotin | −233.3 |
| 6 Ag[GSH]–streptavidin–biotin | −221.1 |
| 31 Ag[GSH]–streptavidin–biotin | −223.6 |
The slight destabilization of protein explains the difference in binding affinity. The tradeoff between the stabilization of the Ag–GSH (by interaction between Ag and the amino acids) with destabilization of protein due to the steric hindrance plays crucial role in this protein biotin interaction. While 31 atom system may sterically destabilize the protein but the silver amino acid interactions counteract the destabilization allowing a successful interaction of the conjugated protein with biotin. Thus, the highlight of this study is that the bioconjugation process still allows the protein to interact with biotin with significant binding affinity which is important for immunoassay applications.
3.4 Fluorescence polarization studies
The confirmation of conjugation was obtained through the fluorescence polarization studies which were performed on the Spectramax M5 plate reader in the fluorescence polarization mode. The correction factor (G) of the plate reader was calculated to be 1. The results of the FP are depicted in the Table 3.The higher value of FP of the streptavidin conjugated FSNPs in comparison to unconjugated FSNPs confirms the bioconjugation of FSNPs to streptavidin. To understand this, we need to have a look at the correlation between fluorescence polarization and size of the fluorophores. The smaller size of unconjugated particles leads to their movement in solutions that results in depolarization of the incident plane polarized excitation energy thus leading to very low polarization value.63,64 However, the conjugation of FSNPs to streptavidin reduces the motion of the particle due to an increase in the size and volume of the particles. This reduces the rotation of particles which causes the excitation energy and the transmitted energy to be in the same state of polarization expressed as a higher magnitude of polarization value.65 This manifests as a higher value of FP for the conjugated particle than that of the unconjugated particle.
3.5 Results of fluorescent silver nanoparticle immunoassay (FSNIA) for the detection of HIV-1 p24
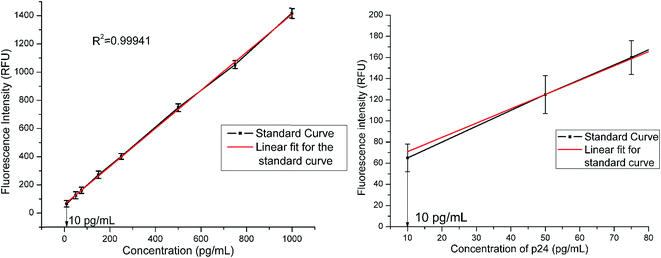
HIV-1 p24 antigen is the target viral protein in this study because of its abundance in the early stages of HIV infection when antibody concentrations are low and not detectable. This in turn helps in early detection of the disease which can allow better management of the disease and antiretroviral treatment. In this study, we adapted p24 antigen to assess the performance of FSNIA. In order to test the sensitivity of the FSNIA, we decided to test its limits in detecting p24 antigen. The immunoassay involved the formation of antibody–antigen–antibody sandwich complex where in the detection step involved interaction between the biotinylated detector antibody and streptavidin conjugated FSNPs. The detection process makes use of the strong non-covalent chemistry between biotin and streptavidin which immobilizes the fluorophore to the sandwich complex. When illuminated by excitation energy, the fluorophore emits and the signal intensity is proportional to the amount of streptavidin conjugated FSNPs. The amount of streptavidin conjugated FSNPs are in turn proportional to amount of p24 present. Thus, the signal intensity is directly proportional to the concentration of p24 present in the sample. The fluorescence signal intensity is then measured using a SpectraMax M5 microplate reader. A calibration curve for the purified HIV-1 p24 antigen is plotted with the measured data.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which was chosen as the measure for determining positive samples.68 The equation of linear correlation was determined to be
1, which was chosen as the measure for determining positive samples.68 The equation of linear correlation was determined to be| FI = 1.34 × C + 55.38 | (3) |
We obtained very good linear correlation in eqn (3) between the concentrations of p24 and the fluorescence signal intensity in FSNIA with a high correlation coefficient of R2 = 0.99941. This also suggests that FSNIA is a linear dose response assay. In this paper, we report that this FSNIA could achieve an analytical sensitivity at the picogram level which is comparable to the sensitivity levels achieved with conventional colorimetric ELISA (Table 4).
