DOI:
10.1039/C5MB00412H
(Paper)
Mol. BioSyst., 2016,
12, 48-58
Insights into the molecular interactions of thymoquinone with histone deacetylase: evaluation of the therapeutic intervention potential against breast cancer†
Received
19th June 2015
, Accepted 12th October 2015
First published on 12th October 2015
Abstract
Many HDAC inhibitors have passed through the gateway of clinical trials. However, they have limited therapeutic implications due to their pleiotropic pharmaceutical properties and off-target effects. In view of this, dietary active phytochemicals were evaluated. Based upon the chemical and structural insights of HDAC active pockets, thymoquinone (TQ) was investigated to uncover its active participation in HDAC inhibition. The synergistic analysis of docking and molecular dynamics simulation disclosed the elementary interaction and stability of TQ with human HDACs. The in silico findings were corroborated with an in vitro analysis, demonstrating the efficient role of TQ in the attenuation of global HDAC activity. Furthermore, TQ also elicited downstream effects of HDAC inhibition: reactivation of HDAC target genes (p21 and Maspin), induction of the pro-apoptotic gene Bax, down regulation of the anti-apoptotic gene Bcl-2 and arrest of the cell cycle at the G2/M phase. Finally, the result of a higher cytotoxicity of TQ towards MCF-7 breast cancer cells in comparison to normal cells indicates the potential of TQ to be an anticancer drug.
1. Introduction
The process for the acetylation and deacetylation of histones presents two sides of the same coin, regulating the activity of chromatin and a plethora of other cellular activities. Several studies have envisaged a role of HDACs in the development and progression of human cancer. These HDACs undergo an active participation in gene silencing in the context of cell cycle regulation, differentiation, apoptosis, angiogenesis, invasion, adhesion and metastasis.1,2 Thus, inhibitors against these enzymes are emerging as a promising class of anticancer agents that have been tested for the treatment of multifarious malignancies. As of now, romidepsin and vorinostat are recognized as US Food and Drug Administration approved drugs for cutaneous T-cell lymphoma, while romidepsin alone is approved for relapsed peripheral T-cell lymphoma. Indeed, most HDAC inhibitors (HDACis) are still under different phases of preclinical and clinical investigations against various relapsed and refractory lymphoid malignancies, myeloid leukemia, and solid tumors.3
Unfortunately, a higher clinical dose of synthetic HDACis induces extensive DNA damage and impairs DNA break repair leading to undesired cytotoxicity.4 The most common toxicities evoked by such HDACis include fatigue, diarrhoea and nausea, while cardiac toxicity is the most prevalent among them all. Nevertheless, the usual doses of synthetic HDACis might also exert adverse effects including vomiting, anorexia, dehydration, myelosuppression, thrombocytopenia, anemia and pulmonary embolism.5 Besides, these inhibitors are expensive and have low specificity. This brings dietary chemopreventive agents into the lime light, with widespread availability, possible incorporation into diets and negligible toxic effects. Of these, some bioactive phytochemicals have the potential to alter the anomalous expression of cancer related genes by modulating epigenetic events. For example, curcumin from turmeric, epigallocatechin-3-gallate (EGCG) from green tea, procyanidine B2 from grape seeds and sulforaphane (SFN) from broccoli are effective phytoconstituents to alter aberrant DNA methylation and histone modification in different malignancies.6,7
Experimental evidence suggests that diallyl disulfide and SFN increase p21waf1/cip1 expression in human colon cancer cells by inhibiting HDACs.2,8 Likewise, polyphenols are one of the important bioactive dietary components having chemopreventive and therapeutic potential against various diseases. Among them, thymoquinone (2-isopropyl-5-methyl-1,4-benzoquinone) (TQ), the active constituent of Nigella sativa (black cumin) holds potential significance. Numerous cell line and animal experiments have demonstrated the anti-inflammatory and chemopreventive nature of TQ in different diseases. It has been reported to have antineoplastic activities in multiple forms of cancer including, breast cancer, osteosarcoma, prostate cancer, myeloblastic leukemia, squamous cell carcinoma and gastric carcinoma.9–12 Additionally, it has also been demonstrated to elicit antitumor activity in mouse tumor models xenografted with breast,13 colon14 and prostate cancer.15 In view of the above perspectives, we intended to investigate the dietary supplement TQ. Several studies have contributed to the explanation of the anticancer effects of TQ, which is attributed to the induction of PPAR-γ (peroxisome proliferator-activated receptor gamma), PTEN (phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase) expression, as well as p53-dependent and independent apoptosis.9,10,16
HDAC inhibitors have been reported to upregulate the expression of multiple genes, including the p21waf1/cip1 gene and GADD45, associated with the inhibition of proliferation, induction of differentiation and/or apoptosis of transformed cells.2,8,17 Remarkably, TQ has also been demonstrated to induce apoptosis via overexpression of the p21waf1/cip1 gene in doxorubicin resistant breast cancer cells and osteosarcoma cells.10,16 In view of this, we sought to investigate the HDAC inhibition activity of TQ. Furthermore, chemical and structural insights of the HDAC active site and the information on existing HDAC inhibitors compelled us to proclaim the inhibitory role of TQ against HDACs. Hence, in the present study, we have attempted to illuminate the elementary interactions of TQ with HDAC by an in silico approach which is complemented by an in vitro cell culture study to investigate the consequence of such interactions. We also aimed to compare the cytotoxic nature of TQ towards breast cancer cells and normal keratinocytes to establish its toxicity to cancer cell lines compared to normal cell lines.
2. Materials and methods
2.1.
In silico approach to study inter-molecular interactions between thymoquinone and HDACs
2.1.1. Multiple sequence alignment.
Multiple sequence alignment was carried out to study the homology of HDAC1 and HDAC2 protein with the model organism Aquifex aeolicus. Amino acid sequences for the respective proteins were retrieved from the Uniprot database bearing the accession numbers Q13547, Q92769 and O67135. The sequence alignment was carried out by the web based application CLUSTALW. The analysis was based on the BLOSUM substitution matrix with a gap penalty of 10 and an extension of 0.2.
2.1.2. Structure preparation for the protein and ligand.
The X-ray crystallographic structures of human HDAC1 and HDAC2 (PDB 4BKX, 3MAX) co-crystallized with Zn2+ and other ligand molecules at resolutions of 3.0 and 2.05 Å, respectively, were retrieved from the RCSB Protein Data Bank.18 The hetero-atoms other than zinc ion (other than the active site) were edited using chimera software. The hydrogen atoms were added to calculate the initial partial Gasteiger charge; however the non-polar hydrogens were merged later on. The structures of both HDACs were optimized using the GROMOS 96.1 (43A2) force field within the GROMACS 4.5 software package.19 The complete protein structures were enclosed in a cubic box with 4.0 Å edge lengths. This cubic box was solvated with a water model and Cl− ions were added for charge neutralization. The solvated box was subjected to an energy minimization of steepest decent for a maximum force of <1000.0 kJ mol−1. The optimized structures of HDACs were subjected to a molecular docking study.
