 Open Access Article
Open Access ArticleHydrogel scaffolds as in vitro models to study fibroblast activation in wound healing and disease
Megan E.
Smithmyer
a,
Lisa A.
Sawicki
a and
April M.
Kloxin
*ab
aChemical & Biomolecular Engineering, University of Delaware, Newark, DE 19716, USA
bMaterials Science & Engineering, University of Delaware, Newark, DE 19716, USA. E-mail: akloxin@udel.edu
First published on 5th March 2014
Abstract
Wound healing results from complex signaling between cells and their environment in response to injury. Fibroblasts residing within the extracellular matrix (ECM) of various connective tissues are critical for matrix synthesis and repair. Upon injury or chronic insult, these cells activate into wound-healing cells, called myofibroblasts, and repair the damaged tissue through enzyme and protein secretion. However, misregulation and persistence of myofibroblasts can lead to uncontrolled accumulation of matrix proteins, tissue stiffening, and ultimately disease. Extracellular cues are important regulators of fibroblast activation and have been implicated in their persistence. Hydrogel-based culture models have emerged as useful tools to examine fibroblast response to ECM cues presented during these complex processes. In this Mini-Review, we will provide an overview of these model systems, which are built upon naturally-derived or synthetic materials, and mimic relevant biophysical and biochemical properties of the native ECM with different levels of control. Additionally, we will discuss the application of these hydrogel-based systems for the examination of fibroblast function and fate, including adhesion, migration, and activation, as well as approaches for mimicking both static and temporal aspects of extracellular environments. Specifically, we will highlight hydrogels that have been used to investigate the effects of matrix rigidity, protein binding, and cytokine signaling on fibroblast activation. Last, we will describe future directions for the design of hydrogels to develop improved synthetic models that mimic the complex extracellular environment.
A. Introduction
Wound healing is the dynamic and multistage process of replacing injured tissue and is characterized by sequential phases of inflammation, proliferation, and remodeling.1–3 Upon tissue injury, clotting occurs, creating a highly-crosslinked provisional matrix containing fibrin and fibronectin. Next, platelets, and then immune cells, secrete growth factors and cytokines, such as transforming growth factor beta 1 (TGFβ1) and platelet derived growth factor (PDGF). This initiates further wound healing processes, including the recruitment and activation of fibroblasts. Fibroblasts respond to many extracellular cues in the wound environment (Fig. 1A) and are key in players in the synthesis and remodeling of the tissue, replacing the provisional matrix with granulation tissue composed of fibronectin, collagen, and various proteoglycans (e.g., hyaluronic acid and heparan sulfate), as well as contracting the wound.4,5 To accomplish these tasks, fibroblasts proliferate, migrate to the wound site, and activate into myofibroblasts, forming highly-organized cytoskeletons containing α-smooth muscle actin (αSMA) stress fibers to enable contraction and wound closure (Fig. 1B).7 The presence of organized αSMA is considered a conclusive indicator of fibroblast activation.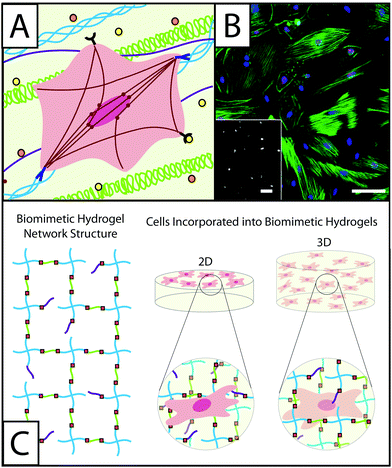 | ||
| Fig. 1 Understanding the role of fibroblasts in healing and disease using hydrogel-based culture models. (A) During normal or misregulated healing in tissues throughout the body, fibroblasts are presented with a variety of signals that affect their function and activation into wound-healing cells, myofibroblasts. These extracellular cues include cytokines, such as TGFβ1 or PDGF (orange and yellow circles), that are released into the ECM by other fibroblasts, immune response cells, and epithelial cells, as well as extracellular matrix proteins, such as collagen (blue triple helix), hyaluronic acid (green), and fibronectin (purple). Interactions between fibroblasts and the temporally changing matrix lead to cytoskeletal reorganization, including expression of α-smooth muscle actin (αSMA) stress fibers (red), as well as alterations in gene expression and secretion of enzymes and proteins for matrix remodeling. The presence of organized αSMA fibers is often used to identify contractile myofibroblasts.5 (B) Traditionally fibroblasts (here, primary rat lung fibroblasts) have been cultured on tissue culture polystyrene, a stiff substrate known to induce activation, as shown here by cells (blue nuclei) that stain positive for αSMA (green stress fibers) and a proliferation marker (red Ki67). Adapted from reference 7. (C) Hydrogel-based culture models afford varying degrees of control of matrix biophysical and biochemical properties to understand their role in healing and disease. Here, a hydrogel with an ideal network structure is depicted with a 4-arm monomer linked by small peptides (green), which can be functionalized to be cell adhesive or proteinase degradable, as well as pendant biofunctional groups (purple). Fibroblasts have been cultured on and within hydrogel scaffolds to understand how individual or combinations of extracellular cues influence their function and fate in healing and disease. | ||
In healthy tissues, myofibroblasts often undergo programmed cell death (apoptosis) after a wound is healed.5 However, in fibrotic tissue, myofibroblast persistence leads to continued ECM deposition, causing tissue stiffening and fibrosis, and eventually loss of organ function.5 While the exact cause of myofibroblast persistence remains unclear, the dynamic interplay between these wound-healing cells and the local ECM has been implicated in fibrosis progression.7 To examine the complex processes of wound healing and fibrosis in vitro, hydrogels have emerged as valuable tools because they allow for the two- and three-dimensional (2D and 3D) culture of fibroblasts in defined extracellular environments with varied degrees of static and temporal property control.
Hydrogels are water-swollen crosslinked polymer networks that are often used as in vitro models to mimic the native ECM. Some hydrogel materials naturally present biological signals inherent to the native ECM while other materials can be engineered to mimic the mechanical properties (e.g., modulus) and biochemical properties (e.g., proteins and peptides) of the ECM. For the culture of fibroblasts in vitro, hydrogels have been formed from naturally-derived native ECM components, such as collagen, fibrin, and hyaluronic acid,8 or synthetic materials functionalized with bioactive moieties, such as polyacrylamide modified with whole ECM proteins9 or poly(ethylene glycol) (PEG) modified with protein mimetic peptides.10 As will be elaborated below (Section B), fibroblasts traditionally have been cultured in fibrin11,12 and type I collagen (collagen I)13,14 to mimic the matrix present during different phases of wound healing. In these models, cell proliferation, activation, matrix synthesis, and contractility have been examined as signs of fibroblast activation. More recently, polysaccharides and synthetic polymers have been functionalized with reactive moieties and bioactive groups to form hydrogels with enhanced control of matrix mechanics and biochemical content (Sections B & C) (Fig. 1C).
The usefulness and versatility of hydrogel in vitro models have allowed for creative and informative studies of fibroblast activation, which are essential for developing an improved understanding of the interplay between the ECM and cell function and fate in healing. For example, the modulus of fibrotic tissue has been observed to be higher than the modulus of healthy tissue in vivo,15 owing to increased deposition and crosslinking of collagen by myofibroblasts; this difference in tissue mechanics is thought to play a role in myofibroblast persistence.7 Recently, several studies in hydrogel model systems have confirmed that matrix stiffness, both initially and over time, does have a significant effect on fibroblast activation within in vitro cell culture, as will be discussed further (Section D). Fibroblasts have responded to increased matrix stiffness by proliferating, secreting collagen, and expressing αSMA fibers.16,17 Conversely, reducing the modulus of a hydrogel from that of diseased tissue (E > 15 kPa) to that of healthy tissue (E < 10 kPa) through matrix degradation resulted in a decreased number of activated fibroblasts.18,19 A variety of hypotheses beyond the role of matrix stiffness have been and can be tested using well-defined hydrogel in vitro models owing to user control over material properties (Sections D & E). In the following sections, we will discuss (B) common materials for the formation and functionalization of hydrogels, and (C) techniques for imparting biologically relevant and biomimetic properties to hydrogel models. Further, (D) recent advances and (E) future directions for the field in utilizing these model systems to understand ECM-related regulation of myofibroblastic activation and persistence will be explored.
