Metal-free and pH-controlled introduction of azides in proteins†
Sanne
Schoffelen
a,
Mark B.
van Eldijk
ab,
Bart
Rooijakkers
a,
Reinout
Raijmakers
c,
Albert J. R.
Heck
c and
Jan C. M.
van Hest
*a
aDepartment of Bioorganic Chemistry, Institute for Molecules and Materials, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands. E-mail: j.vanhest@science.ru.nl; Fax: (+31) 24 3653393; Tel: (+31) 24 3653204
bDutch Polymer Institute, P. O. Box 902, 5600 AX, Eindhoven, The Netherlands
cBiomolecular Mass Spectrometry and Proteomics Group and Netherlands Proteomics Center, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Padualaan 8, 3584 CH, Utrecht, The Netherlands
First published on 14th January 2011
Abstract
Conversion of amines into azides using imidazole-1-sulfonyl azide as a diazotransfer reagent has proven to be a straightforward way to introduce targetable handles into proteins. We explored whether less toxic and milder conditions than described before could be applied. It was shown that the aqueous diazotransfer proceeds without adding Cu(II) not only at pH 11 but also at pH 8.5. The diazotransfer was shown to be selective towards a single amine.
Introduction
Proteins are modified to study or manipulate their function, for conjugation to surfaces or other molecules, and to increase their stability or uptake in single cells or organisms. For many applications, a chemoselective ligation strategy is desired to control the degree and site of modification. A well-known reaction that has proven to be highly suitable for these purposes is the Cu(I)-catalyzed cycloaddition between azides and alkynes.1 Recently, great interest in the copper-free variant of this click reaction has emerged, as it is known that copper is toxic to cells and difficult to remove from proteins.2Azide moieties are introduced in enzymes, antibodies or other proteins in various ways. Via genetic engineering non-natural amino acids such as para-azidophenylalanine and azidohomoalanine have successfully been incorporated at a single site or in a residue-specific manner.3 Alternatively, azides have been installed in glycoproteinsviametabolism of a synthetic azidosugar.4 Using an engineered lipoic acid ligase, an alkyl azide can specifically be attached to proteins containing a short peptide tag.5 Furthermore, native chemical ligation has facilitated functionalization of a protein's N-terminus with an azide moiety.6
Our group recently reported a more straightforward method that does not require any genetic or metabolic manipulation. Free amines were converted into azidesvia an aqueous diazotransfer (Scheme 1).7 The hydrochloric salt of imidazole-1-sulfonyl azide 1 is used as a shelf-stable, non-explosive and water-soluble diazotransfer reagent.8‡ Whereas this approach is much easier to apply, it is less selective. Four and five azides were introduced in the fluorescent protein DsRed and horseradish peroxidase (HRP), respectively. Moreover, a close to equimolar amount of Cu(II) relative to amine was added as a catalyst and by adding potassium carbonate a pH of 11 was created to ensure that all amines were deprotonated. However, not all proteins are stable at high pH9 and Cu(II) may affect the structure or function of the protein or the biological system in which it will subsequently be used.10
Traditionally, amines have been diazotized in solvent mixtures such as water/CH2Cl2 or water/CH2Cl2/MeOH using triflyl azide as a transfer reagent.11 The use of Cu(II), Ni(II) or Zn(II) as a catalyst was initially reported by Alper et al.12 Since then several procedures have been described in which Cu(II) is used as a catalyst in a 1–10 mol% quantity relative to the amine.8,13 Nyffeler and coworkers investigated the diazotransfer reaction in more detail.14 They showed that both copper sulfate and zinc chloride are effective catalysts, but also when no catalyst was present 80% conversion was detected for some of the aminoglycoside substrates after 18 h of reaction.
Here, we demonstrate that in a similar way azides can be introduced in proteins without adding Cu(II). Moreover, lowering the pH to 8.5 leads to selective conversion of the amine with the lowest pKa, the α-amine at the N-terminus of the model proteins.
Results and discussion
Diazotransfer at pH 11 in presence and absence of Cu(II)
Hen egg-white lysozyme C and a recombinant variant of Candida antarcticalipase B (CalB) were used as model enzymes. These proteins contain six and nine lysines, respectively. They were incubated overnight at room temperature in the presence of the diazotransfer reagent 1 (1.75 equivalents relative to the amines) and sodium carbonate as a base, leading to a pH of 11 in the reaction medium. The conversion of amines into azides was analyzed by mass spectrometry.As shown in Fig. 1 and Table 1, amines were indeed converted into azides when no Cu(II) was added. As for lysozyme, on average five azides were introduced, whereas CalB contained mainly two to three azides. In comparison, six azides were present in lysozyme when 1 was added together with Cu(II). Under these conditions up to seven azides were introduced in CalB.