The ligands included in the docking analysis were TSA, SFN and TQ. The 3D co-ordinates of these ligands were obtained from the ChEBI database (http://www.ebi.ac.uk/chebi) (Fig. 1). These ligands hold the CHEBI identification numbers 46024, 47807 and 19371, respectively.
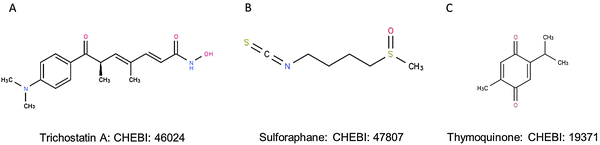 |
| | Fig. 1 Chemical structures of (A) trichostatin A, (B) sulforaphane and (C) thymoquinone retrieved from CHEBI. | |
2.1.3. Autodock.
A molecular docking study was carried out with the help of the automated docking program, Autodock 4.0.20 Docking analysis was based upon the Lamarckian Genetic Algorithm (LGA) and a population size of 150 combinations was considered for grid-energy evaluation and generation of grid maps. In the preliminary step of protein preparation, the associated water molecules were removed and the structure containing a single conformation was taken for further study. Autogrid was set at the docking area for all ligands at grid points of 0.37 Å. Similar structures were grouped into one cluster on the basis of the root mean square deviation (RMSD) and orientation. The docking energy was computed as the sum of the intermolecular and internal energies. In the final instance, the best orientation or the pose with the lowest estimated energy (ΔG) was chosen from each cluster.
2.1.4. Molecular dynamics (MD) simulations.
In order to study the stability of the docked conformation, a molecular dynamics study was carried with the help of the GROMACS 4.5.4 package using the GROMOS96 43a1 force field. The docked complex having the lowest binding energy was subjected to the MD simulation study. The protein topology parameter was assigned based upon the gromacs force field while the ligand topology details were generated using a Dundee PRODRG server.21 HDAC–inhibitor complexes were placed in the cubic box of a simple point charge water molecule. Overall the system containing the HDAC–inhibitor complex was neutralized by adding Cl− ion. The energy minimization was carried out by following the steepest descent and conjugate gradient method in order to release conflicting contacts. The step for energy minimization was followed by an equilibration of the system. The complete system protein–inhibitor complex, counter ions and water molecule were subjected to the position-restrained dynamics simulation (NVT and NPT) at 300 K for 100 ps. Finally the system was subjected to a production run at 300 K temperature and 1 bar pressure for 5000 ps. The co-ordinates of each atom were recorded every 100 ps during the MD simulation analysis of the trajectory.
2.2.
In vitro cell culture analyses
2.2.1. Reagents.
Stock solutions of trichostatin A (TSA), sulforaphane (SFN) and thymoquinone (TQ) (Sigma-Aldrich, St. Louis, MO, USA) were prepared in dimethylsulphoxide (DMSO) (Sigma-Aldrich, St. Louis, MO, USA), stored at −20 °C and diluted in fresh medium just before use. Real time PCR primers of p21, Maspin, Bax and Bcl-2 were obtained from Sigma-Aldrich, St. Louis, MO, USA. For Western blot analysis, the following antibodies were used: rabbit polyclonal anti-p21, rabbit polyclonal anti-Bax, rabbit polyclonal anti-Bcl-2, and mouse monoclonal anti-beta actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA). For immunofluorescence, rabbit polyclonal anti-p21 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used.
2.2.2. Cell culture.
The human breast cancer cell line MCF7 and normal human keratinocytes HaCat were procured from National Centre for Cell Science (NCCS), Pune, India and were cultured in Minimum Essential Eagle's Medium (MEM) and Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS, Invitrogen), 100 IU ml−1 penicillin and 0.1 mg ml−1 streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. The cells were harvested by trypsinization and the number of living cells was calculated by trypan blue staining (0.2% v/v) using a haemocytometer.
2.2.3. HDAC activity assay.
Total HDAC activity was quantified using the commercially available colorimetric HDAC Activity Assay Kit (BioVision, Mountain View, CA) as per the manufacturer's instructions. In brief, MCF-7 cells exposed to TSA and SFN for 24 h and TQ for 24, 48, 72 h were harvested to prepare a whole cell lysate using RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA). Extracts having 100 μg of proteins from vehicle (DMSO) and drug treated cells were incubated with 10 μl 10× HDAC buffer and 5 μl acetylated lysine side chains containing the HDAC colorimetric substrate for 3 h at 37 °C to initiate the HDAC reaction. A lysine developer was then added, and the mixture was incubated for 30 min at 37 °C. The chromophore was then measured using a micro-plate reader spectrophotometer (Perkin-Elmer, Walthman, MA, USA) at 405 nm. The colorimetric signal produced is directly proportional to the HDAC activity.
2.2.4. Total RNA extraction and analysis of mRNA expression.
The MCF-7 breast cancer cells were treated with TSA, SFN and TQ at their respective IC50 for 24 h. Total RNA was then extracted with TriReagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's recommendations. After that, total RNA (1 μg) was reverse-transcribed with oligodT using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative reverse transcription PCR (qRT-PCR) was performed with a Realplex4 Eppendorf system using SYBR® Green JumpStart™ Taq ReadyMix. After an initial denaturation step at 95 °C for 6 min, 40 cycles were performed including a denaturation step at 95 °C for 15 s, annealing at 53 °C (p21, Maspin) and 59.8 °C (Bax, Bcl-2) for 30 s, and extension at 72 °C for 60 s. GAPDH amplification was used as a qualitative control. The primer sequences for qRT-PCR analysis of p21, Maspin, Bax, Bcl-2 and GAPDH genes are given in Table 1.
Table 1 List of sequence and product length of the real-time PCR primers
| Gene |
Primer sequence |
Amplicon size (bp) |
| p21 |
5′-TGAGCCGCGACTGTGATG-3′ |
82 |
| 5′-GTCTCGGTGACAAAGTCGAAGTT-3′ |
|
|
| Maspin |
5′-GGAATGTCAGAGACCAAGGGA -3′ |
139 |
| 5′-GGTCAGCATTCAATTCATCCTT-3′ |
|
|
| Bax |
5′-TTCATCCAGGATCGAGCAG-3′ |
94 |
| 5′-CGCTCAGCTTCTTGGTGG-3′ |
|
|
| Bcl-2 |
5′-CCTGTGGATGACTGAGTACC-3′ |
128 |
| 5′-GAGACAGCCAGGAGAAATCA-3′ |
|
|
| GAPDH |
5′-GGAGCGAGATCCCTCCAAAAT-3′ |
197 |
| 5′-GGCTGTTGTCATACTTCTCATGG-3′ |
2.2.5. Protein extraction and Western blot analysis.
80–85% confluent MCF-7 cells were used to prepare cell lysates using RIPA buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP40 and 0.1% sodium dodecyl sulfate) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA) as described previously.22 The protein concentration in the supernatants was estimated by the Bradford method. Aliquots (50 μg) of total protein were subjected to electrophoresis through a 12% SDS-polyacrylamide gel at a constant voltage of 120 V in running buffer. Resolved proteins were then transferred to PVDF membranes (Millipore). The protein containing membranes were blocked with 3% bovine serum albumin (BSA) in phosphate buffered saline containing 0.1% Tween-20 (PBST) for 2 h. Immunostaining was carried out with primary antibody incubation overnight. The blots were then subjected to washing and incubated with the appropriate secondary antibody coupled to horseradish peroxidase (HRP). The SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) was used for visualization of the secondary antibody. The signals from the blots were scanned using the Gel Documentation system (Bio-Rad) and analyzed by ImageJ software. Beta actin was used to confirm equal protein loading.