B. Base materials for the construction of hydrogels for fibroblast culture
A variety of materials have been used as the base component for hydrogels within in vitro culture models. These materials can be roughly divided into two categories, naturally-derived and synthetic. Generally, naturally-derived materials mimic more of the biophysical and biochemical complexity of tissues than synthetic materials. While it is informative to use complex models, their complexity can present challenges in individually tuning matrix structure (e.g., fibrillar structure), mechanical properties (e.g., modulus), and biological cues (e.g., integrin binding sites) to assess the individual or synergistic effects of each property on fibroblast function.8 Additionally, the purification of ECM components can be difficult, leading to variability between batches of an isolated protein, or undesirable biological signals that can remain in the material as a result of incomplete purification. Synthetic materials offer a higher degree of control, as they are initially biologically inert and can be modified to present relevant physical and chemical ECM cues. However, these materials must contain peptides or proteins that promote cell adhesion and survival, and the resulting synthetic matrix may still lack key elements of the native ECM (e.g., heterogeneous structure, particular integrin binding sites, or appropriate polarization).20 Naturally-derived and synthetic base materials also offer different degrees of property control in time or in space depending on their modification with reactive functionalities. This section will cover a variety of hydrogels based on different naturally-derived or synthetic structural components that have been used in vitro to study fibroblast function and activation in wound healing or fibrosis, including collagen I, fibrin, hyaluronic acid, polyacrylamide, and poly(ethylene glycol) (PEG).Collagen
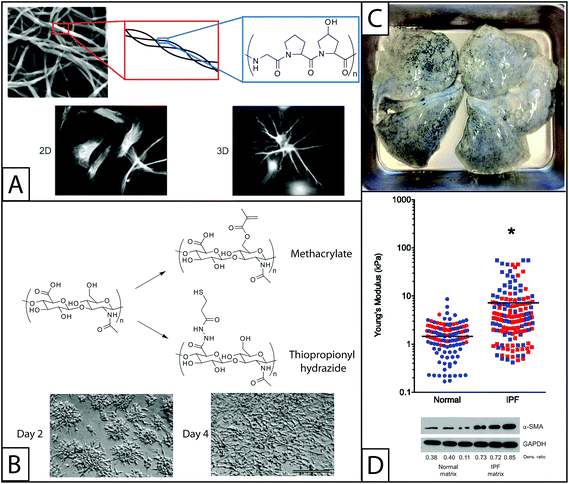 | ||
| Fig. 2 Naturally derived base materials for the construction of hydrogels used in fibroblast culture. (A) Collagen I forms hydrogels through self-assembly at a physiological pH. Here, SEM was used to observe assembled collagen fibrils (top).145 These fibrils are composed of triple helical polymers formed from repeating amino acids sequences, such as GPO (glycine-proline-hydroxyproline) monomer. Collagen has been widely used to coat materials like polyacrylamide for 2D fibroblast studies or as the major component in hydrogels for 3D fibroblast culture. Here, fibroblasts are shown cultured on a coated coverslip (2D) or encapsulated within the gel (3D) (bottom).146 Adapted from references 145 and 146. (B) The pendant carboxylic acid groups of hyaluronic acid are typically functionalized with thiols or methacrylates for polymerization and hydrogel formation for fibroblast culture. The modulus of hyaluronic acid gels has been controlled by altering the concentration of functionalized HA or crosslinker in solution. Fibroblasts cultured on methacrylate functionalized HA hydrogels and are observed to adhere and spread over the course of 4 days (scale bar, 100 μm).52 Adapted from reference 52. (C) For naturally derived hydrogel scaffolds, tissues have been isolated from healthy and diseased patients and stripped of cells (decellularized), to provide a base material with appropriate mechanics, structure, and some biochemical cues for fibroblast cell culture. Here, lung tissue was isolated from a patient with idiopathic pulmonary fibrosis (IPF), decellularized, and subsequently seeded with fibroblasts for in vitro culture studies. Adapted from reference 15. (D) The Young's modulus (E) of normal and IPF decellularized lungs was analyzed using atomic force microscopy. The moduli of samples taken from the lungs of four individuals (two healthy and two with IPF) are shown, with each color representing a different individual. The average modulus of IPF lungs (E ∼ 16 kPa) was significantly higher than that of normal lungs (E ∼ 2 kPa) (p < 0.0001). Normal human pulmonary fibroblasts were seeded onto these decellularized normal or diseased lungs. Cells cultured on decellularized IPF lung expressed αSMA to a greater degree than cells cultured on decellularized normal lung, indicating increased cell activation into wound-healing myofibroblasts.15 Adapted from reference 15. Reprinted with permission of the American Thoracic Society. Copyright © 2014 American Thoracic Society. Cite: Booth et al. (2012) Acellular Normal and Fibrotic Human Lung Matrices as a Culture System for In Vitro Investigation. Am J Respir Crit Care Med186 866–876. Official Journal of the American Thoracic Society. | ||
Collagen I deposition and crosslinking significantly increase during wound healing and fibrosis.24 As noted in the Introduction, when fibroblasts are activated into myofibroblasts, they secrete large quantities of collagen I that are deposited into the ECM to form a highly crosslinked environment. Collagen I also plays a key role in cell adhesion to the ECM by presenting several integrin-binding sites, including the glycine-phenylalanine-hydroxyproline-glycine-glutamic acid-arginine (GFOGER) sequence25 and the well-known arginine-glycine-aspartic acid (RGD) sequence. Fibroblasts can respond to changes in the modulus of the extracellular matrix by exerting force on the matrix through these integrin binding sites. These cell–matrix interactions are involved in mechanotransduction pathways that contribute to the acquisition of the myofibroblast phenotype.26
Collagen hydrogels also are useful for studying the effects of physical activation triggers. Fibroblasts cultured in mechanically-loaded collagen gels formed from 0.3 mg mL−1 rat tail collagen I in 100 mm diameter cell culture dishes were observed to enter an activated state while fibroblasts cultured in unloaded collagen gels enter a quiescent state.14,35 Here, the term ‘mechanically-loaded’ refers to hydrogels that are somehow attached to a surface, commonly a cell culture plate so that the gels resist forces exerted by fibroblasts. Alternatively, free floating gels do not resist the forces exerted by encapsulated fibroblasts cultured within them.14,27,28 The mechanisms by which these fibroblasts translate ‘mechanical loading’ to activation have been the subject of further study.36 This phenomenon is particularly interesting given more recent results linking the differences in the stiffness of synthetic hydrogel matrices to fibroblast activation,16,31,37 and the observed increase in stiffness in fibrotic tissue compared to healthy tissue.15
Fibrin
Traditionally, fibrin has been a popular material for tissue engineering and regenerative medicine applications. It has been used in approaches aimed at regenerating a number of tissues in vitro, or to mimicking tissues in biological implants44 and in wound healing applications. Fibrin also has been used as a sealant for open wounds,44 to deliver proteins such as thrombin or cytokines associated with wound healing,45 or cells like fibroblasts or cardiomyocytes46,47 to promote the wound healing process. This wide range of healing-related applications has motivated additional studies on fibroblast culture in hydrogels. Fibroblast migration to fibrin clots is a critical step in the wound healing process and for this reason fibrin clots are sometimes included within in vitro models. These models have been used to understand what drives migration and to elucidate the causes of improper wound healing. Fibronectin and PDGF play a role in fibroblast migration to fibrin clots in collagen hydrogels.48 Further, plasminogen expression aids in fibroblast invasion of fibrin, but may not be essential. Evidence suggests that the expression of other proteinases can overcome plasminogen deficiency.49
Hyaluronic acid
HA also has been modified with various reactive functional groups to allow formation of covalently crosslinked hydrogels.56,57 Synthetically modified HA enables the formation of hydrogels with controllable modulus58 and the potential for in situ photopatterning,59 affording control of matrix properties in space and in time. For example, HA has been modified with thiol groups by reaction with dithiobis(propanoic dihydrazide) or dithiobis(butyric dihydrazide) followed by reduction with dithiothreitol.60 This method has been used to modify up to 70% of the available carboxylic acid groups with thiols that then participate in a number of reactions to form hydrogels, including a Michael-type addition reaction with acrylates or a free-radical initiated chain polymerization with acrylates or methacrylates.58,61 As an alternative to the addition of thiol groups, HA has been reacted with methacrylic anhydride under basic conditions (pH ∼ 8) to add methacrylate groups. These free radical chain polymerization reactions enable gel formation and modification upon application of cytocompatible doses of long wavelength UV light in the presence of a photoinitiator.32,52 HA gels have been used to culture a number of fibroblast cell lines including human dermal fibroblasts,62 vocal fold fibroblasts,32 and valvular interstitial cells (VICs).63 For example, elastin synthesis by vocal fold fibroblasts was found to be inversely related to the molecular weight of HA included in HA-polyacrylamide gels.32
Other natural materials
Many additional components of the ECM offer favorable hydrogel properties for fibroblast in vitro culture beyond the more commonly used scaffold materials that have been described in detail. For example, the protein elastin lends the native ECM much of its elasticity. While native elastin is highly crosslinked and can be difficult to handle in vitro, recombinantly expressed elastin and electrospun elastin have been used to form hydrogels and could be promising for fibroblast culture.64,65 Similarly, elastin-like polypeptides have been used to form hydrogels for the culture of fibroblasts, although specific investigations of wound healing related properties are currently limited.66Decellularized tissue is another interesting material that has been recently used to culture fibroblasts. Decellularized tissue consists of tissue samples that have been stripped of cells such that only extracellular matrix components remain, including structural proteins, glycosaminoglycans, and cytokines. While perhaps not traditionally considered a hydrogel, these complex crosslinked protein scaffolds closely resemble the native ECM experienced by fibroblasts in vivo. Characterizing the mechanical properties and protein composition of normal and fibrotic decellularized tissues has been a helpful step toward identifying pro-fibrotic signals. In particular, decellularized fibrotic lungs were found to have a significantly higher modulus than decellularized healthy lungs (E ∼ 15 kPa vs. 3 kPa, respectively), and human pulmonary fibroblasts reseeded on these fibrotic matrices showed an increase in αSMA expression as compared to cells seeded on normal lungs (Fig. 2D).15 This result supports the hypothesis that signals from the extracellular environment, especially changes in the structure and concentrations of ECM proteins, promote activation of fibroblasts and significantly contribute to fibrosis. While these are informative studies, decellularized tissue can be difficult to obtain, purify, and characterize, limiting its broad use as a culture model.