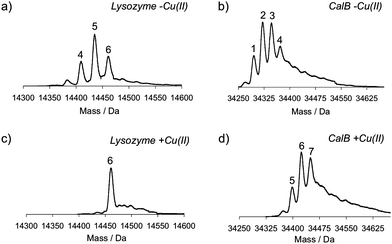 | ||
| Fig. 1 Mass spectra of lysozyme and CalB after diazotransfer at pH 11, which was performed without or in the presence of Cu(II). a) Lysozyme without Cu(II), b) CalB without Cu(II), c) lysozyme with Cu(II), and d) CalB with Cu(II). Numbers above the peaks indicate the number of azides that are present in the protein. | ||
| Protein | Unmodified (Da)a | Cu(II) | Detected (Da)b |
|---|---|---|---|
| a Molecular weight of the unmodified protein, calculated/detected. b Molecular weight of the protein after diazotransfer using 1.75 equivalents (lysozyme and CalB) or 175 equivalents (ELP-lys1 and ELP-lys3) of diazotransfer reagent. | |||
| Lysozyme | 14305.1/14305.2 | − | 14382.4, 14409.3, 14435.2, 14461.1 |
| + | 14461.1 | ||
| CalB | 34269.7/34269.0 | − | 34294.9, 34320.9, 34346.2, 34371.9 |
| + | 34372.3, 34397.9, 34423.8, 34449.0 | ||
| ELP-lys1 | 39670.9/39671.7 | − | 39723.5 |
| + | 39722.8 | ||
| ELP-lys3 | 39927.2/39929.9 | − | 40032.3 |
| + | 40031.2 |
Elastin-like polypeptide (ELP), a structural protein that has previously been engineered in our lab,15 was used as a third model. Two variants of ELP were investigated with either one (ELP-lys1) or three lysines (ELP-lys3). To these proteins a large excess of 1 (175 equivalents relative to the amines) was added. Using this excess all amines in both variants were diazotized (see Table 1), whether Cu(II) was added or not.
As stated before, Cu(II), Ni(II) or Zn(II) are known to catalyze the diazotransfer reaction. Even though 1 was synthesized without using any metal salts, the imidazole ring present in this compound might have complexed some of these metal ions. To verify whether this was the case, 1 was analyzed by inductively coupled plasma mass spectrometry (ICP-MS). The amount of Cu(II) that was detected per μmol diazotransfer reagent varied between 2 and 5 pmol. The Ni(II) and Zn(II) contents were less than 2.5 pmol and 20 pmol per μmol diazotransfer reagent, respectively. These values correspond to trace amounts of 0.0002–0.002 mol% catalyst relative to amines, and indicate that metal ions do not play a significant role in the diazotransfer reaction and are well below values known to affect protein activity.
Introduction of azides at pH 9.0 and 8.5
In addition to excluding Cu(II) from the reaction, we investigated the effect of lowering the pH. The diazotransfer was performed on lysozyme and CalB in diethanolamine buffered solutions of pH 8.5 and 9.0, using 1.75 equivalents of 1, first still in the presence of Cu(II). Mass analysis on the whole proteins confirmed that also under these conditions azide moieties were introduced. The number of amines that are converted into azides decreases upon lowering the pH to 9.0 and 8.5 (Fig. 2).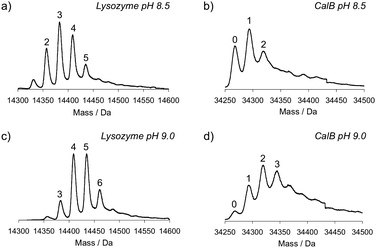 | ||
| Fig. 2 Mass spectra of lysozyme and CalB after diazotransfer at pH 8.5 and pH 9.0 in the presence of Cu(II). a) Lysozyme at pH 8.5, b) CalB at pH 8.5, c) lysozyme at pH 9.0, and d) CalB at pH 9.0. Numbers indicate the amount of azides that are present. | ||
When the diazotransfer was performed both at lower pH and in absence of Cu(II), the conversion efficiency was further reduced. However, increasing the amount of 1 from 1.75 to 17.5 equivalents relative to amines had a positive effect (Fig. 3). It was remarkable to see that in CalB only one azide was introduced at pH 8.5 even when 17.5 equivalents of 1 were added. Using the same conditions also in ELP-lys1 and ELP-lys3 only one azide was introduced, whereas two or four azides were introduced at pH 11 and/or in presence of Cu(II).