2.2.6. Immunofluorescence microscopy.
MCF-7 breast cancer cells were plated on sterilized glass coverslips in 6-well dishes and treated with TSA, SFN and TQ at their respective IC50 for 24 h. The cells were then fixed in ice cold methanol and were permeabilized by 0.25% Triton X-100 in phosphate buffered saline (PBST). After blocking in 1% BSA in PBST for 30 min, the cells were incubated overnight at 4 °C in primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in phosphate buffered saline, 1% bovine serum albumin for all antibodies). The cells were rinsed with PBS then incubated with a FITC-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotech).
100 dilution in phosphate buffered saline, 1% bovine serum albumin for all antibodies). The cells were rinsed with PBS then incubated with a FITC-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotech).
2.2.7. Cell-cycle analysis.
MCF-7 cells were plated at 5 × 105 cells per 60 mm dish. The cells were treated with TSA, SFN and TQ at their respective doses for 24 h. After the treatment, cells were harvested by trypsinization and fixed in 70% ethanol. The cells were then collected by centrifugation at 600 × g for 5 min at 4 °C and washed twice with PBS. This was followed by the resuspension of cells in PBS containing 250 U ml−1 RNase for 20 min at room temperature and staining with 100 μg ml−1 propidium iodide (PI). The DNA content was examined with FACSAria (Becton Dickinson, San Jose, CA, USA). Cell-cycle distribution was analyzed using FlowJo (Becton Dickinson) software.
2.2.8. Evaluation of the cytotoxicity of TSA, SFN and TQ using the MTT assay.
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay is a colorimetric detection method. It is a sensitive measure of the cytotoxic effects induced by cancer chemotherapeutic drugs in vitro. MCF7 and HaCat cells (80% confluent) were seeded into 96 well culture plates (8 × 103 cells per well for MCF-7 and 5 × 103 cells per well for HaCat). The drugs TSA (25–200 nM), SFN (1–25 μM) and TQ (10–50 μM) were then incubated in both cell lines. After 24 hours of drug treatment, 100 μl of the MTT solution (0.8 mg ml−1, diluted in serum free culture medium) was added and allowed to incubate for 4 hours in the dark at 37 °C. The black formazans that formed were dissolved with DMSO and were allowed to incubate for 15 minutes in the dark. The absorbance was measured at 570 nm by a micro-plate reader spectrophotometer (Perkin-Elmer, Walthman, MA, USA) and the results were expressed as the mean of three replicates as a percentage of the control (taken as 100%).
2.2.9. Cell morphology and imaging.
MCF7 cells in the logarithmic growth phase were seeded at 3 × 105 cells per ml and treated with TSA, SFN and TQ at their sub-lethal concentration (IC50), in the control group DMSO (0.01%) was added, and incubated for 24 h. Cell morphology was observed and phase contrast images of the cells were taken using an inverted microscope (Olympus IX71, USA).
2.2.10. Chromatin condensation assay.
For the chromatin condensation assay, MCF-7 and HaCat cells (105 cells per well) were seeded in 6 well culture plates and allowed to grow for one day. Then cells were treated with drugs (TSA, SFN and TQ) at their respective IC50 values for 24 h. The cells were stained with Hoechst 33342 dye and images were taken under UV filter using an epi-fluorescent microscope (Olympus IX71, USA) at 400× magnification. Condensed nuclei were counted against the total number of nuclei in the field, and the percentage of apoptotic nuclei were calculated and plotted graphically.
2.2.11. Measurement of DNA damage using the comet assay.
Comet assays were performed under alkaline conditions to determine the amount of double-strand DNA breaks. MCF-7 cells and HaCat cells were subjected to a treatment with TSA, SFN and TQ at their respective IC50 values for 24 h. Then cells were harvested and added to preheated (37 °C) low-melting point agarose. The solution was pipetted onto slides precoated with 1% agarose. The slides were allowed to lyse overnight at 4 °C in an alkaline lysis solution (1.2 M NaCl, 100 mM Na2EDTA, 0.1% sodium lauryl sarcosinate, 0.26 M NaOH, pH > 13) prior to immersion in an alkaline electrophoresis solution (0.03 M NaOH, 2 mM Na2EDTA, pH ∼ 12.3). After 30 min, the slides were placed into horizontal electrophoresis chamber samples for 25 min (0.6 V cm−1). The slides were washed with deionized H2O to remove the alkaline buffer, stained with propidium iodide (10 μg ml−1 stock) and incubated for 20 min. The slides were then washed with water and examined using an epi-fluorescent microscope (Olympus IX71, USA).
2.2.12. Cell migration analysis using the wound-healing assay.
MCF-7 cells and HaCat cells were seeded to form a nearly confluent cell monolayer on the culture surface. Wounds were then made using sterile 200 μl pipette tips and the cell debris was removed by washing with PBS. The wounded cell monolayer was then incubated with TSA, SFN and TQ at their respective IC50 values. After the wound in the control was healed, photographs were taken.
2.2.13. Clonogenic assay.
MCF-7 cells and HaCat cells were plated at a low density (500 cells per well) in 6 well plates and allowed to attach for 24 hours prior to treatment with the drugs. Then cells were treated with TSA, SFN and TQ at their respective doses. Cells were grown until distinct colonies were formed in the untreated controls. Then they were fixed and stained with a mixture of 6.0% glutaraldehyde and 0.5% crystal violet, air dried, photographed and evaluated for colony estimation.
2.2.14. Statistical analyses.
Statistical analyses of the data were performed by t-test using SPSS software. Differences were analyzed for significance by one way ANOVA with a post hoc Tukey HSD (Honestly Significant Difference) test, which tests each sample against the respective control. All data are presented as the mean ± SD. A variation with p < 0.05 was considered to be significant for all experiments. In case of the HDAC activity, a variation with p < 0.01 and p < 0.05 were considered to be significant.
3. Results
3.1.
In silico investigations
3.1.1. HDAC1 and HDAC2 establish homology with Aquifex aeolicus.
The homology in the deacetylase domain for human HDAC1 and HDAC2 proteins was established with the model organism Aquifex aeolicus. In Fig. 2, the highlighted regions in blue indicate the presence of conserved domains between the HDAC proteins and the model organism. The amino-acid sequence highlighted in yellow indicates the presence of similar amino acids in the domain. Owing to the higher homology with the model organism, human HDAC1 and HDAC2 proteins were considered for further investigations in our study.