| Method | Analyte | Concentration (pg mL−1) | Ref. |
|---|---|---|---|
| Conventional ELISA | p24 antigen | 10–15 | 70 |
| EIS | Arsenic | 13 | 71 |
| CLMI | Cysteine | 25 | 72 |
| Multiarray-LSPR | Proteins | 100 | 73 |
| Colorimetric | Cancer biomarker cyclin A2 | 30 | 74 |
| SPRI | IgG | 7 | 75 |
| RLS | Alpha feto protein | 40 | 76 |
| Lab on chip | Staphylococcal enterotoxin B | 100 | 77 |
| Lab on disc | Anti-hepatitis B virus | 510 | 78 |
| SERS | DNA | 50 | 79 |
| FSNIA | HIV-1 p24 | 10 | Present study |
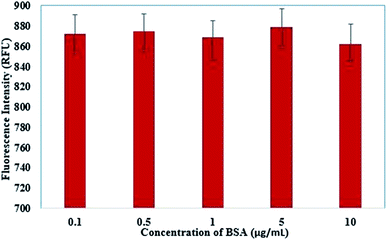
3.6 Control experiment
3.7 Optimization of FSNIA
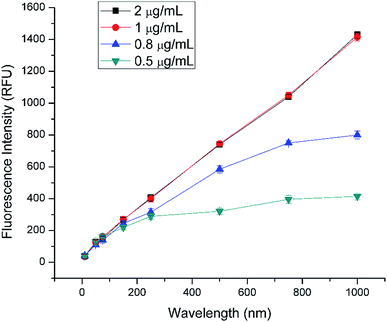
The optimization of immunoassay was performed by optimizing two critical parameters in the immunoassay: capture antibody concentration and FSN-SA concentration.3.7.1.1 Optimization of the capture antibody concentration. A very important parameter that affects the sensitivity of the immunoassay is the concentration of capture antibodies. There is an optimal concentration of capture antibody at which the signal will be highest; at higher concentrations the background signal can also be higher and concentrations below this, the signal will be weaker.69 Optimization of concentration of capture antibody required for the assay was done by testing different concentrations of 0.5 μg mL−1, 0.8 μg mL−1, 1 μg mL−1 and 2 μg mL−1. As shown in Fig. 8, for 0.5 μg mL−1 and 0.8 μg mL−1 the signal response was poor and there was no good correlation between the observed signal and antibody concentration. However, at 1 μg mL−1, there was a strong response observed. For concentration higher than 1 μg mL−1, there was no change in the response which can be attributed to the saturation of the detection area with capture antibodies at 1 μg mL−1. As a result, 1 μg mL−1 was chosen as the optimal concentration of capture antibody for further experimentation.
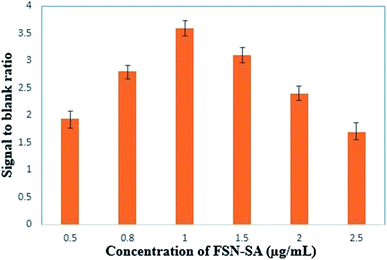
3.7.2.1 Optimization of the streptavidin conjugated FSNPs concentration. Concentration of FSN- SA greatly affects the performance of the assay. Six concentrations of FSN-SA: 0.5 mg mL−1, 0.8 mg mL−1, 1 mg mL−1, 1.5 mg mL−1, 2 mg mL−1, 2.5 mg mL−1, were taken to study its effect on the signal to blank ratio (S/B ratio) in FSNIA. The blank measurements were obtained by performing the immunoassay in absence of the antigen. The optimization of FSN-SA was carried out by measuring and calculating the S/B ratio for concentration of 250 pg mL−1 of p24 antigen. It is clear from Fig. 9 that no linear relation exists between the FSN-SA concentration and the S/B ratio. At a concentration 1.5 mg mL−1 of FSN-SA the highest S/B ratio was observed. With an increase in FSN-SA concentration beyond the optimal concentration the blank intensity was also observed to have increased rapidly when compared to the signal intensity. This increase of blank intensity could be attributed to the non-specific absorption of FSN-SA.