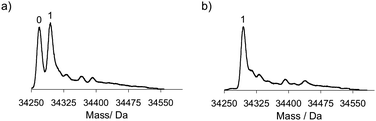 | ||
| Fig. 3 Mass spectra of CalB after diazotransfer at pH 8.5 in absence of Cu(II) using 1.75 (a) and 17.5 (b) equivalents of diazotransfer reagent. | ||
Identification of the single reactive amine
It was speculated that the selectivity was caused by differences in reactivity of the amines. The reactivity will depend on the pKa, which is influenced by the microenvironment of the amine. The pKa values of amines in a protein can vary significantly. For lysozyme for example, the amine with the lowest value is the α-amine (pKa is 7.0), while the two most basic amines have a pKa of 11.2.16 In general, the α-amine at the N-terminus of a protein will often have the lowest pKa. However, it is also the solvent accessibility of the amines that will influence their reactivity.Mono-functionalized CalB and ELP were treated with trypsin to determine which amine had been converted. The digests were analysed by mass spectrometry. A peak was detected at 1544.8 Da instead of 1518.7 Da (Fig. 4a). So, the N-terminal fragment of CalB (amino acids 1–15) had shifted 26 Da in mass, indicating that the α-amine at the N-terminus of CalB had been converted into an azide. Additional LC-MS/MS analysis using trypsin, LysN and chymotrypsin demonstrated that all other lysines had remained unmodified, with the exception of a very low level of conversion of Lys210 (see ESI†).
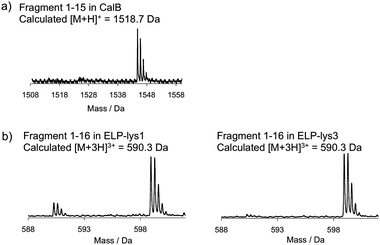 | ||
| Fig. 4 Mass spectra of the N-terminal fragments of mono-functionalized (a) CalB, (b) ELP-lys1 and ELP-lys3. Calculated masses are those of unmodified peptides. | ||
In the tryptic digests of ELP-lys1 and ELP-lys3 a peak of the triply charged fragment was detected that corresponded to amino acids 1–16 containing an azide, i. e. [M + 3H]3+ being 598.9 Da instead of 590.3 Da (Fig. 4b). For ELP-lys1 a small fraction of the unreacted fragment was detected as well. The peak corresponding to amino acids 17–24 of ELP-lys1 and ELP-lys3 containing a converted lysine at position 23 was only present when Cu(II), a base and/or 175 equivalents of diazotransfer reagent were added. Furthermore, a protein fragment was detected that confirmed that the two additional lysines in ELP-lys3 were unchanged in the mono-functionalized sample (see ESI†). It was therefore concluded that in both ELP variants it was the N-terminal amine as well that had been converted at pH 8.5 in the absence of Cu(II).
To confirm that selectivity is correlated to the pKa of an amine, the pH-controlled, copper-free diazotransfer was performed on the tripeptide Lys-Phe-Phe. While the α-amine at the N-terminus of the tripeptide is expected to have a pKa value of approximately 8.0, the ε-amine in the lysine side-chain will have a pKa around 9.5.17 The diazotransfer was performed both at pH 11 in the presence of Cu(II) and at pH 8.5 without adding Cu(II). Whereas the first reaction conditions resulted in a product containing two azides, only one amine was converted at pH 8.5 in the absence of Cu(II). 13C NMR proved that the N-terminal amine had been diazotized (ESI†).
Lysozyme was subjected to proteolytic digestion and LC-MS/MS analysis as well. Even though the N-terminus has the lowest pKa of all amines present in the protein,16 it was not modified after diazotransfer at pH 8.5. This can be explained by the extremely low surface accessibility of the N-terminus, as can be deduced from its crystal structure.18 At pH 8.5 in absence of Cu(II), lysine 13 and lysine 116 turned out to be partially modified (see ESI†). When Cu(II) was present during the modification reaction at pH 8.5, additional amines were converted, at lysine 96 and/or 97 and the N-terminal lysine residue of the protein.