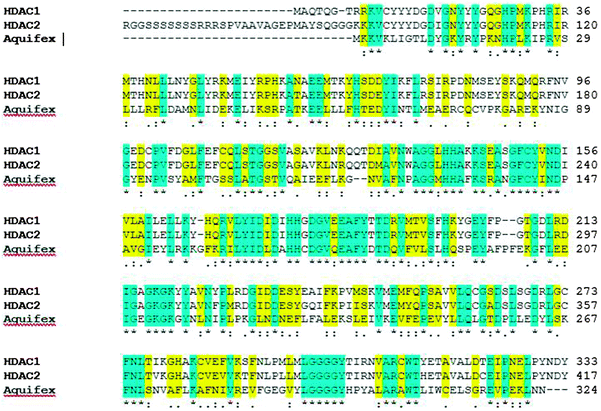 |
| | Fig. 2 Depiction of conserved deacetylase domains among human HDAC proteins (HDAC1 and HDAC2) and with the model organism, Aquifex aeolicus. Blue highlighted regions are conserved domains within human HDAC1, HDAC2 and HDACA. The amino-acid sequence highlighted in yellow indicates the presence of similar amino acids in the domain. | |
3.1.2. Molecular interaction and binding energy of TQ with HDAC1 and HDAC2.
Each classical HDAC shares a common deacetylase domain of approximately 390 amino acids constituting the catalytic site. A narrow cylindrical structure of a length equivalent to that of a 4–6 carbon straight chain with Zn2+ near its base, forms the enzyme pocket of the catalytic domain. The inhibitory potential of TQ was analyzed at the Zn2+ binding site of the HDAC catalytic pocket.23 The interaction with TQ was evaluated in reference to TSA and SFN, two well-known HDAC inhibitors. The binding energy was calculated based upon intermolecular and torsional free energies and the best possible orientation of the ligand was identified. In an interaction with the HDAC1 (4BKX) enzyme, 100% of the conformation of TQ showed an average binding energy of −6.98 kcal mol−1. In contrary, only 50% of the total conformation of TSA bound to the active site pocket with an average binding energy of −7.2 kcal mol−1 (Fig. 3, panel [I] C). Similarly, 80% of the conformation of TQ holds a mean binding energy of −7.2 kcal mol−1, while only 55% of TSA interacts with the active site pocket of the HDAC2 (3MAX) enzyme with a binding energy of −9.2 kcal mol−1 (Fig. 3, panel [II] C). The binding energy of SFN is comparatively less than TQ and TSA (Table 2). The elevation in the average binding energy is characterized by non-covalent interactions, mainly electrostatic, hydrogen bonding and van der Waals interactions. While TSA establishes an average of 7 hydrogen bonds with HDAC1, the higher binding energy with HDAC2 protein is due to its non-covalent interactions of hydrogen bonding, electrostatic and van der Waals interactions. TQ interacts via 3 hydrogen bonds in the active site pocket of both HDAC1 and HDAC2. SFN exhibits less binding due to a distant association of hydrogen bonds and van der Waals interactions. Histidine and aspartate amino residues are identified to actively participate in the interaction. The detailed molecular interactions of the inhibitors with the respective HDACs can be visualized in Fig. 3, panels [I] and [II] B. Owing to the detailed study of enzyme–inhibitor complexes (Fig. 3, panels [I] and [II] A), the stability of complexes was predicted based upon molecular dynamics simulation studies.
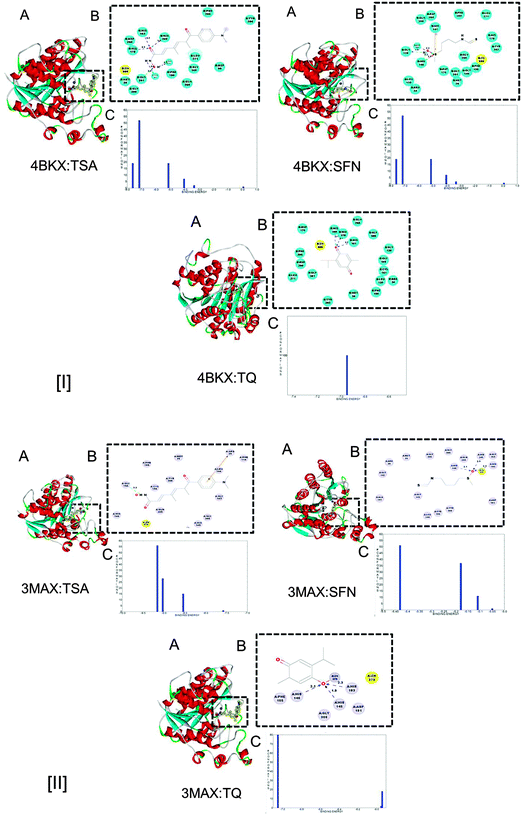 |
| | Fig. 3 (A) Molecular interactions of HDAC1 (4BKX) and HDAC2 (3MAX) with known HDAC inhibitors (TSA, SFN) and the candidate inhibitor (TQ). (B) Detailed molecular interactions between active site amino acid residues of HDAC 1 and 2 with TSA, SFN and TQ. The hydrogen bonds are shown by dotted lines. (C) Binding energy analysis of the docked complex of HDAC1 and 2 with the respective known and candidate inhibitors. | |
Table 2 Binding energy (kcal mol−1) details of HDAC–inhibitor complexes
| Protein |
Ligand |
Binding energy (kcal mol−1) |
| HDAC1 (4BKX) |
TSA |
−7.2 |
| TQ |
−6.98 |
| SFN |
−5.8 |
|
|
| HDAC2 (3MAX) |
TSA |
−9.2 |
| TQ |
−7.2 |
| SFN |
−5.45 |
3.1.3. Characterization of the stability of HDAC–inhibitor complexes by MD simulation.
The molecular dynamics simulation study was carried out to investigate the dynamic behaviour of the ligands (TSA, SFN and TQ) within the pocket of the receptor HDAC and to predict the most reliable receptor–ligand interaction mechanism. The stability of the docked conformation was analysed with respect to the root mean square deviation (RMSD) and average hydrogen-bonding between HDAC–inhibitor complexes. Overall, HDAC1–inhibitor complexes remained stable up to 2 ns, while mean fluctuations of 2.14 Å and 2.43 Å were observed when TSA and TQ interact with the HDAC1 protein. Similarly, TSA and TQ, upon interaction with HDAC2, attain equilibrium with average fluctuations of 2.06 and 2.55 Å. When SFN interacts with HDAC proteins, average deviations of 2.74 and 2.75 Å are attained by the respective protein backbone (Fig. 4) (Table 3). Thus, from the findings, it can be inferred that TQ holds a similar stability with HDACs as that of SFN.