3.8 Precision and recovery rate of the assay
Our next objective was to study the applicability of FSNIA in real world scenario by testing human plasma samples. The first step was to determine the sensitivity of the assay which was done by testing 50 plasma samples of healthy individuals. All the samples tested negative and no false positives were observed. The next step was to further evaluate the analytical sensitivity of the assay and this was done by taking known concentrations of p24 antigen and spiking the HIV negative plasma samples collected from healthy individuals (HIV negative sample). The plasma samples were diluted 100 times which in turn reduced the sample amount required for analysis. To the diluted plasma sample, purified p24 antigen concentrations of 10, 50, 150, 250 and 500 pg mL−1 were added. All experiments were conducted in triplicate and the concentration was calculated by taking the average signal response and comparing it with the standard curve (Fig. 5a). It is clear from Table 5 that the concentrations of p24 antigen calculated from the results of FSNIA are comparable to the actual amounts spiked into sample. Thus FSNIA can quantitatively determine the concentration of HIV-1 p24 antigen in clinical plasma samples.| S. no | p24 spiked (pg mL−1) | p24 detected (pg mL−1) | Recovery (%) |
|---|---|---|---|
| 1 | 10 | 9.2 ± 0.5 | 92.0 ± 5.0 |
| 2 | 50 | 47.6 ± 2.1 | 95.2 ± 4.2 |
| 3 | 75 | 71.3 ± 4.5 | 95.0 ± 4.3 |
| 4 | 150 | 145.1 ± 5.8 | 96.7 ± 3.9 |
| 5 | 250 | 242.7 ± 6.5 | 97.1 ± 2.7 |
| 6 | 500 | 482.3 ± 15.9 | 96.4 ± 3.3 |
| 7 | 750 | 740.0 ± 19.6 | 98.6 ± 2.8 |
| 8 | 1000 | 989.2 ± 39.4 | 98.9 ± 3.9 |
3.9 Clinical serum sample analysis
To further establish the effectiveness of FSNIA, we tested the specificity of FSNIA using samples from infected individuals (HIV positive samples). Plasma samples were diluted 100 times prior to the analysis. We tested 50 HIV positive samples (which were confirmed by 3rd and 4th generation commercial ELISA kits) using FSNIA and no false negatives were observed. To test FSNIA for cross reactivity we also evaluated the assay with 10 HBV positive/HIV negative and 10 HCV positive/HIV negative plasma samples for cross reactivity. We observed that the signal intensity from these samples were as low as the negative control. The comparison of the signals from different plasma samples is presented in Fig. 10a and b. Thus, we could confirm that the assay is very specific to p24 antigen and is not prone to cross reactivity effects from other biomolecules. As clearly evident from the Table 6 the FSNIA has very high specificity and low cross reactivity.| S. no | Sample type | No. of samples | No. of samples tested to be HIV positive | No. of samples tested to be HIV negative |
|---|---|---|---|---|
| 1 | HIV positive | 50 | 50 | NIL |
| 2 | HIV negative | 50 | NIL | 50 |
| 3 | HIV negative/HBV positive | 10 | NIL | 10 |
| 4 | HIV negative/HCV positive | 10 | NIL | 10 |
Thus, we have successfully demonstrated the detection of picogram level concentration of HIV-1 p24 antigen using FSNIA. Our findings confirm that fluorescent nanoparticle based immunoassays could dramatically increase detection sensitivity and have applications in improved diagnosis and point-of-care use. Due to high specificity of streptavidin conjugated silver nanoparticles which have been confirmed by the in silico studies, FSNIA could be developed into a universal labeling technology by replacing capture and detection antibodies for detection of various other antigens. There is no need for specific training of the technicians to handle FSNIA as it is similar to traditional ELISA which is widely practiced across various laboratories and clinics. Upon further optimization and simplification, this could be developed into a rapid and ultrasensitive testing platform for clinical diagnosis and laboratory research in resource limited settings.