Polyacrylamide
Polyacrylamide gels with stiffness gradients, typically achieved by varying crosslink density and modulus, have been particularly useful for assessing the effects of matrix stiffness on fibroblast activation.16,69 Notably, a polyacrylamide gel with a gradient in stiffness and coated with collagen I was used to examine the effect of matrix rigidity on the activation of human pulmonary fibroblasts. Human pulmonary fibroblasts proliferated to a greater extent on the stiffer portion of the gradient (Fig. 3A). Additionally, on discrete stiffness gels, little to no procollagen I and low levels of αSMA expression were observed on lower stiffness materials (Young's modulus E < 0.5 kPa), while greater levels of procollagen I and αSMA expression were observed on high stiffness materials (E > 10 kPa).16 This is one of several studies utilizing polyacrylamide37,70 that indicates that the mechanics of the extracellular environment play a significant role in fibroblast activation, furthering our understanding of the complex signals that contribute to fibrosis and wound healing.
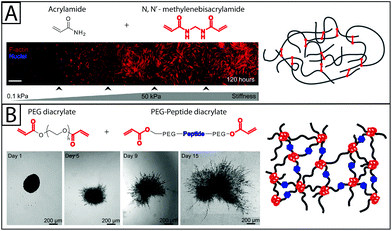 | ||
| Fig. 3 Synthetic base materials for the construction of hydrogels used in fibroblast culture. (A) Polyacrylamide hydrogels (right) are commonly formed from acrylamide and N,N′-methylenebisacrylamide and often used in 2D fibroblast culture. For example, human pulmonary fibroblasts (left) have been observed to proliferate on a collagen-coated polyacrylamide substrate with a stiffness gradient. As the modulus of the polyacrylamide substrate increased, fibroblasts proliferated to a greater degree and had highly organized cytoskeletons (red f-actin).16 Adapted from references 16 and 147. (B) Polyethylene glycol (PEG) has been used in 2D and 3D fibroblast culture. PEG hydrogels, like the one shown on the right, can be formed by functionalizing PEG with acrylate end groups and then polymerizing with an acrylate-functionalized peptide.147 Proteinase-degradable peptides often have been used in PEG hydrogels for 3D cell culture because they allow the matrix to be degraded and invaded by the encapsulated fibroblasts. Here, 3T3 fibroblasts are observed spreading in degradable PEGDA hydrogel containing basic fibroblast growth factor (bFGF) (time lapse images). bFGF was found to induce cell spreading within the gel over the course of 15 days.81 Adapted from references 81 and 147. | ||
Poly(ethylene glycol)
PEG hydrogels have been used to culture several types of fibroblasts, including VICs,18 human foreskin fibroblasts (HFF),76,79 and pulmonary fibroblasts.17 These hydrogels have been used to study the effects of matrix stiffness,17,74,77,80 cell adhesion sites,77 and growth factor presentation75,81 on fibroblast adhesion and activation. For example, PEG hydrogels have been used to study the individual and synergistic effects of matrix modulus and TGFβ1 on fibroblast activation. It has been observed that pulmonary fibroblasts cultured on lower modulus PEG hydrogels (E ∼ 20 kPa) do not express αSMA fibers when TGFβ1 is introduced, even at concentrations as high as 100 ng mL−1. On more rigid substrates (E ∼ 900 kPa), αSMA expression can be induced through the addition of TGFβ1, but higher concentrations are needed on less rigid substrates.17 The conclusion of this study, that low modulus substrate can inhibit αSMA expression, is supported by a number of studies.77,82
In another investigation into the effect of matrix stiffness, PEG hydrogels were used to study fibroblast activation in response to dynamic changes in substrate modulus during culture. Changes in gene expression, proliferation, and expression of αSMA have been observed in VICs grown on photodegradable PEG substrates where modulus was decreased after three days of culture. These changes suggest that, on the time scale observed (5 total days of culture), decreases in modulus from that of fibrotic tissue (E ∼ 32 kPa) to that of healthy tissue (E ∼ 7 kPa) led to fibroblast de-activation and quiescence,18,74 indicating that matrix modulus alone can alter fibroblast fate over relatively short times. Additionally, growth factors like FGF-1 have been shown to increase fibroblast invasion into PEG hydrogels (Fig. 3B)81 of varying moduli.
Other synthetic materials
Polyacrylamide and PEG are the most commonly used synthetic hydrogel scaffold materials for in vitro studies of healing and disease. However, there are many other synthetic materials that have been used to form hydrogels for general cell culture. For an excellent review of materials for hydrogel formation and their properties see Slaughter et al.10 Briefly, fibroblasts have been successfully cultured on poly(vinyl alcohol) (PVA), which offers many of the benefits of PEG hydrogels, but is difficult to control, on the microscale.84,85 Poly(2-hydroxyethyl methacrylate) (PHEMA) historically has been used in biological implants10 and with limited success to culture fibroblasts in vitro.86 Additionally, poly(dimethyl siloxane) (PDMS), which can be used to form polymer networks with soft tissue-like moduli, has been applied in fibroblast culture to study cell contraction. Wrinkling of the PDMS substrate has been correlated to the contraction force that the fibroblast or myofibroblast exerts on its environment, where cells cultured on stiff surfaces (E ∼ 100 kPa) exhibit enhanced wrinkle formation, indicating greater cell contraction and pro-fibrotic behavior.7C. Biological functionalization of hydrogels for fibroblast culture
The bioinert nature of synthetic hydrogels allows user control over the addition of various biochemical ECM cues to enable the examination of their effects on fibroblast proliferation, migration, and activation in 2D and 3D culture and to facilitate appropriate cell adhesion. In this section, materials that are commonly incorporated into hydrogel models of wound healing or fibrosis for the purpose of biological functionalization are overviewed, including whole ECM proteins, protein functional domains, protein mimetic peptides, and growth factors. This section also will discuss common methods for bioconjugation of the hydrogel base materials discussed in Section B.Whole ECM proteins and protein functional domains
In addition to promoting cell adhesion, the Extra Type III A (EDA) domain of fibronectin has been implicated in fibroblast activation to myofibroblasts.89,90 Primary lung fibroblasts isolated from patients with idiopathic pulmonary fibrosis (IPF) express EDA at a higher concentration (as observed by RT PCR and western blotting) than primary lung fibroblasts isolated from healthy patients.90 Further, bleomycin treated mice, a common in vivo pulmonary fibrosis model, do not develop fibrosis when they are EDA deficient.90 Dermal fibroblasts cultured on gelatin expressed lower levels of αSMA when treated with an anti-EDA antibody and TGFβ1, which is known to activate fibroblasts, when compared to a control fibroblasts cultured without the antibody.89 These results suggest that, while fibronectin is often included in hydrogel in vitro models to promote cell adhesion, fibronectin also contributes to TGFβ1-induced fibroblast activation.