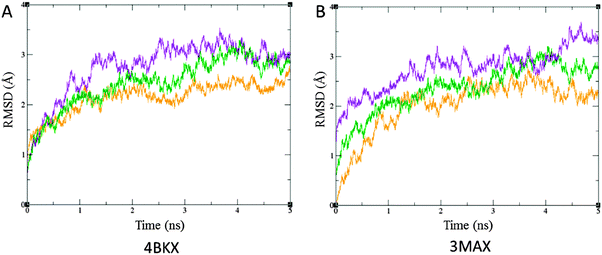 |
| | Fig. 4 RMSD plot with respect to time in ns upon interaction of TSA (orange), SFN (violet) and TQ (green) with, (A) 4BKX and (B) 3MAX in Å. RMSD is calculated for heavy atoms with reference to their respective orientation in the crystal structures. TQ exhibits a similar deviation upon interaction with HDAC and forms a highly stable complex as that of SFN. | |
Table 3 Molecular dynamics simulation analysis in terms of root mean square deviation in (Å) and average hydrogen bonding
| Protein |
Ligand |
RMSD (Å) |
Average hydrogen bonding |
| HDAC1 (4BKX) |
TSA |
2.14 |
3.47 |
| TQ |
2.43 |
2.35 |
| SFN |
2.74 |
2.12 |
|
|
| HDAC2 (3MAX) |
TSA |
2.06 |
5.2 |
| TQ |
2.55 |
2.5 |
| SFN |
2.75 |
2.2 |
The stability of HDAC–inhibitor complexes was also characterised by the binding mode of inhibitors (TSA, SFN and TQ) inside the active site pocket. An average hydrogen bond formation was estimated between HDAC–inhibitor complexes. It is evident from our investigation that TQ forms a higher number of hydrogen bonds with the cognate donor/receptor in the proximity of the active site pocket of the HDAC enzyme. On average 3.47 and 5.2 hydrogen bonds are formed when TSA makes a complex with HDAC1 and HDAC2, respectively. Similarly, TQ interacts with the respective protein with an average of 2.35 and 2.5 hydrogen bonds. SFN also holds a similar mode of interaction as that of TQ (Fig. S1, ESI†) (Table 3).
Thus, MD simulation studies based upon RMSD and average hydrogen bonding affirm that the stability of HDAC–TQ complexes is similar to that of SFN. Later, an in vitro characterization was carried out to uncover the ability of TQ in HDAC inhibition and successfully restore the expression of tumor suppressor genes.
3.2.
In vitro analyses
3.2.1. TQ inhibits HDAC activity in MCF-7 breast cancer cells.
The well-established HDAC inhibitors including TSA, butyrate and SFN modulate histone acetylation levels by inhibiting the activity of HDAC. To examine whether TQ has any role in HDAC inhibition, we analyzed TQ treated MCF7 cells for in vitro HDAC activity. Corroborating our in silico findings, TQ was observed to exhibit HDAC inhibition at its IC50 (34 μM) in a time dependent manner. MCF-7 cells treated with TQ for 24 h were shown to significantly attenuate HDAC activity compared to control (p < 0.05). However, a drastic change in HDAC activity is observed post-treatment with TQ for 48 and 72 h. TSA and SFN exert a similar change in HDAC activity following treatment for 24 h. This change in enzyme activity is found to be significant with a p-value < 0.01 (Fig. 5).
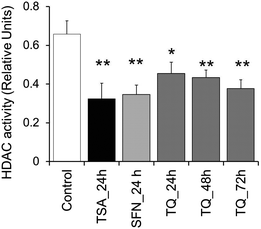 |
| | Fig. 5 TQ exhibits HDAC inhibition activity in MCF7 cells in a time dependent manner. MCF-7 cells were treated with TQ for 24, 48 and 72 h and with TSA and SFN for 24 h. Whole cell protein lysates were prepared and incubated with the acetylated substrate peptide provided by the manufacturer. Statistical analysis of the data was performed using a one way ANOVA with a post hoc Dunnett multiple comparisons test, which compares each sample against the control. Data are expressed as the mean ± S.D., n = 3. Single asterisk (p < 0.05) and double asterisk (p < 0.01) indicate the level of significance. | |
3.2.2. Expression analysis of HDAC target and apoptosis related genes (p21, Maspin, Bax and Bcl-2).
The cell cycle regulatory gene, p21waf1/cip1, and the anti-cell proliferative and apoptosis inducing gene, Maspin, have been shown to be epigenetically governed via histone acetylation.24,25 It has been widely reported that the promoter region of p21waf1/cip1 is negatively controlled by HDAC recruitment at its Sp1 sites.1,26 According to Juan et al., HDAC regulates the acetylation status of p53 which in turn, results in decreased expression of Bax. However, in the presence of the HDAC inhibitor, Bax is overexpressed.27 Hence, it is hypothesized that TQ induces transcription at the p21, Maspin and Bax promoter through down-regulation of HDAC activity similar to other HDAC inhibitors. To examine this, we executed quantitative RT PCR and Western blot analyses. As expected, we found an increased transcript level of p21, Maspin and Bax in TQ treated MCF-7 cells (Fig. 6). The result obtained from Western blot and immunofluorescence microscopy also clearly demonstrated the role of TQ in promoting the expression of p21 at the protein level (Fig. 7 and 8). Moreover, TQ treatment promoted pro-apoptotic Bax expression, while attenuating anti-apoptotic Bcl-2 expression, at both the transcript and protein level compared to control (Fig. 6 and 7).
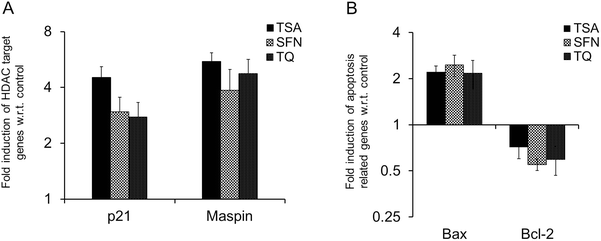 |
| | Fig. 6 Real time RT-PCR analysis of HDAC target genes (A) p21 and Maspin, and apoptosis related genes (B) Bax and Bcl-2 after treatment with TSA, SFN and TQ in MCF-7 cells. The transcript level of p21, Maspin and Bax are upregulated after treatment with TQ as are known HDAC inhibitors (TSA and SFN), while Bcl-2 is downregulated. Data are expressed as the mean ± S.D., n = 3, and p < 0.05. | |
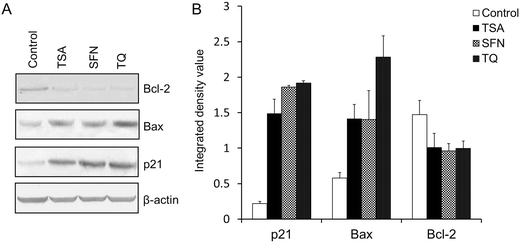 |
| | Fig. 7 (A) Western blot analysis of p21, Bax and Bcl-2 in MCF-7 cells treated with TSA or SFN for 24 h and TQ for 48 h. The protein level of p21 and Bax is enhanced after treatment with TQ like the known HDAC inhibitors (TSA and SFN), while Bcl-2 exhibits a diminished expression. β-Actin was used as the internal control to confirm equal protein loading. (B) A histogram depicting the integrated density value for different bands obtained from the Western blot using ImageJ software. Data are expressed as the mean ± S.D., n = 3, and p < 0.05. | |
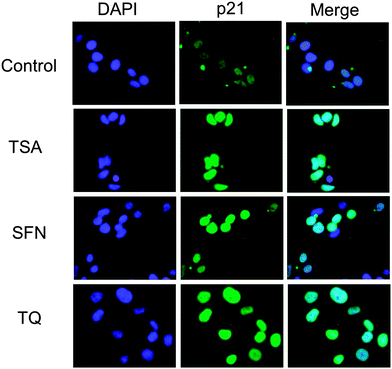 |
| | Fig. 8 Immunofluorescence microscopy was used to detect p21 expression in untreated and TSA, SFN and TQ treated MCF-7 cells. The expression of p21 is enhanced after treatment with TQ. Each of the images is representative of 30 images obtained from three independent experiments. | |
In addition, the HDAC inhibitor dependent increased expression of p21 can also arrest the cell cycle at the G1 and/or G2 phase.