4 Conclusion
Our work has shown the use of fluorescent silver nanoparticles in infectious disease assay development can result in enhanced assay sensitivity. Although there are similar assays, this assay is capable of detecting HIV p24 antigen in the pg mL−1 range which will be useful in detecting early HIV infection. The findings described in this paper are important for those developing highly sensitive assays for the estimation of HIV incidence and detection of acute infection cases as part of the strategy towards an AIDS-free generation. This highly sensitive p24 assay can also be adapted to microfluidic platforms and be made available as point of care technology which can help improve blood safety by reducing the antibody negative window period in blood donors in resource limited settings where nucleic acid testing is not practical or feasible.Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Food and Drug Administration, U.S. Department of Health and Human Services.Statement of informed consent
The authors declare that all the experiments that were performed are in compliance with the guidelines of the Institutional Ethics Committee (IEC) of Sri Sathya Sai Institute of Higher Learning, Prasanthi Nilayam. The study was approved by the IEC under the study id SSSIHL/IEC/PSN/BS/2012/01. The plasma samples used in the study were obtained from the patients with their informed consent.Acknowledgements
All authors are grateful to Bhagawan Sri Sathya Sai Baba for his constant inspiration and guidance. Aditya Kurdekar thanks DST for their support through the DST-INSPIRE Fellowship program, Ministry of Science and Technology, Government of India. L. A. Avinash Chunduri acknowledges UGC, Government of India for BSR fellowship. The authors thank the Sri Sathya Sai Institute of Higher Medical Sciences for providing us the plasma samples for clinical analysis. They also wish to acknowledge with gratitude Dr B. K. Panigrahi for providing the TEM facility for characterization.References
- Infectious diseases: Global Killers [Internet], 2016 [cited 2016 Sep 9], Available from: http://needtoknow.nas.edu/id/threats/global-killers/.
- S. T. Sanjay, G. Fu, M. Dou, F. Xu, R. Liu and H. Qi, et al., Biomarker detection for disease diagnosis using cost-effective microfluidic platforms, Analyst, 2015, 140(21), 7062–7081 RSC.
- AIDS FACT SHEET 2016 (Internet), 2016 [cited 2016 Sep 9], available from: http://www.unaids.org/en/resources/fact-sheet.
- WHO HIV/AIDS Fact sheet [Internet], 2016 [cited 2016 Jan 1], available from: http://www.who.int/mediacentre/factsheets/fs360/en/.
- M. Février, K. Dorgham and A. Rebollo, CD4+ T Cell Depletion in Human Immunodeficiency Virus (HIV) Infection: Role of Apoptosis, Viruses, 2011, 3(12), 586–612 CrossRef PubMed.
- R. D. Moore and R. E. Chaisson, Natural history of HIV infection in the era of combination antiretroviral therapy, AIDS, 1999, 13(14), 1933–1942 CrossRef CAS PubMed.
- J. Hurst, M. Hoffmann, M. Pace, J. P. Williams, J. Thornhill and E. Hamlyn, et al., Immunological biomarkers predict HIV-1 viral rebound after treatment interruption, Nat. Commun., 2015, 6, 8495 CrossRef CAS PubMed.
- J. D. Stekler, P. D. Swenson, R. W. Coombs, J. Dragavon, K. K. Thomas and C. A. Brennan, et al., HIV Testing in a High-Incidence Population: Is Antibody Testing Alone Good Enough?, Clin. Infect. Dis., 2009, 49(3), 444–453 CrossRef PubMed.
- F. Oehlenschläger, P. Schwille and M. Eigen, Detection of HIV-1 RNA by nucleic acid sequence-based amplification combined with fluorescence correlation spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(23), 12811–12816 CrossRef.
- C. D. Pilcher, K. A. Christopoulos and M. Golden, Public health rationale for rapid nucleic acid or p24 antigen tests for HIV, J. Infect. Dis., 2010, 201(1), S7–S15 CrossRef CAS PubMed.
- R. Hans and N. Marwaha, Nucleic acid testing-benefits and constraints, Asian J. Transfus. Sci., 2014, 8(1), 2 CrossRef PubMed.
- S. Tang, J. Zhao, J. J. Storhoff, P. J. Norris, R. F. Little and R. Yarchoan, et al., Nanoparticle-Based Biobarcode Amplification Assay (BCA) for Sensitive and Early Detection of Human Immunodeficiency Type 1 Capsid (p24) Antigen, JAIDS, J. Acquired Immune Defic. Syndr., 2007, 46(2), 231–237 CrossRef PubMed.
- E. S. Daar, T. Moudgil, R. D. Meyer and D. D. Ho, Transient high levels of viremia in patients with primary human immunodeficiency virus type 1 infection, N. Engl. J. Med., 1991, 324(14), 961–964 CrossRef CAS PubMed.