Growth factor sequestration may explain heparin's observed antifibrotic effects in vivo, and its simultaneous ability to stimulate fibroblast activation in vitro. Similarly, heparin growth factor sequestration affects fibroblast adhesion onto hydrogel scaffolds containing heparin. Human aortic adventitial fibroblasts adhered at higher rates to hydrogels formed from PEG conjugated with RGD peptides than the same base hydrogel formed with LMWH (Fig. 4A).94 This result indicates that LMWH had an adverse effect on cell adhesion. However, this adverse effect was counteracted by the addition of bFGF.94
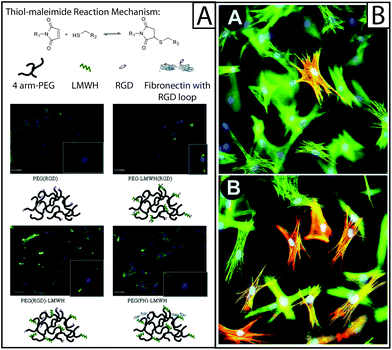 | ||
| Fig. 4 Approaches to biological functionalization of hydrogel culture systems. (A) Heparan sulfate is found throughout the ECM and is known to be an important mediator of cytokine binding and presentation; to mimic this, heparin has been incorporated within hydrogel-based culture systems for the purpose of biological functionalization. For example, four-arm PEG has been functionalized with an RGD adhesive peptide sequence (PEG(RGD)) or fibronectin (PEG(FN)) and crosslinked with low molecular weight heparin (LMWH) or LMWH functionalized with RGD (LMWH(RGD)). Hydrogels were formed from these materials using thiol–maleimide click chemistry (top). The resulting hydrogels were used to culture fibroblasts, where fibroblasts were observed to spread preferentially on hydrogels containing fibronectin, as determined by immunostaining for f-actin (green). The addition of LMWH to PEG(RGD) gels also increased fibroblast spreading, but the addition of LMWH(RGD) did not, indicating that the process of conjugating RGD to LMWH may have inhibited protein function.94 Adapted from reference 94. (B) Cytokines and growth factors are secreted by resident cells, as well as recruited immune cells, during healing. To capture this within model systems, these soluble proteins have been added to the culture medium or functionalized to the scaffold. Here, rat subcutaneous fibroblasts (green f-actin) were cultured on collagen lattices (inset A) and TGFβ1 was added (inset B), significantly increasing the number of activated cells (red αSMA).6 Adapted from reference 6. | ||
Protein mimetic peptides
Protein mimetic peptides can be synthetically produced and modified with various functional chemistries, allowing for the construction of well-defined and highly controlled biomimetic hydrogels. Specifically, peptides have been synthesized to contain terminal thiol groups that are then reacted with ‘ene’-functionalized PEG monomers.17,77 Peptides also have been functionalized with acrylate groups for reaction with acrylate functionalized PEG101 as well as acrylamide. As will be discussed below, protein mimetic peptides are most often incorporated into synthetic hydrogel systems to enable appropriate fibroblast adhesion, although they also can be used to elicit specific cellular responses, including activation. Protein mimetic peptides allow the isolation of integrin binding from the other functions of whole proteins, such as growth factor sequestration, and enabling the testing of hypotheses concerning integrin binding without the addition of confounding factors.17
| Sequence | Mimicked protein | Associated integrin | Cell line | Observed cell response | Reference |
|---|---|---|---|---|---|
| a Most strongly bound amongst several integrins, which include α5β1, α8β1, αvβ1, αvβ5, αvβ6, & αvβ8. | |||||
| RGD | Fibronectin, Collagen I, Laminin α1, Vitronectin | αVβ3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
Human pulmonary fibroblast | Adhesion | 17 |
| Human skin fibroblast | Collagen I synthesis | 105 | |||
| 104,137–139 | VIC | Adhesion | 77 | ||
| Human periodontal ligament fibroblast | Adhesion | 140 | |||
| GFOGER-(POG)n | Collagen I | α2β1, α1β1 | Human dermal fibroblast | Collagen I synthesis | 141 |
| 23,142 | |||||
| DGEA | Collagen I | α2β1, α3β1 | Human pulmonary fibroblast | Adhesion | 17 |
| 143 | 143 | ||||
| YIGSR | Laminin β1 | α3β1, α4β1, α6β1 | Human periodontal ligament fibroblast | Adhesion | 106 |
| 144 | Human dermal fibroblast | Adhesion | 140 | ||
| IKVAV | Laminin α1 | α3β1, α4β1, α6β1 | Human pulmonary fibroblast | Adhesion | 17 |
| 144 | Human periodontal ligament fibroblast | Adhesion | 140 | ||
| PHSRN(SG)5RGDSP | Fibronectin | α5β1 | 3T3 fibroblasts | Adhesion | |
| 104,138 | |||||
| P15 | Collagen I | α2β1 | VIC | αSMA expression | 77 |
| 73 | 73 | ||||
| VGVAPG | Elastin | affinity binding protein | VIC | αSMA expression | 77 |
| 73 | |||||
In addition to promoting cell adhesion, some protein mimetic peptides have been observed to affect fibroblast behavior. For example, RGD was found to increase collagen synthesis in human skin fibroblasts cultured on PDMS, a function associated with activated fibroblasts during wound healing.105 Similarly, the sequence YIGSR (tyrosine-isoleucine-glycine-serine-arginine) which is derived from laminin has been shown to promote collagen synthesis in human dermal fibroblasts.106 In another example, VAPG (valine-alanine-proline-glycine), a peptide derived from elastin, was found to enhance expression of αSMA by VICs cultured on PEG hydrogels, indicating increased activation.77
Cytokines
D. Increasing dimensionality towards a better understanding of activation
Designing materials that closely mimic the native ECM, from its 3D structure to dynamic nature, is critical in the development of hydrogel models to understand wound healing and disease. The move from hard plastic culture dishes to hydrogels has already advanced the study of wound healing and disease by revealing how matrix modulus plays a role in fibroblast activation. The move from 2D models to 3D models is allowing researchers to investigate how polarization and cell shape may influence fibroblast behavior in vitro and in vivo. The development of temporally changing models is also improving our understanding of the role of dynamic signaling in the wound healing process and the development of fibrosis. This section will discuss the exciting discoveries that have resulted from creating models with increased dimensionality starting with the use of models with varied stiffness and ending with temporal signaling.2D: the move to biomimetic surfaces
‘Stiff’ or high modulus substrates have been observed to induce the expression of αSMA in a number of different cell–matrix systems: these include human lung fibroblasts and rat hepatic fibroblasts cultured on polyacrylamide coated with collagen I16,31,70 and PEG hydrogels modified with RGD, DGEA, and IKVAV peptides,17 as well as VICs cultured on PEG hydrogels modified with fibronectin20 or RGDS, VGVAPG or P15.73 For example, rat portal (hepatic) fibroblasts cultured on discrete stiffness gels ranging from E ∼ 400 Pa to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Pa exhibit increasing αSMA stress fiber expression with increasing stiffness (Fig. 5A). This result was quantified on the transcriptional level using RT-PCR.70 Additionally, work done in collagen hydrogels suggests that ECM protein secretion is higher when gels are ‘mechanically-loaded,’28 suggesting that fibroblast activation is influenced by matrix resistance to fibroblast contractile forces. Further, fibroblasts that have been cultured on stiff substrates for various periods of time express high levels αSMA when cultured on soft substrates compared to cells that were never cultured on a stiff substrate, indicating an important temporal component in cell response to matrix mechanical properties where cells have an increasing ‘memory’ of activation over time.7
000 Pa exhibit increasing αSMA stress fiber expression with increasing stiffness (Fig. 5A). This result was quantified on the transcriptional level using RT-PCR.70 Additionally, work done in collagen hydrogels suggests that ECM protein secretion is higher when gels are ‘mechanically-loaded,’28 suggesting that fibroblast activation is influenced by matrix resistance to fibroblast contractile forces. Further, fibroblasts that have been cultured on stiff substrates for various periods of time express high levels αSMA when cultured on soft substrates compared to cells that were never cultured on a stiff substrate, indicating an important temporal component in cell response to matrix mechanical properties where cells have an increasing ‘memory’ of activation over time.7
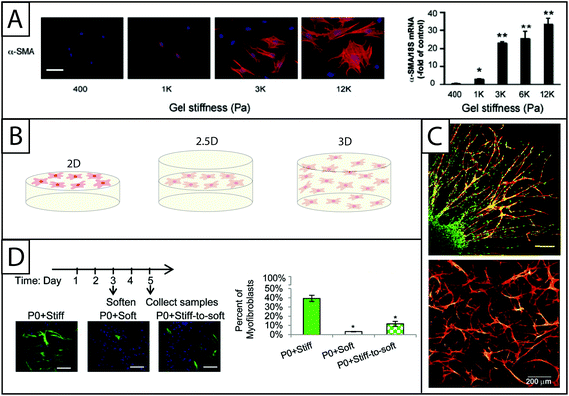 | ||
| Fig. 5 Increasing dimensionality: Examining fibroblast response within static and dynamic model systems from two to three dimensions. (A) Polyacrylamide gels of various moduli modified with collagen I have been a dependable system for examining the effect of modulus on fibroblasts in 2D culture. Increasing the modulus of the hydrogels increases expression of αSMA (here, rat hepatic fibroblasts, αSMA immunostaining (left) and qRT-PCR (right)).70 This result supports the growing body of evidence that substrate stiffness has a significant impact on fibroblast activation, corresponding with in vivo observations of myofibroblast persistence with increased tissue stiffening. Adapted from reference 70. (B) While many seminal studies have been made in 2D culture, cells natively are surrounded by matrix. Towards addressing this, fibroblasts have been cultured in 2D, 2.5D, and 3D geometries to understand the effect of polarization on cell behavior. 2D cell culture geometry, where fibroblasts are seeded on top of hydrogels, has been commonly used to study fibroblasts in vitro (left). 2.5D cell culture geometry is an emerging technique that allows for a decrease in polarization while maintaining the ability to image cells, a key to assessing αSMA stress fiber formation and cell activation (middle). 3D cell culture geometry is used to accurately mimic the lack of cell polarization within the in vivo environment (right); however, cells must be able to degrade and spread in the matrix of interest. (C) For example, in 3D culture, human foreskin fibroblasts (top) have been cultured within PEG gels that were rendered degradable by the incorporation of MMP-degradable peptide sequences. Fibroblasts encapsulated within a fibrin clot were shown invading the synthetic hydrogel by degrading and migrating through the network (scale bar, 150 μm). Human foreskin fibroblasts directly encapsulated in these PEG hydrogels (bottom) formed a network with single cells migrating considerably within the gel over 30 days.76,79 Adapted from references 76 and 79. (D) Towards capturing temporal changes in the cell microenvironment, in vitro culture systems whose properties can be changed in time utilizing external triggers have been created. For example, photodegradable hydrogels have been utilized to trigger a temporal decrease in matrix modulus with light. Here, when the modulus of the hydrogel was reduced after three days of cell culture, fewer valve fibroblasts (VICs) expressed αSMA stress fibers (green) compared to VICs continually cultured on stiff hydrogels; however, the number of αSMA positive cells was still higher than VICs continually cultured on soft substrates, indicating that some portion of the fibroblasts can be de-activated with changes in matrix modulus at this time point in culture.19 Adapted from reference 19. | ||
ECM topography also has been observed to affect activation. In one study, PEG microrods were patterned on a 2D PEG-coated silicon wafer, allowing fibroblasts to attach and align along the rods without adhesive ligands. Microrods of varied moduli down-regulated expression of αSMA, cyclin D1, and integrin α3, and decreased proliferation, indicating that topography plays a role in fibroblast activation.114 Additionally, studies examining the combined effects of matrix rigidity and patterning of ECM proteins on fibroblast adhesion and organization have been performed to create better mimics of the in vivo microenvironment. For example, PDMS micropost arrays coated with ECM proteins were created for 2D culture.115 Fibroblasts seeded on top of the patterned surface exerted forces on the posts and spread over the surface of the material. The formation of focal adhesions on micropost arrays and flat surfaces varied depending on both the substrate stiffness (where shorter posts correspond to a more rigid substrate) and ECM protein pattern. Fibroblasts cultured on uniform ECM-coated flat surfaces exhibited the most spreading. The rigidity of the substrate and the patterning of the posts also affected cytoskeletal organization, as qualitatively observed by F-actin staining.