3.2.3. TQ arrests the G2/M phase of the cell cycle.
HDAC inhibitors, including SFN have been demonstrated to disrupt checkpoints during the G2/M phase of the cell cycle.28 In view of this, the effect of TSA, SFN and TQ on breast cancer cell cycle distribution was analyzed by FACS. It is observed that the percentage of cells in the G2M phase reached a value of 33.2% after a 48 h exposure to TQ, which is nearly equal to that of TSA (33.9%) with respect to the control (17.7%) (Fig. 9). This substantiates the active role of TQ in HDAC inhibition.
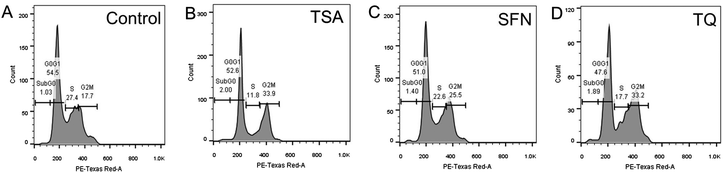 |
| | Fig. 9 Cell cycle analysis of MCF-7 cells exposed to TSA and SFN for 24 h and TQ for 48 h at their respective IC50 in parallel with control (DMSO treated) cells. TQ arrests the MCF-7 cells at the G2/M interphase of cell cycle like TSA and SFN. Shown are the representative images of two independent experiments. | |
Furthermore, in the upcoming analyses, we have demonstrated the cytotoxic effect of TQ in metastatic breast adenocarcinoma MCF-7 cells.
3.2.4. TQ treatments alter cancer cell morphology.
HDACs are important regulators of cell proliferation and differentiation. Interestingly, phase-contrast light microscopy reveals that TQ treatment of MCF-7 cells changed their morphology. Generally, untreated MCF-7 cells have a slightly polygonal and cobblestone-like phenotype. Following incubation with different HDAC inhibitors, the cells undergo distinct morphological changes. During TSA treatment, the cells underwent shrinkage with prominent extensions, while SFN had no considerable phenotypic alterations. Intriguingly, TQ leads to the formation of cytoplasmic vacuoles and the rounding of cells in MCF-7 cells (Fig. S2A, ESI†) in contrast to normal cells (HaCat) (Fig. S2B, ESI†). Moreover, a decreased cell number and the appearance of floating cells revealed the cytotoxic effect of TQ against MCF-7 cells.
3.2.5. TQ exerts a higher cytotoxic effect on breast cancer cells than normal cells.
The cytotoxic property of TSA, SFN and TQ was examined in breast cancer cells as well as in normal keratinocytes (HaCat). A remarkable decline in cell viability was noticed with an increasing concentration of these drugs in MCF-7 cells. Fortunately, in the case of normal cells, TQ elicits no lethal effect as that of TSA and almost all cells remained viable even at 100 μM TQ. For MCF-7 cells, the sub-lethal concentration (IC50) of TSA, SFN and TQ was identified to be 180 nM, 10 μM and 34 μM, respectively. Of note, we observed that SFN at its IC50 has some cytotoxic effect on normal cells, while for TQ there was no trace of such an effect (Fig. S3, ESI†).
3.2.6. TQ induces chromatin condensation and DNA damage in breast cancer cells.
The chromatin condensation assay aids in determining the cell death inducing ability of a drug, which is indicated by the formation of highly condensed and fragmented apoptotic bodies. The number of condensed nuclei in TQ is found to be almost similar to that of SFN in MCF-7 cells in contrast to untreated cells (Fig. S4, ESI†). Moreover, the non-toxic effect of TQ towards normal cells is clearly visible due to the smallest number of condensed nuclei in comparison to TSA.
Furthermore, the comet assay indicates the extent of DNA damage due to apoptosis upon application of any insults to the cells. The tail length denotes the apoptosis inducing ability of drugs. The increased tail length is observed in TQ treated MCF-7 cells similar to TSA and SFN. Conversely, in normal cells TQ induces less DNA damage, as revealed by the short tail length (Fig. S5, ESI†).
3.2.7. TQ inhibits migration and proliferation of breast cancer cells.
The cytotoxic property of the inhibitors is also characterized by their anti-migratory and anti-proliferative potency. In view of this we have seen that TQ holds considerable efficiency to inhibit the migration of MCF-7 cells similar to that of TSA and SFN. In contrast, it does not affect the migration of normal cells (Fig. S6, ESI†).
Similarly, the clonogenic assay reveals the anti-proliferative effect of TQ in MCF-7 cells. It has been observed that TQ effectively inhibits the proliferation of MCF-7 cells, while it is not effective in normal cells. This is clearly revealed by the comparatively lower number of colonies formed in MCF-7 than in HaCaT cells (Fig. S7, ESI†).
Based upon the above findings it is evident that TQ is non-toxic to normal cells and can be successfully implemented in clinical applications.
4. Discussion and conclusion
DNA methylation mediated gene silencing is invariably associated with histone deacetylase activity.29,30 The recognition that HDACs are causally implicated in cancer has exhilarated the development of small-molecule inhibitors as anticancer drugs. Despite the identification of structurally diverse classes of HDAC inhibitors, several of them hold considerable toxicity, unintended off-target effects and poor bioavailability leading to their limited therapeutic implications.31 This led us to explore the possible role of the most commonly consumed dietary phytochemical, thymoquinone (TQ), in HDAC inhibition. Previous studies have explored the anticancer activity of TQ through p53-dependent and independent apoptosis, involving the induction of PPAR-γ, PTEN, NF-κB and AKT.9,10,16,32,33 Gurung et al. explained the anticancer effect of TQ in terms of its ability to induce DNA damage and telomere attrition, by inhibiting telomerase, and cell death.34 However, the molecular mechanism of TQ induced growth inhibition and its pro-apoptotic effects in cancer are still in its infancy.14,34 The present investigation, for the first time, illustrates a plausible insight into this question by demonstrating that TQ inhibits HDAC activity in human breast adenocarcinoma MCF-7 cells.