- S. Laperche, M. Maniez-Montreuil and A. M. Couroucé, Screening tests combined with p24 antigen and anti-HIV antibodies in early detection of HIV-1, Transfusion Clinique et Biologique: Journal de La Societe Francaise de Transfusion Sanguine, 2000, 7(1), 18s–24s CrossRef.
- J. M. Lange, D. A. Paul, H. G. Huisman, F. de Wolf, H. van den Berg and R. A. Coutinho, et al., Persistent HIV antigenaemia and decline of HIV core antibodies associated with transition to AIDS, Br. Med. J., 1986, 293(6560), 1459–1462 CrossRef CAS PubMed.
- S. A. Spector, C. Kennedy, J. A. McCutchan, S. A. Bozzette, R. G. Straube and J. D. Connor, et al., The antiviral effect of zidovudine and ribavirin in clinical trials and the use of p24 antigen levels as a virologic marker, J. Infect. Dis., 1989, 159(5), 822–828 CrossRef CAS PubMed.
- E. T. N. Abraham, The use of viral culture and p24 antigen testing to diagnose Human Immunodeficiency virus in neonates, N. Engl. J. Med., 2013, 346(24), 1845–1853 CrossRef PubMed.
- J. Böni, M. Opravil, Z. Tomasik, M. Rothen, L. Bisset and P. J. Grob, et al., Simple monitoring of antiretroviral therapy with a signal-amplification-boosted HIV-1 p24 antigen assay with heat-denatured plasma, AIDS, 1997, 11(6), F47–F52 CrossRef.
- R. M. Lequin, Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA), Clin. Chem., 2005, 51(12), 2415–2418 CAS.
- S. Pang, J. Smith, D. Onley, J. Reeve, M. Walker and C. Foy, A comparability study of the emerging protein array platforms with established ELISA procedures, J. Immunol. Methods, 2005, 302(1–2), 1–12 CrossRef CAS PubMed.
- G. P. Rai and K. S. Venkateswaran, Limitations and Practical Problems in Enzyme Linked Immunosorbent Assays, Def. Sci. J., 1992, 42(2), 71–84 CrossRef CAS.
- L.-M. Lei, Y.-S. Wu, N.-Q. Gan and L.-R. Song, An ELISA-like time-resolved fluorescence immunoassay for microcystin detection, Clin. Chim. Acta, 2004, 348(1–2), 177–180 CrossRef CAS PubMed.
- J.-Y. Hou, T.-C. Liu, G.-F. Lin, Z.-X. Li, L.-P. Zou and M. Li, et al., Development of an immunomagnetic bead-based time-resolved fluorescence immunoassay for rapid determination of levels of carcinoembryonic antigen in human serum, Anal. Chim. Acta, 2012, 734, 93–98 CrossRef CAS PubMed.
- T. B. Martins, N. H. Augustine and H. R. Hill, Development of a multiplexed fluorescent immunoassay for the quantitation of antibody responses to group A streptococci, J. Immunol. Methods, 2006, 316(1–2), 97–106 CrossRef CAS PubMed.
- Direct vs indirect immunofluorescence [Internet], [cited 2016 Sep 15], available from: http://www.abcam.com/secondary-antibodies/direct-vs-indirect-immunofluorescence.
- K. Murray, Y.-C. Cao, S. Ali and Q. Hanley, Lanthanide doped silica nanoparticles applied to multiplexed immunoassays, Analyst, 2010, 135(8), 2132 RSC.
- Y. Tao, M. Li, J. Ren and X. Qu, Metal nanoclusters: novel probes for diagnostic and therapeutic applications, Chem. Soc. Rev., 2015, 44, 8636–8663 RSC.
- X. Yuan, Z. Luo, Y. Yu, Q. Yao and J. Xie, Luminescent Noble Metal Nanoclusters as an Emerging Optical Probe for Sensor Development, Chem.–Asian J., 2013, 8(5), 858–871 CrossRef CAS PubMed.
- N. Goswami, Q. Yao, Z. Luo, J. Li, T. Chen and J. Xie, Luminescent Metal Nanoclusters with Aggregation-Induced Emission, J. Phys. Chem. Lett., 2016, 7(6), 962–975 CrossRef CAS PubMed.
- C. Biology, L. Shang and G. Ulrich, Metal nanoclusters: Protein corona formation and implications for biological applications, Int. J. Biochem. Cell Biol., 2016, 75, 175–179 CrossRef PubMed.