These results concerning the role of matrix stiffness and topography are informative, but often these studies are conducted on 2D substrates. In these cases, fibroblasts interact with the matrix on one side of the cell. This inappropriate polarization of the cell body may alter the cytoskeletal organization of the cell and consequently activation. For this reason, there have been many efforts to study fibroblast behavior in their native orientation in three dimensions.
2.5D and 3D cell culture
The move to 3D cell culture has taken two distinct forms (Fig. 5B): (i) ‘sandwich studies’ or 2.5D studies in which hydrogels are stacked on top of one another with a layer of cells in between them and (ii) 3D studies in which fibroblasts are suspended in a hydrogel precursor solution and encapsulated within the hydrogel network upon polymerization. The 2.5D cell culture geometry is relatively new for fibroblast culture, although relevant protocols have been published.71,116 The technique previously has been used to successfully culture salivary gland cells in hyaluronic acid hydrogels117 and breast epithelial cells in an ECM derived matrix.118 Additionally, human umbilical vein endothelial cells have been cultured in an interesting derivative of the 2.5D sandwich method where cells were seeded on polyacrylamide coated with collagen and then covered with another layer of assembled collagen. Although the layer above the cells has a much lower modulus, the cells can still interact with the collagen substrate on all sides, reducing polarization.116 The benefit of the 2.5D cell culture geometry is that it allows for cell imaging without more specialized equipment (e.g., confocal microscope). Since the most commonly recognized marker of fibroblast activation is the organization of αSMA stress fibers, imaging is a critical component of cell characterization and provides a strong argument for the utilization of this cell culture geometry. Another benefit to the 2.5D cell culture technique is that it can be used with polymerization techniques that are cytotoxic, as noted earlier with 2.5D polyacrylamide hydrogels for fibroblast culture.67While there are benefits to ‘sandwich’ studies, the 3D cell culture geometry more closely mimics the in vivo environment. 3D cell culture can be achieved when using a cytocompatible material and polymerization chemistry. Historically, fibroblasts have been encapsulated within collagen hydrogels for 3D cultures because they offer the natural benefits of a cytocompatible polymerization (self-assembly), fibroblast adhesion sequences, and proteinase degradable properties for fibroblast driven remodeling.14 Cells have been suspended in collagen in slightly acidic conditions and encapsulated in collagen hydrogels as the pH is raised and hydrogels are formed.28 More recently, 3D cell culture studies of fibroblasts have been conducted using PEG hydrogels.32,119 For example, PEG-hyaluronic acid gels were formed using a photoinitiator and acrylate functionalized monomers to encapsulate vocal fold fibroblasts. Fibroblast response to HA of various molecular weights was assessed by measuring the synthesis of ECM proteins (elastin and collagen), proliferation, and αSMA expression. The molecular weight of the HA included in the gel was observed to affect fibroblast behavior, with intermediate molecular weight HA inducing activation.32 In addition to cell encapsulation, other methods of achieving 3D-like cell culture have been employed, including inducing fibroblast migration into the hydrogel81 and patterning hydrogels with pores into which fibroblasts are seeded.120 In order to conduct 3D studies using encapsulation and migration techniques, cellular remodeling of the hydrogel must be possible. One approach to engineering this property into gel systems is the incorporation of MMP degradable peptide linkers into the hydrogel network as described in Section B.77,79 For example, VICs have been cultured in PEG-MMP degradable peptide hydrogels formed by thiol–ene click reactions. In this gel system, crosslink density and RGD incorporation (100 μM to 2000 μM) was found to significantly affect cell morphology in long term culture (>10 days).82 In another example, the incorporation of MMP degradable peptide linkers allowed human foreskin fibroblast migration out of a fibrin clot into PEG hydrogel. Fibroblast migration is an important step in the wound healing process and must be enabled in 3D studies which are designed to better understand this stage of the wound healing process.76
Incorporating temporal signaling
The models discussed thus far have incorporated static signals that affect fibroblast phenotype and function in hydrogel-based cell culture (e.g., initial modulus or continuous presentation of protein or binding sequence). However, in vivo, fibroblasts respond to dynamic signaling events. Earlier in this section, a study in which fibroblasts were grown first on stiff substrates and then soft substrates was mentioned. This study indicated that fibroblasts possess a “mechanical memory,” indicating that fibroblast response to a signal of interest must be interpreted in the context of its prior history.7 The incorporation of temporal property changes within hydrogel-based culture models enables critical studies to examine fibroblast response to changing microenvironment properties.Several studies have examined the effects of temporal changes in hydrogel modulus on fibroblast proliferation and phenotype.18,68,74 The inclusion of select photodegradable groups, such as o-nitrobenzyl ether groups, allows for the formation of hydrogels that are sensitive to cytocompatible doses of UV, visible, and two-photon irradiation, enabling user-directed gel degradation and triggered changes in the matrix modulus.18,68 Decreases in overall substrate modulus have been shown to decrease the expression of αSMA stress fibers, the percentage of activated fibroblasts, and proliferation of VICs cultured on these hydrogels (Fig. 5D).74 Using this dynamic culture approach, the P13 K/AKT pathway recently was implicated as a mechanism by which fibroblasts respond to changes in matrix rigidity.19 In addition to changing the overall modulus of the hydrogel, UV light or two-photon lasers can be used to pattern hydrogels with biochemical moieties,121–123 which may be a promising approach to similarly examine changes in activation in response to temporal changes in matrix composition.
E. Future directions
The sections above describe materials for hydrogel synthesis and functionalization that have been used in wound healing and disease models. This section will focus on novel materials and techniques that can be used to increase the complexity of these in vitro models. Three general topics will be covered: spatial patterning within hydrogels, temporal hydrogel stiffening, and novel materials for incorporation into hydrogel models.Spatial patterning of hydrogels
Fibrotic tissue is significantly more heterogeneous than healthy tissue, due to the formation of fibrotic foci that unevenly deposit ECM within the tissue.15 The role of heterogeneous protein deposition has not been widely explored within the literature because technologies enabling spatial control of protein conjugation within hydrogels are only recently emerging. With newer technologies, whole proteins, peptide sequences, and cytokines can be conjugated to a specific location within a 3D hydrogel using a variety of methods. This technology could be used to examine the effect of heterogeneity on fibroblast activation. Also, spatial patterning of hydrogels can be employed to exert a high degree of control over cell migration, cell–protein interactions, and cell–growth factor interactions. Photoreactive chemistries and photomasked UV light or two-photon absorption photolithography can be applied to control the site of the reaction, as demonstrated in previous work where patterns of conjugated peptides124,125 and whole proteins126 have been created within hydrogels. These techniques also can be used to change substrate properties or control the internal geometry of the hydrogels.18,121,127,148 In the future, these technologies may prove useful to study fibroblast activation within patterned hydrogels to elucidate the role of ECM heterogeneity or spatially controlled integrin binding.Temporal hydrogel stiffening
In Section D, fibroblast response to a decrease in substrate modulus was highlighted. However, the temporal nature of fibroblast response to such decreases has not been fully explored. There have been recent efforts in designing materials that stiffen in response to an external trigger like UV light.83 HA modified with methacrylates was used to initially form gels crosslinked by dithiothreitol (DTT). Subsequent stiffening was induced after the application of UV light and the polymerization of remaining methacrylate groups to form new crosslinks within the gel.83 Stiffening materials such as this could prove useful in new studies to mimic tissue stiffening during healing or fibrosis to better understand fibroblast activation in response to temporal changes in matrix mechanics.Novel materials
Self-assembling peptides are exciting materials for producing hydrogels that enable the culture of cells within a dynamic structure. These peptides have the benefit of mimicking the temporally changing and heterogeneous structure of the native ECM while maintaining a high degree of control over the formation and structure of soft hydrogels. Peptide-based hydrogels have been used for cell culture, but few studies have utilized them for wound healing or fibrosis models.128 Similarly, engineered proteins or protein mimics are interesting tools for the formation of well-defined and tightly controlled hydrogel models that retain some of the complexity of the native ECM.129In addition to new synthetic materials, several natural materials that have not been previously included in hydrogel in vitro models may be of interest in future studies. For example, biglycan, a small leucine rich proteoglycan that interacts with ECM proteins and several growth factors, including TGFβ1, is suspected to play a role in wound healing, interacting with immune cells to recruit inflammatory cells to the site of injury.130 The role of biglycan in wound healing and fibrosis is supported by a number of studies, which show increased concentrations of biglycan in fibrotic disease; however, it has not yet been incorporated into hydrogel models.131–133 Biglycan is just one example of several proteins and cytokines that have been shown to play a role in the development of fibrosis through in vivo models, yet their exact roles are difficult to decipher within the complex in vivo environment. Other such proteins include surfactant protein A,134 serum amyloid P,135 and adenyl cylase 6.136 The incorporation of these and other relevant cues could build upon prior work using hydrogel in vitro models and help bridge the gap between in vitro and in vivo studies of fibrosis.