We began our investigation with an in silico approach using docking analyses followed by a molecular dynamics (MD) simulation study. Known inhibitors of HDACs, TSA and SFN were considered as reference molecules in our analyses. The higher binding affinity of TQ with HDAC1 and HDAC2 compared to SFN and its apparent similarity to that of TSA reveal its potential to inhibit HDACs (Fig. 3, panels [I] and [II] C). Furthermore, the stability of TQ inside the binding pocket of HDACs has been validated by RMSD and average hydrogen bonding (Fig. 4 and Fig. S1, ESI†). The formation of such a stable complex is also underpinned by the fact that TQ undergoes covalent bond formations with the nucleophilic amino acid side chains of Cys and His residues. It occurs through a highly reactive α,β-unsaturated carbonyl group (a Michael acceptor) of TQ.35,36 This clearly shows that TQ forms a stable and energetically favored complex with HDACs. Such property of TQ can be successfully employed in the development of irreversible drugs for the treatment of cancer.37 Corroborating our in silico findings, the in vitro examination explains the potential role of TQ in HDAC inhibition. Generally, enhanced expression of HDACs has been reported in various cancers38 and inhibitors have been identified to successfully impede their activity.1,23 Previously, the phytochemicals, such as butyrate and SFN are known to inhibit enzymatic activity of HDAC.2 Intriguingly, our study has revealed TQ as a potential candidate that ameliorated the total cellular HDAC activity in MCF-7 cells in a time dependent manner. Our observation that TQ is capable of decreasing HDAC activity in treated cells is consistent with the existing HDAC inhibitors (Fig. 5). This finding is further substantiated by the enhanced expression of HDAC target genes upon application of TQ. Previous reports have shown that the p21 promoter is epigenetically repressed by HDAC activity via their Sp1 sites and HDAC inhibitors, such as TSA, apicidin, phenylbutyrate, SAHA and Helminthosporium carbonum toxin, could induce p21 expression in MCF-7 cells.1,26,39 Similarly, the histone hypoacetylation mediated epigenetic silencing of Maspin can be successfully reversed by TSA in MCF-7 cells.25 In agreement with this, we observed that TQ markedly enhanced the expression of p21 and Maspin both at the transcript and protein level. Moreover, we demonstrated that the ability of TQ to restore the expression of these HDAC target genes is almost similar to TSA and SFN treatment. As previously seen by other groups, it was found that upon treatment with known HDAC inhibitors (TSA and SFN), the expression of pro-apoptotic Bax is upregulated and that of anti-apoptotic Bcl-2 is down-regulated.40,41 A consistent expression profile of Bax and Bcl-2 was observed in the case of TQ treated MCF-7 cells (Fig. 6–8). The differential expression profile of the HDAC target and apoptosis related genes are the universal consequences of the downstream effects of the HDAC inhibitor. In our investigation, TQ elicits a similar expression profile; this apparently reflects the HDAC inhibition potential of TQ.
To further support our findings, we analyzed the cell cycle progression of MCF-7 cells treated with TSA, SFN and TQ. In line with previous reports of HDAC inhibitor induced G2/M phase arrest in cancer cells,42,43 we found an increased percentage of cells in the G2/M interface following TSA, SFN and TQ treatment, compared to the untreated control (Fig. 9). It is well known that p21 is implicated in the induction of G1/S phase arrest through its interactions with cyclin E/cdk2, however, in p53 wild type cells, such as LnCaP prostate cancer cells, p21 induction by an HDAC inhibitor has also been demonstrated to induce a G2/M arrest.43 Collectively, TQ mediated HDAC activity attenuation leads to p21 induction, which in turn arrests MCF-7 cells at the G2/M interface ultimately resulting in reduced cell proliferation.
Identification of HDAC inhibitors with a high therapeutic efficacy and minimal cytotoxicity continues to be an unmet challenge. In a quest to address this, after unraveling the role of TQ in HDAC inhibition, our next attempt was to inspect its cytotoxic nature towards normal cells. TQ, at its sub-lethal concentration (IC50 34 μM) induces the formation of cytoplasmic vacuoles in MCF-7 breast cancer cells, while HaCat cells exhibit no obvious abnormalities (Fig. S2, ESI†). This may stem from the cytotoxic property of TQ, as shown previously by Racoma et al. in glioblastoma cells.44 It is also apparent from the cytotoxic MTT assay that TQ, unlike TSA, possesses no cytotoxic effect in normal HaCat cells while it exerts an extensive toxic effect in MCF-7 cells (Fig. S3, ESI†). Supporting these findings, in normal cells, TQ was found to induce less chromatin condensation and DNA damage (Fig. S4 and S5, ESI†), which are hallmarks of apoptosis. In agreement with this finding, TQ was also demonstrated to exert a decreased anti-proliferative and anti-migratory effect in HaCat cells compared to MCF-7 cells (Fig. S6 and S7, ESI†). Thus, our findings illustrate well that TQ exhibits specific cytotoxicity in MCF-7 cells, while eliciting no such effect in normal cells.
In conclusion, this investigation reveals a new role for TQ in inducing an anticancer threat on metastatic breast adenocarcinoma cells via the attenuation of global HDAC activity and the ensuing prevalent epigenetic changes. Indeed, such changes are implicated in the regulation of genes, including p21, Maspin, Bax and Bcl-2, which eventually elicits a strong anticancer effect. Moreover, TQ is a natural dietary component with no obvious cytotoxicity to normal cells. In line with this, further investigations may be executed in experimental models and clinical settings to enlist TQ as a chemo-preventive drug, which may lead to a lasting cure for breast cancer.
Conflicts of interest
The authors declare no competing financial interest.
Acknowledgements
S. Parbin is thankful to DST, Govt. of India for INSPIRE fellowship awarded to her. A. Shilpi, S. Kar, D. Sengupta, M. Deb and S. K. Rath are thankful to NIT-Rourkela for granting them fellowships under the Institute Research Scheme. We apologize to those, whose works and related publications we have not been able to discuss and cite due to space limitations.
References
- M. A. Glozak and E. Seto, Oncogene, 2007, 26, 5420–5432 CrossRef CAS PubMed.
- M. C. Myzak and R. H. Dashwood, Curr. Drug Targets, 2006, 7, 443–452 CrossRef CAS PubMed.
- M. Dokmanovic, C. Clarke and P. A. Marks, Mol. Cancer Res., 2007, 5, 981–989 CrossRef CAS PubMed.
- N. Azad, C. A. Zahnow, C. M. Rudin and S. B. Baylin, Nat. Rev. Clin. Oncol., 2013, 10, 256–266 CrossRef CAS PubMed.
- A. A. Lane and B. A. Chabner, J. Clin. Oncol., 2009, 27, 5459–5468 CrossRef CAS PubMed.
- S. Shukla, S. M. Meeran and S. K. Katiyar, Cancer Lett., 2014, 355, 9–17 CrossRef CAS PubMed.
- A. Shilpi, S. Parbin, D. Sengupta, S. Kar, M. Deb, S. K. Rath, N. Pradhan, M. Rakshit and S. K. Patra, Chem.-Biol. Interact., 2015, 233, 122–138 CrossRef CAS PubMed.
- N. Druesne, A. Pagniez, C. Mayeur, M. Thomas, C. Cherbuy, P. H. Duee, P. Martel and C. Chaumontet, Carcinogenesis, 2004, 25, 1227–1236 CrossRef CAS PubMed.
- C. C. Woo, S. Y. Loo, V. Gee, C. W. Yap, G. Sethi, A. P. Kumar and K. H. Tan, Biochem. Pharmacol., 2011, 82, 464–475 CrossRef CAS PubMed.
- M. Roepke, A. Diestel, K. Bajbouj, D. Walluscheck, P. Schonfeld, A. Roessner, R. Schneider-Stock and H. Gali-Muhtasib, Cancer Biol. Ther., 2007, 6, 160–169 CrossRef CAS PubMed.