- G. Doria, J. Conde, B. Veigas, L. Giestas, C. Almeida and M. Assunção, et al., Noble Metal Nanoparticles for Biosensing Applications, Sensors, 2012, 12(2), 1657–1687 CrossRef CAS PubMed.
- M. Swierczewska, G. Liu, S. Lee and X. Chen, High-sensitivity nanosensors for biomarker detection, Chem. Soc. Rev., 2012, 41(7), 2641–2655 RSC.
- R. El-Dessouky, M. Georges and H. M. E. Azzazy Silver Nanostructures: Properties, Synthesis, and Biosensor Applications, in Functional Nanoparticles for Bioanalysis, Nanomedicine, and Bioelectronic Devices Volume 1, 2012, pp. 14–359 Search PubMed.
- S. Oldenburg. Silver Nanoparticles: Properties and Applications [Internet], [cited 2016 Sep 15], available from: http://www.sigmaaldrich.com/materials-science/nanomaterials/silver-nanoparticles.html.
- A. Travan, E. Marsich, I. Donati and S. Paoletti, Silver Nanocomposites and Their Biomedical Applications, in Nanotechnologies for the Life Sciences, 2007 Search PubMed.
- P. Dallas, V. K. Sharma and R. Zboril, Silver polymeric nanocomposites as advanced antimicrobial agents: Classification, synthetic paths, applications, and perspectives, Adv. Colloid Interface Sci., 2011, 166(1–2), 119–135 CrossRef CAS PubMed.
- S. A. Latif-ur-Rahman, S. B. Khan, A. M. Asiri, H. Hussain and C. Han, et al., Synthesis, characterization, and application of Au–Ag alloy nanoparticles for the sensing of an environmental toxin, pyrene, J. Appl. Electrochem., 2015, 45(5), 463–472 CrossRef.
- M. Meena Kumari, J. Jacob and D. Philip, Green synthesis and applications of Au–Ag bimetallic nanoparticles, Spectrochim. Acta, Part A, 2015, 137, 185–192 CrossRef CAS PubMed.
- S. Hu, B. Ye, X. Yi, Z. Cao, D. Wu, C. Shen, et al., Dumbbell-shaped metallothionein-templated silver nanoclusters with applications in cell imaging and Hg 2p sensing, Elsevier, Talanta, 2016;vol. 155, pp. 272–7 Search PubMed.
- Y. Yuan, K. Huang, M. Chang, C. Qin and S. Zhang, Dual emission fluorescent silver nanoclusters for sensitive detection of the biological coenzyme NAD +/NADH, Anal. Biochem., 2016, 494, 46–48 CrossRef CAS PubMed.
- X. Yuan, M. I. Setyawati, A. S. Tan, C. N. Ong and D. T. Leong, Highly luminescent silver nanoclusters with tunable emissions: cyclic reduction – decomposition synthesis and antimicrobial properties, NPG Asia Mater., 2013, 5(2), e39–8 CrossRef CAS.
- Z. Parang, A. Keshavarz, S. Farahi, S. M. Elahi, M. Ghoranneviss and S. Parhoodeh, Fluorescence emission spectra of silver and silver/cobalt nanoparticles, Sci. Iran., Trans. A, 2012, 19(3), 943–947 CrossRef CAS.
- M. Padma, B. Govindh and B. V. Rao, Synthesis & Characterization of Fluorescent Silver Nanoparticles stabilized by Tinospora Cordifolia leaf Extract-A Green Procedure, Int. J. Eng. Res. Ind. Appl., 2014, 4(9), 100–107 Search PubMed.
- N. Khatoon, R. Ahmad and M. Sardar, Robust and fluorescent silver nanoparticles using Artemisia annua: Biosynthesis, characterization and antibacterial activity, Biochem. Eng. J., 2015, 102, 91–97 CrossRef CAS.
- L. Shang, S. Dong and G. U. Nienhaus, Ultra-small fluorescent metal nanoclusters: Synthesis and biological applications, Nano Today, 2011 Aug, 6(4), 401–418 Search PubMed.
- F. Qu, Q. Li and J. You, Fluorescent silver nanoclusters capped by polyethyleneimine with different molecular weights: Universal synthesis and application as a temperature sensor, J. Lumin., 2016, 177, 133–138 CrossRef CAS.