F. Conclusions
Advances in our understanding of the mechanisms of wound healing and disease have made it apparent that the extracellular environment plays an essential role in these biological processes. In vitro models have been essential to developing this understanding and will continue to be important tools for the study of fibroblast function and phenotype in healing and disease. In this article, we have reviewed natural and synthetic materials that have been used within in vitro hydrogel-based cell culture models. We also have discussed materials and chemistries for biological functionalization of materials and emerging techniques that may prove useful in the development and advancement of new in vitro systems to further mimic key complexities of in vivo healing and disease progression.Acknowledgements
The authors gratefully acknowledge support, for related work in their laboratories, from a National Science Foundation CAREER Award (DMR-1253906), the Pew Charitable Trusts (00026178), the National Science Foundation Systems Biology of Cells in Engineered Environments IGERT Program, and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P20GM103541). Additionally, the authors would like to thank Prathamesh Kharkar and Matthew Rehmann for feedback on earlier versions of this manuscript.References
- M. A. Carlson and M. T. Longaker, Wound Repair Regen., 2004, 12, 134–147 CrossRef PubMed.
- W. K. Stadelmann, A. G. Digenis and G. R. Tobin, Am. J. Surg., 1998, 176, 26S–38S CrossRef CAS.
- K. S. Midwood, L. V. Williams and J. E. Schwarzbauer, Int. J. Biochem. Cell Biol., 2004, 36, 1031–1037 CrossRef CAS PubMed.
- E. Crouch, Am. J. Physiol., 1990, 259, L159–L184 CAS.
- B. Hinz, J. Invest. Dermatol., 2007, 127, 526–537 CrossRef CAS PubMed.
- B. Hinz, G. Celetta, J. J. Tomasek, G. Gabbiani and C. Chaponnier, Mol. Biol. Cell, 2001, 12, 2730–2741 CrossRef CAS.
- J. L. Balestrini, S. Chaudhry, V. Sarrazy, A. Koehler and B. Hinz, Integr. Biol., 2012, 4, 410–421 RSC.
- M. W. Tibbitt and K. S. Anseth, Biotechnol. Bioeng., 2009, 103, 655–663 CrossRef CAS PubMed.
- K. A. Beningo, C.-M. Lo and Y.-L. Wang, Methods Cell Biol., 2002, 69, 325–339 CrossRef CAS.
- B. V. Slaughter, S. S. Khurshid, O. Z. Fisher, A. Khademhosseini and N. A. Peppas, Adv. Mater., 2009, 21, 3307–3329 CrossRef CAS PubMed.
- T. L. Tuan, A. Song, S. Chang, S. Younai and M. E. Nimni, Exp. Cell Res., 1996, 223, 127–134 CrossRef CAS PubMed.
- P. A. Janmey, R. G. Wells, R. K. Assoian and C. A. McCulloch, Differentiation, 2013, 86, 112–120 CrossRef CAS PubMed.
- Y. Adachi, T. Mio, K. Takigawa, I. Striz, D. J. Romberger, J. R. Spurzem and S. I. Rennard, In Vitro Cell. Dev. Biol. Anim., 1998, 34, 203–210 CrossRef CAS.
- F. Grinnell, Trends Cell Biol., 2003, 13, 264–269 CrossRef CAS.
- A. J. Booth, R. Hadley, A. M. Cornett, A. A. Dreffs, S. A. Mattes, J. L. Tsui, K. Weiss, J. C. Horowitz, V. F. Fiore, T. H. Barker, B. B. Moore, F. J. Martinez, L. E. Niklason and E. S. White, Am. J. Respir. Crit. Care Med., 2012, 186, 866–876 CrossRef CAS PubMed.
- F. Liu, J. D. Mih, B. S. Shea, A. T. Kho, A. S. Sharif, A. M. Tager and D. J. Tschumperlin, J. Cell Biol., 2010, 190, 693–706 CrossRef CAS PubMed.
- H. N. Chia, M. Vigen and A. M. Kasko, Acta Biomater., 2012, 8, 2602–2611 CrossRef CAS PubMed.
- A. M. Kloxin, J. A. Benton and K. S. Anseth, Biomaterials, 2010, 31, 1–8 CrossRef CAS PubMed.
- H. Wang, M. W. Tibbitt, S. J. Langer, L. A. Leinwand and K. S. Anseth, Proc. Natl. Acad. Sci. U. S. A., 2013, 11, 19336–19341 CrossRef PubMed.
- M. C. Cushing, P. D. Mariner, J.-T. Liao, E. A. Sims and K. S. Anseth, FASEB J., 2008, 22, 1769–1777 CrossRef CAS PubMed.
- W. Lee, T. G. Lee and W. G. Koh, J. Ind. Eng. Chem., 2007, 13, 1195–1200 CAS.
- L. Harvey, A. Berk, C. A. Kaiser, M. Krieger, M. P. Scott, A. Bretscher, M. Ploegh and P. Matsudaira, Molecular Cell Biology, New York, 4th edn, 2000 Search PubMed.
- R. W. Farndale, T. Lisman, D. Bihan, S. Hamaia, C. S. Smerling, N. Pugh, A. Konitsiotis, B. Leitinger, P. G. de Groot, G. E. Jarvis and N. Raynal, Biochem. Soc. Trans., 2008, 36, 241–250 CrossRef CAS PubMed.
- R. Raghow, FASEB J., 1994, 8, 823–831 CAS.
- A. M. Wojtowicz, A. Shekaran, M. E. Oest, K. M. Dupont, K. L. Templeman, D. W. Hutmacher, R. E. Guldberg and A. J. García, Biomaterials, 2010, 31, 2574–2582 CrossRef CAS PubMed.
- B. Hinz and G. Gabbiani, Curr. Opin. Biotechnol., 2003, 14, 538–546 CrossRef CAS PubMed.
- M. S. Hahn, B. A. Teply, M. M. Stevens, S. M. Zeitels and R. Langer, Biomaterials, 2006, 27, 1104–1109 CrossRef CAS PubMed.
- K. Mochitate, P. Pawelek and F. Grinnell, Exp. Cell Res., 1991, 193, 198–207 CrossRef CAS.
- E. Tamariz and F. Grinnell, Mol. Biol. Cell, 2002, 13, 3915–3929 CrossRef CAS PubMed.
- K. K. Sethi, I. V. Yannas, V. Mudera, M. Eastwood, C. McFarland and R. A. Brown, Wound Repair Regen., 2002, 10, 397–408 CrossRef.
- A. Marinković, F. Liu and D. J. Tschumperlin, Am. J. Respir. Cell Mol. Biol., 2013, 48, 422–430 CrossRef PubMed.
- D. J. Munoz-Pinto, A. C. Jimenez-Vergara, L. M. Gelves, R. E. McMahon, V. Guiza-Arguello and M. S. Hahn, Biotechnol. Bioeng., 2009, 104, 821–831 CAS.
- Y. K. Zhu, X. D. Liu, M. C. Sköld, T. Umino, H. Wang, D. J. Romberger, J. R. Spurzem, T. Kohyama, F. Q. Wen and S. I. Rennard, Am. J. Respir. Cell Mol. Biol., 2001, 25, 245–253 CrossRef CAS PubMed.
- T. Kohyama, X. Liu, F. Wen, Y. K. Zhu, H. Wang, H. J. Kim, H. Takizawa, L. B. Cieslinski, M. S. Barnette and S. I. Rennard, Am. J. Respir. Cell Mol. Biol., 2002, 26, 694–701 CrossRef CAS PubMed.
- D. Kessler, S. Dethlefsen, I. Haase, M. Plomann, F. Hirche, T. Krieg and B. Eckes, J. Biol. Chem., 2001, 276, 36575–36585 CrossRef CAS PubMed.
- J. Fringer and F. Grinnell, J. Biol. Chem., 2003, 278, 20612–7 CrossRef CAS PubMed.
- X. Huang, N. Yang, V. F. Fiore, T. H. Barker, Y. Sun, S. W. Morris, Q. Ding, V. J. Thannickal and Y. Zhou, Am. J. Respir. Cell Mol. Biol., 2012, 47, 340–348 CrossRef CAS PubMed.
- M. C. Cushing and K. S. Anseth, Science, 2007, 316, 1133–1134 CrossRef CAS PubMed.
- D. Velegol and F. Lanni, Biophys. J., 2001, 81, 1786–1792 CrossRef CAS.
- S. Nakagawa, P. Pawelek and F. Grinnell, J. Invest. Dermatol., 1989, 93, 792–798 CAS.
- D. D. Allison, K. R. Braun, T. N. Wight and K. J. Grande-Allen, Acta Biomater., 2009, 5, 1019–1026 CrossRef CAS PubMed.
- J. W. Weisel, Adv. Protein Chem., 2005, 70, 247–299 CrossRef CAS.
- S. Cox, M. Cole and B. Tawil, Tissue Eng., 2004, 10, 942–954 CrossRef CAS PubMed.
- T. A. E. Ahmed, E. V. Dare and M. Hincke, Tissue Eng. Part B. Rev., 2008, 14, 199–215 CrossRef CAS PubMed.
- O. Ziv-Polat, H. Skaat, A. Shahar and S. Margel, Int. J. Nanomedicine, 2012, 7, 1259–1274 CrossRef CAS PubMed.
- K. Yuan Ye, K. E. Sullivan and L. D. Black, J. Vis. Exp., 2011, 55, 1–6 Search PubMed.