- X. Lei, X. Lv, M. Liu, Z. Yang, M. Ji, X. Guo and W. Dong, Biochem. Biophys. Res. Commun., 2012, 417, 864–868 CrossRef CAS PubMed.
- S. Das, K. K. Dey, G. Dey, I. Pal, A. Majumder, S. MaitiChoudhury, S. C. kundu and M. Mandal, PLoS One, 2012, 7, e46641 CAS.
- C. C. Woo, A. Hsu, A. P. Kumar, G. Sethi and K. H. Tan, PLoS One, 2013, 8, e75356 CAS.
- H. Gali-Muhtasib, M. Ocker, D. Kuester, S. Krueger, Z. El-Hajj, A. Diestel, M. Evert, N. El-Najjar, B. Peters, A. Jurjus, A. Roessner and R. Schneider-Stock, J. Cell. Mol. Med., 2008, 12, 330–342 CrossRef PubMed.
- A. O. Kaseb, K. Chinnakannu, D. Chen, A. Sivanandam, S. Tejwani, M. Menon, Q. P. Dou and G. P. Reddy, Cancer Res., 2007, 67, 7782–7788 CrossRef CAS PubMed.
- S. A. Arafa el, Q. Zhu, Z. I. Shah, G. Wani, B. M. Barakat, I. Racoma, M. A. El-Mahdy and A. A. Wani, Mutat. Res., 2011, 706, 28–35 CrossRef PubMed.
- Z. Chen, S. Clark, M. Birkeland, C. M. Sung, A. Lago, R. Liu, R. Kirkpatrick, K. Johanson, J. D. Winkler and E. Hu, Cancer Lett., 2002, 188, 127–140 CrossRef CAS PubMed.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I.
N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- S. Pronk, S. Pall, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess and E. Lindahl, Bioinformatics, 2013, 29, 845–854 CrossRef CAS PubMed.
-
G. M. Morris, R. Huey and A. J. Olson, in Current protocols in bioinformatics, ed. A. D. Baxevanis, et al., 2008, ch. 8, unit 8.14 Search PubMed.
- A. W. Schuttelkopf and D. M. van Aalten, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 1355–1363 CrossRef PubMed.
- M. Deb, D. Sengupta, S. K. Rath, S. Kar, S. Parbin, A. Shilpi, N. Pradhan, S. K. Bhutia, S. Roy and S. K. Patra, Biochim. Biophys. Acta, 2015, 1852, 1630–1645 CrossRef CAS PubMed.
- S. Parbin, S. Kar, A. Shilpi, D. Sengupta, M. Deb, S. K. Rath and S. K. Patra, J. Histochem. Cytochem., 2014, 62, 11–33 CrossRef PubMed.
- A. L. Gartel and A. L. Tyner, Exp. Cell Res., 1999, 246, 280–289 CrossRef CAS PubMed.
- X. H. Liao, Y. Q. Li, N. Wang, L. Zheng, W. J. Xing, D. W. Zhao, T. B. Yan, Y. Wang, L. Y. Liu, X. G. Sun, P. Hu, H. Zhou and T. C. Zhang, Cell. Signalling, 2014, 26, 1335–1346 CrossRef CAS PubMed.
- R. Varshochi, F. Halim, A. Sunters, J. P. Alao, P. A. Madureira, S. M. Hart, S. Ali, D. M. Vigushin, R. C. Coombes and E. W. Lam, J. Biol. Chem., 2005, 280, 3185–3196 CrossRef CAS PubMed.
- L. J. Juan, W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin and C. W. Wu, J. Biol. Chem., 2000, 275, 20436–20443 CrossRef CAS PubMed.
- S. V. Singh, A. Herman-Antosiewicz, A. V. Singh, K. L. Lew, S. K. Srivastava, R. Kamath, K. D. Brown, L. Zhang and R. Baskaran, J. Biol. Chem., 2004, 279, 25813–25822 CrossRef CAS PubMed.
- S. K. Patra and M. Szyf, FEBS J., 2008, 275, 5217–5235 CrossRef CAS PubMed.
- S. Kar, S. Parbin, M. Deb, A. Shilpi, D. Sengupta, S. K. Rath, M. Rakshit, A. Patra and S. K. Patra, Cell. Mol. Life Sci., 2014, 71, 1017–1032 CrossRef CAS PubMed.
- O. H. Kramer, M. Gottlicher and T. Heinzel, Trends Endocrinol. Metab., 2001, 12, 294–300 CrossRef CAS PubMed.
- G. Sethi, K. S. Ahn and B. B. Aggarwal, Mol. Cancer Res., 2008, 6, 1059–1070 CrossRef CAS PubMed.
- T. Yi, S. G. Cho, Z. Yi, X. Pang, M. Rodriguez, Y. Wang, G. Sethi, B. B. Aggarwal and M. Liu, Mol. Cancer Ther., 2008, 7, 1789–1796 CrossRef CAS PubMed.
- R. L. Gurung, S. N. Lim, A. K. Khaw, J. F. Soon, K. Shenoy, S. Mohamed Ali, M. Jayapal, S. Sethu, R. Baskar and M. P. Hande, PLoS One, 2010, 5, e12124 Search PubMed.
- J. M. Chalker, G. J. Bernardes, Y. A. Lin and B. G. Davis, Chem. – Asian J., 2009, 4, 630–640 CrossRef CAS PubMed.
- C. Liao, J. E. Park, J. K. Bang, M. C. Nicklaus and K. S. Lee, ACS Med. Chem. Lett., 2010, 1, 110–114 CrossRef CAS PubMed.
- A. Wissner and T. S. Mansour, Arch. Pharm., 2008, 341, 465–477 CrossRef CAS PubMed.
- S. K. Patra, A. Patra and R. Dahiya, Biochem. Biophys. Res. Commun., 2001, 287, 705–713 CrossRef CAS PubMed.
- V. M. Richon, T. W. Sandhoff, R. A. Rifkind and P. A. Marks, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 10014–10019 CrossRef CAS PubMed.
- L. Wang, D. Liu, T. Ahmed, F. L. Chung, C. Conaway and J. W. Chiao, Int. J. Oncol., 2004, 24, 187–192 Search PubMed.
- J. Ma, X. Guo, S. Zhang, H. Liu, J. Lu, Z. Dong, K. Liu and L. Ming, Mol. Med. Rep., 2015, 11, 4525–4531 CAS.
- S. J. Jackson and K. W. Singletary, J. Nutr., 2004, 134, 2229–2236 CAS.
- S. Roy, K. Packman, R. Jeffrey and M. Tenniswood, Cell Death Differ., 2005, 12, 482–491 CrossRef CAS PubMed.
- I. O. Racoma, W. H. Meisen, Q. E. Wang, B. Kaur and A. A. Wani, PLoS One, 2013, 8, e72882 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mb00412h |
|
| This journal is © The Royal Society of Chemistry 2016 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in phosphate buffered saline, 1% bovine serum albumin for all antibodies). The cells were rinsed with PBS then incubated with a FITC-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotech).
100 dilution in phosphate buffered saline, 1% bovine serum albumin for all antibodies). The cells were rinsed with PBS then incubated with a FITC-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotech).