- J. Xu and C. Wei, The aptamer DNA-templated fluorescence silver nanoclusters: ATP detection and preliminary mechanism investigation, Biosens. Bioelectron., 2016, 422–427 Search PubMed.
- A. Contino, G. Maccarrone, M. Zimbone, R. Reitano, P. Musumeci and L. Calcagno, et al., Tyrosine capped silver nanoparticles: A new fluorescent sensor for the quantitative determination of copper (II) and cobalt (II) ions, J. Colloid Interface Sci., 2016, 462, 216–222 CrossRef CAS PubMed.
- C. Wang, L. Xu, Y. Wang, D. Zhang, X. Shi and F. Dong, et al., Fluorescent Silver Nanoclusters as Effective Probes for Highly Selective Detection of Mercury(II) at Parts-per-Billion Levels, Chem.–Asian J., 2012, 7(7), 1652–1656 CrossRef CAS PubMed.
- P. Yousefpour, F. Atyabi, R. Dinarvand and E. Vasheghani-Farahani, Preparation and comparison of chitosan nanoparticles with different degrees of glutathione thiolation, Daru, 2011, 19(5), 367–375 CAS.
- Molecular Operating Environment [Internet], 2016 [cited 2016 Sep 9], available from: https://www.chemcomp.com/MOE-Molecular_Operating_Environment.htm.
- Hex Protein Docking [Internet], 2013 [cited 2016 Sep 6], available from: http://hex.loria.fr/.
- D. M. Jameson and J. A. Ross, Fluorescence Polarization/Anisotropy in Diagnostics and Imaging, Chem. Rev., 2010, 110(5), 2685–2708 CrossRef CAS PubMed.
- L. Maretti, P. S. Billone, Y. Liu, J. C. Scaiano. Facile Photochemical Synthesis and Characterization of Highly Fluorescent Silver Nanoparticles. 2009;(28):13972–13980.
- K. Jia, P. Wang and X. Liu, Facile synthesis of luminescent silver nanoparticles and fluorescence interactions with blue-emitting polyarylene ether nitrile, J. Mater. Chem. C, 2015, 3(15), 3522–3529 RSC.
- B. A. Ashenfelter, A. Desireddy, S. H. Yau, T. Goodson and T. P. Bigioni, Fluorescence from Molecular Silver Nanoparticles, J. Phys. Chem. C, 2015, 119(35), 20728–20734 CAS.
- S. Pattanayak, A. Priyam and P. Paik, Facile tuning of plasmon bands in hollow silver nanoshells using mild reductant and mild stabilizer, Dalton Trans., 2013, 42(29), 10597–10607 RSC.
- C. Zhu, J. Zhai and S. Dong, Bifunctional fluorescent carbon nanodots: green synthesis via soy milk and application as metal-free electrocatalysts for oxygen reduction, Chem. Commun., 2012, 48(75), 9367 RSC.
- S. K. Balavandy, K. Shameli, D. R. B. A. Biak and Z. Z. Abidin, Stirring time effect of silver nanoparticles prepared in glutathione mediated by green method, Chem. Cent. J., 2014, 8(1), 11 CrossRef PubMed.
- K. Shameli, A. M. Bin, S. D. Jazayeri, S. Sedaghat, P. Shabanzadeh and H. Jahangirian, et al., Synthesis and characterization of polyethylene glycol mediated silver nanoparticles by the green method, Int. J. Mol. Sci., 2012, 13(6), 6639–6650 CrossRef CAS PubMed.
- M. J. E. Fischer, in Amine Coupling Through EDC/NHS: A Practical Approach BT - Surface Plasmon Resonance: Methods and Protocols, ed. J. N. Mol and E. M. J. Fischer, Humana Press, Totowa, NJ, 2010, pp. 55–73 Search PubMed.
- G. T. Hermanson, in Bioconjugate Techniques, Academic Press, Boston, 3rd edn, 2013, p. xvii. available from: http://www.sciencedirect.com/science/article/pii/B9780123822390000388 Search PubMed.
- W. A. Lea and A. Simeonov, Fluorescence polarization assays in small molecule screening, Expert Opin. Drug Discovery, 2011, 6(1), 17–32 CrossRef CAS PubMed.