- J. E. Mogford, B. Tawil, S. Jia and T. A. Mustoe, Wound Repair Regen., 2009, 17, 405–410 CrossRef PubMed.
- D. Greiling and R. A. Clark, J. Cell Sci., 1997, 110(Pt 7), 861–870 CAS.
- K. B. Hotary, I. Yana, F. Sabeh, X.-Y. Li, K. Holmbeck, H. Birkedal-Hansen, E. D. Allen, N. Hiraoka and S. J. Weiss, J. Exp. Med., 2002, 195, 295–308 CrossRef CAS.
- N. C. Hunt and L. M. Grover, Biotechnol. Lett., 2010, 32, 733–742 CrossRef CAS PubMed.
- D. Jiang, J. Liang and P. W. Noble, Annu. Rev. Cell Dev. Biol., 2007, 23, 435–461 CrossRef CAS PubMed.
- K. S. Masters, D. N. Shah, G. Walker, L. A. Leinwand and K. S. Anseth, J. Biomed. Mater. Res., Part A, 2004, 71, 172–180 CrossRef PubMed.
- S. T. Kreger and S. L. Voytik-Harbin, Matrix Biol., 2009, 28, 336–346 CrossRef CAS PubMed.
- A. J. E. Farran, S. S. Teller, A. K. Jha, T. Jiao, R. A. Hule, R. J. Clifton, D. P. Pochan, R. L. Duncan and X. Jia, Tissue Eng. Part A, 2010, 16 Search PubMed.
- Y. Li, D. Jiang, J. Liang, E. B. Meltzer, A. Gray, R. Miura, L. Wogensen, Y. Yamaguchi and P. W. Noble, J. Exp. Med., 2011, 208, 1459–1471 CrossRef CAS PubMed.
- J. A. Burdick and G. D. Prestwich, Adv. Mater., 2011, 23, H41–H56 CrossRef CAS PubMed.
- X. Xu, A. K. Jha, D. A. Harrington, M. C. Farach-Carson and X. Jia, Soft Matter, 2012, 8, 3280–3294 RSC.
- F. Rehfeldt, A. E. X. Brown, M. Raab, S. Cai, A. L. Zajac, A. Zemel and D. E. Discher, Integr. Biol., 2012, 4, 422–430 RSC.
- S. C. Owen, S. A. Fisher, R. Y. Tam, C. M. Nimmo and M. S. Shoichet, Langmuir, 2013, 29, 7393–7400 CrossRef CAS PubMed.
- X. Z. Shu, Y. Liu, Y. Luo, M. C. Roberts and G. D. Prestwich, Biomacromolecules, 2002, 3, 1304–1311 CrossRef CAS PubMed.
- Y. Dong, A. O. Saeed, W. Hassan, C. Keigher, Y. Zheng, H. Tai, A. Pandit and W. Wang, Macromol. Rapid Commun., 2011, 120–126 CAS.
- K. Ghosh, X.-D. Ren, X. Z. Shu, G. D. Prestwich and R. A. F. Clark, Tissue Eng., 2006, 12, 601–613 CrossRef CAS PubMed.
- K. S. Masters, D. N. Shah, L. A. Leinwand and K. S. Anseth, Biomaterials, 2005, 26, 2517–2525 CrossRef CAS PubMed.
- J. R. McDaniel, D. C. Radford and A. Chilkoti, Biomacromolecules, 2013, 14, 2866–2872 CrossRef CAS PubMed.
- J. Rnjak, Z. Li, P. K. M. Maitz, S. G. Wise and A. S. Weiss, Biomaterials, 2009, 30, 6469–6477 CrossRef CAS PubMed.
- S. E. Grieshaber, A. J. E. Farran, S. Bai, K. L. Kiick and X. Jia, Biomacromolecules, 2012, 13, 1774–1786 CrossRef CAS PubMed.
- C. E. Kandow, P. C. Georges, P. A. Janmey and K. A. Beningo, Methods Cell Biol., 2007, 83, 29–46 CAS.
- M. T. Frey and Y.-L. Wang, Soft Matter, 2009, 5, 1918–1924 RSC.
- N. A. Hale, Y. Yang and P. Rajagopalan, ACS Appl. Mater. Interfaces, 2010, 2, 2317–2324 CAS.
- Z. Li, J. A. Dranoff, E. P. Chan, M. Uemura, J. Sévigny and R. G. Wells, Hepatology, 2007, 46, 1246–1256 CrossRef CAS PubMed.
- K. A. Beningo and Y. Wang, in Methods in Molecular Biology, 2007, vol. 370, pp. 203–211 Search PubMed.
- P. M. Kharkar, K. L. Kiick and A. M. Kloxin, Chem. Soc. Rev., 2013, 42, 7335–7372 RSC.
- S. T. Gould and K. S. Anseth, J. Tissue Eng. Regen. Med., 2013 Search PubMed.
- H. Wang, S. M. Haeger, A. M. Kloxin, L. A. Leinwand and K. S. Anseth, PLoS One, 2012, 7, e39969 CAS.
- A. S. Gobin and J. L. West, Biotechnol. Prog., 2003, 19, 1781–1785 CrossRef CAS PubMed.
- M. P. Lutolf, J. L. Lauer-Fields, H. G. Schmoekel, A. T. Metters, F. E. Weber, G. B. Fields and J. A. Hubbell, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5413–5418 CrossRef CAS PubMed.
- S. T. Gould, N. J. Darling and K. S. Anseth, Acta Biomater., 2012, 8, 3201–3209 CrossRef CAS PubMed.
- M. W. Tibbitt, A. M. Kloxin, L. A. Sawicki and K. S. Anseth, Macromolecules, 2013, 46, 2785–2792 CrossRef CAS PubMed.
- G. P. Raeber, M. P. Lutolf and J. A. Hubbell, Acta Biomater., 2007, 3, 615–629 CrossRef CAS PubMed.
- C. A. Durst, M. P. Cuchiara, E. G. Mansfield, J. L. West and K. J. Grande-Allen, Acta Biomater., 2011, 7, 2467–2476 CrossRef CAS PubMed.
- S. Sokic and G. Papavasiliou, Acta Biomater., 2012, 8, 2213–2222 CrossRef CAS PubMed.
- J. A. Benton, B. D. Fairbanks and K. S. Anseth, Biomaterials, 2009, 30, 6593–6603 CrossRef CAS PubMed.
- M. Guvendiren and J. A. Burdick, Nat. Commun., 2012, 3, 792 CrossRef PubMed.
- W. Y. Chuang, T. H. Young, C. H. Yao and W. Y. Chiu, Biomaterials, 1999, 20, 1479–1487 CrossRef CAS.
- T. Koyano, N. Minoura, M. Nagura and K. Kobayashi, J. Biomed. Mater. Res., 1998, 39, 486–490 CrossRef CAS.
- S. Hattori, J. D. Andrade, J. B. Hibbs, D. E. Gregonis and R. N. King, J. Colloid Interface Sci., 1985, 104, 72–78 CrossRef CAS.
- M. R. Hynd, J. P. Frampton, N. Dowell-Mesfin, J. N. Turner and W. Shain, J. Neurosci. Methods, 2007, 162, 255–263 CrossRef CAS PubMed.
- C. L. Carraher and J. E. Schwarzbauer, J. Biol. Chem., 2013, 288, 14805–14814 CrossRef CAS PubMed.
- G. Serini, M. L. Bochaton-Piallat, P. Ropraz, A. Geinoz, L. Borsi, L. Zardi and G. Gabbiani, J. Cell Biol., 1998, 142, 873–881 CrossRef CAS.
- A. F. Muro, F. A. Moretti, B. B. Moore, M. Yan, R. G. Atrasz, C. A. Wilke, K. R. Flaherty, F. J. Martinez, J. L. Tsui, D. Sheppard, F. E. Baralle, G. B. Toews and E. S. White, Am. J. Respir. Crit. Care Med., 2008, 177, 638–645 CrossRef CAS PubMed.
- R. Timpl, W. Rohde, P. G. Robey, S. I. Rennard, J.-M. Foidart and G. R. Martin, J. Biol. Chem., 1979, 254, 9933–9937 CAS.
- J. R. Couchman, M. Höök, D. A. Rees and R. Timpl, J. Cell Biol., 1983, 96, 177–183 CrossRef CAS.
- V. P. Terranova, M. Aumailley, L. H. Sultan, G. R. Martin and H. K. Kleinman, J. Cell. Physiol., 1986, 127, 473–479 CrossRef CAS PubMed.
- T. Nie, R. E. Akins and K. L. Kiick, Acta Biomater., 2009, 5, 865–875 CrossRef CAS PubMed.
- A. Utani, Y. Momota, H. Endo, Y. Kasuya, K. Beck, N. Suzuki, M. Nomizu and H. Shinkai, J. Biol. Chem., 2003, 278, 34483–34490 CrossRef CAS PubMed.
- C. Mcgann and K. Kiick, in Engineering Biomaterials for Regenerative Medicine: Novel Technologies for Clinical Applications, ed. S. K. Bhatia, Springer, New York, NY, 2012, pp. 225–250 Search PubMed.
- B. Casu, A. Naggi and G. Torri, Matrix Biol., 2011, 29, 442–452 CrossRef PubMed.
- J.-H. Lee, H. Lee, Y. K. Joung, K. H. Jung, J.-H. Choi, D.-H. Lee, K. D. Park and S.-S. Hong, Biomaterials, 2011, 32, 1438–1445 CrossRef CAS PubMed.
- M. C. Cushing, P. D. Mariner, J.-T. Liao, E. A. Sims and K. S. Anseth, FASEB J., 2008, 22, 1769–1777 CrossRef CAS PubMed.
- M. C. Cushing, J.-T. Liao, M. P. Jaeggli and K. S. Anseth, Biomaterials, 2007, 28, 3378–3387 CrossRef CAS PubMed.
- D. L. Hern and J. A. Hubbell, J. Biomed. Mater. Res., 1998, 39, 266–276 CrossRef CAS.
- R. H. Schmedlen, K. S. Masters and J. L. West, Biomaterials, 2002, 23, 4325–4332 CrossRef CAS.
- J. M. Goffin, P. Pittet, G. Csucs, J. W. Lussi, J.-J. Meister and B. Hinz, J. Cell Biol., 2006, 172, 259–268 CrossRef CAS PubMed.
- S. E. Ochsenhirt, E. Kokkoli, J. B. McCarthy and M. Tirrell, Biomaterials, 2006, 27, 3863–3874 CrossRef CAS PubMed.
- B. Li, J. Chen and J. H. Wang, J. Biomed. Mater. Res., Part A, 2006, 79A, 989–998 CrossRef CAS PubMed.
- J. H. Yoon, J. Kim, H. Lee, S. Y. Kim, H.-H. Jang, S. H. Ryu, B. J. Kim and T. G. Lee, Biochem. Biophys. Res. Commun., 2012, 428, 416–421 CrossRef CAS PubMed.
- T. M. Maher, I. C. Evans, S. E. Bottoms, P. F. Mercer, A. J. Thorley, A. G. Nicholson, G. J. Laurent, T. D. Tetley, R. C. Chambers and R. J. McAnulty, Am. J. Respir. Crit. Care Med., 2010, 182, 73–82 CrossRef CAS PubMed.
- R. A. Evans, Y. C. Tian, R. Steadman and A. O. Phillips, Exp. Cell Res., 2003, 282, 90–100 CrossRef CAS.
- A. Saxena, W. Chen, Y. Su, V. Rai, O. U. Uche, N. Li and N. G. Frangogiannis, J. Immunol., 2013, 191, 4838–4848 CrossRef CAS PubMed.
- Y. Akasaka, I. Ono, A. Tominaga, Y. Ishikawa, K. Ito, T. Suzuki, R. Imaizumi, S. Ishiguro, K. Jimbow and T. Ishii, Wound Repair Regen., 2007, 15, 378–389 CrossRef PubMed.
- B. K. Mann, R. H. Schmedlen and J. L. West, Biomaterials, 2001, 22, 439–444 CrossRef CAS.
- K. G. Robinson, T. Nie, A. D. Baldwin, E. C. Yang, K. L. Kiick and R. E. Akins, J. Biomed. Mater. Res., Part A, 2012, 100, 1356–1367 CrossRef PubMed.
- J. E. Saik, D. J. Gould, E. M. Watkins, M. E. Dickinson and J. L. West, Acta Biomater., 2011, 7, 133–143 CrossRef CAS PubMed.
- P. Ayala, J. I. Lopez and T. A. Desai, Tissue Eng., Part A, 2010, 16, 2519–2527 CrossRef CAS PubMed.
- S. Weng and J. Fu, Biomaterials, 2011, 32, 9584–9593 CrossRef CAS PubMed.
- R. S. Fischer, K. A. Myers, M. L. Gardel and C. M. Waterman, Nat. Protocols, 2012, 7, 2056–2066 CAS.
- S. Pradhan-bhatt, D. A. Harrington, R. L. Duncan, X. Jia, R. L. Witt and M. C. Farach-carson, Tissue Eng., Part A, 2013, 19, 1610–1620 CrossRef CAS PubMed.
- G. Y. Lee, P. A. Kenny, E. H. Lee and M. J. Bissell, Nat. Protoc., 2007, 4, 359–365 CAS.
- E. J. Yun, B. Yon, M. K. Joo and B. Jeong, Biomacromolecules, 2012, 13, 1106–1111 CrossRef CAS PubMed.
- Y.-H. Lee, J.-R. Huang, Y.-K. Wang and K.-H. Lin, Integr. Biol., 2013, 5, 1447–1455 RSC.
- A. M. Kloxin, A. M. Kasko, C. N. Salinas and K. S. Anseth, Science, 2009, 324, 59–63 CrossRef CAS PubMed.
- C. A. Goubko, A. Basak, S. Majumdar and X. Cao, J. Biomed. Mater. Res., Part A, 2013, 1–11 Search PubMed.
- R. G. Wylie and M. S. Shoichet, Biomacromolecules, 2011, 12, 3789–3796 CrossRef CAS PubMed.
- M. S. Hahn, J. S. Miller and J. L. West, Adv. Mater., 2006, 18, 2679–2684 CrossRef CAS.
- J. H. Wosnick and M. S. Shoichet, Chem. Mater., 2008, 20, 55–60 CrossRef CAS.
- K. A. Mosiewicz, L. Kolb, A. J. van der Vlies, M. M. Martino, P. S. Lienemann, J. A. Hubbell, M. Ehrbar and M. P. Lutolf, Nat. Mater., 2013, 12, 1072–1078 CrossRef CAS PubMed.
- C. A. DeForest, B. D. Polizzotti and K. S. Anseth, Nat. Mater., 2009, 8, 659–664 CrossRef CAS PubMed.
- J. E. Gough, A. Saiani and A. F. Miller, ICE Publ., 2011, 1, 2–12 Search PubMed.
- E. Cosgriff-Hernandez, M. S. Hahn, B. Russell, T. Wilems, D. Munoz-Pinto, M. B. Browning, J. Rivera and M. Höök, Acta Biomater., 2010, 6, 3969–3977 CrossRef CAS PubMed.
- M. V. Nastase, M. F. Young and L. Schaefer, J. Histochem. Cytochem., 2012, 60, 963–975 CrossRef PubMed.
- M. B. Stokes, S. Holler, Y. Cui, K. L. Hudkins, F. Eitner, A. Fogo and C. E. Alpers, Kidney Int., 2000, 57, 487–498 CrossRef CAS PubMed.
- K. Moreth, R. V. Iozzo and L. Schaefer, Cell Cycle, 2012, 11, 2084–2091 CrossRef CAS PubMed.
- T. Ebihara, N. Venkatesan, R. Tanaka and M. S. Ludwig, Am. J. Respir. Crit. Care Med., 2000, 162, 1569–1576 CrossRef CAS PubMed.
- H. Goto, J. G. Ledford, S. Mukherjee, P. W. Noble, K. L. Williams and J. R. Wright, Am. J. Respir. Crit. Care Med., 2010, 181, 1336–1344 CrossRef CAS PubMed.
- L. A. Murray, R. Rosada, A. P. Moreira, A. Joshi, M. S. Kramer, D. P. Hesson, R. L. Argentieri, S. Mathai, M. Gulati, E. L. Herzog and C. M. Hogaboam, PLoS One, 2010, 5, e9683 Search PubMed.
- X. Liu, F. Li, S. Q. Sun, M. Thangavel, J. Kaminsky, L. Balazs and R. S. Ostrom, Am. J. Physiol., 2010, 819–829 Search PubMed.
- E. Ruoslahti, Annu. Rev. Cell Dev. Biol., 1996, 12, 697–715 CrossRef CAS PubMed.
- J. A. Craig, E. L. Rexeisen, A. Mardilovich, K. Shroff and E. Kokkoli, Langmuir, 2008, 24, 10282–10292 CrossRef CAS PubMed.
- Y. Yamada, K. Hozumi and M. Nomizu, Chem.–Eur. J., 2011, 17, 10500–10508 CrossRef CAS.
- Y. Kumada and S. Zhang, PLoS One, 2010, 5, e10305 Search PubMed.
- C. M. Yamazaki, Y. Kadoya, K. Hozumi, H. Okano-Kosugi, S. Asada, K. Kitagawa, M. Nomizu and T. Koide, Biomaterials, 2010, 31, 1925–1934 CrossRef CAS PubMed.
- C. D. Reyes and A. J. García, J. Biomed. Mater. Res., Part A, 2003, 65, 511–523 CrossRef PubMed.
- V. P. Ivanova, Z. V. Kovaleva, V. V. Anokhina and A. I. Krivchenko, Cell Tissue Biol., 2013, 7, 21–28 CrossRef.
- J. E. Frith, R. J. Mills, J. E. Hudson and J. J. Cooper-White, Stem Cells Dev., 2012, 21, 2442–2456 CrossRef CAS PubMed.
- C. B. Raub, V. Suresh, T. Krasieva, J. Lyubovitsky, J. D. Mih, A. J. Putnam, B. J. Tromberg and S. C. George, Biophys. J., 2007, 92, 2212–2222 CrossRef CAS PubMed.
- S. Rhee, Exp. Mol. Med., 2009, 41, 858–865 CrossRef CAS PubMed.
- A. M. Kloxin, C. J. Kloxin, C. N. Bowman and K. S. Anseth, Adv. Mater., 2010, 22, 3484–3494 CrossRef CAS PubMed.
- M. Guvendiren, M. Perepelyuk, R. G. Wells and J. A. Burdick, J. Mech. Behav. Biomed. Mater., 2013, 34, 1–11 Search PubMed.
| This journal is © The Royal Society of Chemistry 2014 |