- Fluorescence Polarization [Internet], 2010 [cited 2016 Feb 9], available from: https://www.thermofisher.com/in/en/home/references/molecular-probes-the-handbook/technical-notes-and-product-highlights/fluorescence-polarization-fp.html.
- A. Bruno, C. de Lisio and P. Minutolo, Time resolved fluorescence polarization anisotropy of carbonaceous particles produced in combustion systems, Opt. Express, 2005, 13(14), 5393 CrossRef CAS PubMed.
- E. W. Fiebig, D. J. Wright, B. D. Rawal, P. E. Garrett, R. T. Schumacher and L. Peddada, et al., Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection, AIDS, 2003, 17(13), 1871–1879 CrossRef PubMed.
- C. L. Saltzman, J. E. Herzenberg and W. A. Phillips, What nononcologists need to know about MEN IIb, Oncology, 1988, 2(6), 15 CAS.
- A. Shrivastava and V. Gupta, Methods for the determination of limit of detection and limit of quantitation of the analytical methods, Chron. Young Sci., 2011, 2(1), 21 CrossRef.
- J. Liu, B. Du, P. Zhang, M. Haleyurgirisetty, J. Zhao and V. Ragupathy, et al., Development of a microchip Europium nanoparticle immunoassay for sensitive point-of-care HIV detection, Biosens. Bioelectron., 2014, 61, 177–183 CrossRef CAS PubMed.
- S. Tang and I. Hewlett, Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen, J. Infect. Dis., 2010, 201(1), S59–S64 CrossRef CAS PubMed.
- R. A. Dar, N. G. Khare, D. P. Cole, S. P. Karna and A. K. Srivastava, Green synthesis of a silver nanoparticle–graphene oxide composite and its application for As(III) detection, RSC Adv., 2014, 4(28), 14432 RSC.
- L. Shang and S. Dong, Sensitive detection of cysteine based on fluorescent silver clusters, Biosens. Bioelectron., 2009, 24(6), 1569–1573 CrossRef CAS PubMed.
- T. Endo, K. Kerman, N. Nagatani, H. M. Hiepa, D.-K. Kim and Y. Yonezawa, et al., Multiple Label-Free Detection of Antigen−Antibody Reaction Using Localized Surface Plasmon Resonance-Based Core−Shell Structured Nanoparticle Layer Nanochip, Anal. Chem., 2006, 78(18), 6465–6475 CrossRef CAS PubMed.
- X. Wang, L. Wu, J. Ren, D. Miyoshi, N. Sugimoto and X. Qu, Label-free colorimetric and quantitative detection of cancer marker protein using noncrosslinking aggregation of Au/Ag nanoparticles induced by target-specific peptide probe, Biosens. Bioelectron., 2011, 26(12), 4804–4809 CrossRef CAS PubMed.
- S. Paul, D. Paul, G. R. Fern and A. K. Ray, Surface plasmon resonance imaging detection of silver nanoparticle-tagged immunoglobulin, J. R. Soc., Interface, 2011, 8(61), 1204–1211 CrossRef CAS PubMed.
- Z. Chen, Y. Lei and X. Chen, Immunoassay for serum alpha-fetoprotein using silver nanoparticles and detection via resonance light scattering, Microchim. Acta, 2012, 179(3–4), 241–248 CrossRef CAS.
- S. Sun, M. Yang, Y. Kostov and A. Rasooly, ELISA-LOC: lab-on-a-chip for enzyme-linked immunodetection, Lab Chip, 2010, 10(16), 2093 RSC.
- B. S. Lee, J.-N. Lee, J.-M. Park, J.-G. Lee, S. Kim and Y.-K. Cho, et al., A fully automated immunoassay from whole blood on a disc, Lab Chip, 2009, 9(11), 1548 RSC.
- M. Liu, Z. Wang, S. Zong, R. Zhang, D. Zhu and S. Xu, et al., SERS-based DNA detection in aqueous solutions using oligonucleotide-modified Ag nanoprisms and gold nanoparticles, Anal. Bioanal. Chem., 2013, 405(18), 6131–6136 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Chemistry of synthesis of FSNPs, preliminary computational studies on bioconjugation and components of immunoassay. See DOI: 10.1039/c6ra28737a |
| This journal is © The Royal Society of Chemistry 2017 |