PEGylation and activity of azide-functionalized proteins
It was demonstrated that the azides that were introduced in CalB and lysozyme were available for bioconjugation (Fig. 5). Polyethyleneglycol (PEG) functionalized with an aza-dibenzocyclooctyne19 was added to facilitate a copper-free click reaction. This compound was added to the model proteins after dialysis against phosphate buffer (pH 7) to remove the excess of diazotransfer reagent and, if present, Cu(II). Upon overnight incubation conjugation was visualized by gel electrophoresis. The degree of PEGylation correlated to the number of azides that had been introduced.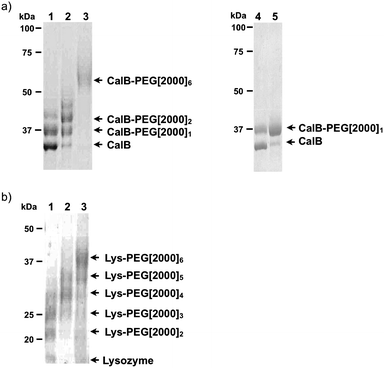 | ||
| Fig. 5 SDS-PAGE gel after PEGylation of (a) CalB and (b) lysozyme, after diazotransfer using 1.75 eq of 1 in the presence of Cu(II) at pH 8.5 (lane 1), pH 9.0 (lane 2) and pH 11 (lane 3), and after diazotransfer without Cu(II) at pH 8.5 using 1.75 eq (lane 4) or 17.5 eq (lane 5) of 1. | ||
The activities of both diazotized CalB and lysozyme were determined (see ESI†). Mono-functionalized CalB was nearly as active as the untreated control. With respect to lysozyme, when the sample incubated at pH 11, and especially in the presence of Cu(II) was compared to lysozyme treated under mild diazotation conditions (pH 8.5, no Cu(II)), a significant decrease in activity was detected. An additional reduction of activity was observed upon introduction of multiple azides. This effect was particularly large when an increasing number of azides was introduced, suggesting that under those conditions lysines are converted which are crucial for catalytic activity. This result indicates that selective modification is desired to limit loss in activity.
Conclusions
We show that azides can be introduced in proteinsvia an aqueous diazotransfer on amines without adding Cu(II) or any other transition metal salt. Furthermore, it is shown that the reaction proceeds not only at pH 11 but also at pH 8.5. The copper-free reaction at pH 8.5 leads to a selective diazotransfer in which only amines with the lowest pKa, such as the α-amine at the N-terminus, react.The selectivity at lower pH will be especially useful when site-specific functionalization is desired or when lysines play a role in maintaining structure or activity of the protein. It is expected that for individual proteins, the exact conditions for site-specific modification will depend on the surface accessibility and pKa values of the amines in the protein.
Longer reaction times and, if possible, a slightly increased reaction temperature will enable the efficient introduction of azides at even lower pH. With respect to CalB, for example, a single azide could already be introduced at pH 8 while using 17.5 equivalents of 1 relative to the amine. Approximately 60% conversion was obtained under those conditions. As for lysozyme, incubation at 30 °C and 37 °C at pH 8.5 was shown to improve the degree of diazotation. Extension of the reaction time to 60 h had a beneficial effect on the introduction of azides as well. Furthermore, reaction times of only a few hours instead of overnight incubation may be applied. The most reactive amines will be converted in a few hours, as has been observed in our laboratory for several peptides as well as for ELP.
Using these non-toxic and mild conditions, the diazotransfer reaction will be an excellent candidate for modification of proteins, which are unstable at high pH or inactivated by binding of Cu(II) ions. The strategy will be useful for the site-specific immobilization of enzymes, for example inside microchannels or onto nanostructured materials such as polymeric vesicles and DNA scaffolds. So far, a small number of relatively stable biocatalysts, like lipases and a few oxidoreductases have been used in these applications.20 For synthetic purposes it will be desirable to expand the set of biocatalysts that can be exploited.
Experimental section
Diazotransfer to proteins
Stock solutions of diazotransfer reagent 1 (2 mg mL−1, 9.5 mM), Na2CO3 (10 mg mL−1, 94.3 mM) and CuSO4·5H2O (1 mg mL−1, 4 mM) in MQ were prepared. To 100 μg protein in MQ (concentrations varying from 4 to 10 mg mL−1) 1.75 equivalents of diazotransfer reagent per amine were added. 5.9 μL Na2CO3 and 5.0 μL CuSO4·5H2O were added where intended. MQ was added to a final volume of 100 μL. To the reactions performed at pH 8.5 or 9 no Na2CO3 was added and an aqueous solution of 50 mM diethanolamine (pH 8.5 or 9) was used instead of MQ to obtain a final volume of 100 μL. In case the reactions were performed in presence of 17.5 or 175 equivalents of diazotransfer reagent, a 20 mg mL−1 stock solution of 1 was prepared. NaOH was added to the stock solutions of 1 to neutralize the pH to 7. All reactions were incubated overnight at room temperature while shaken on an Eppendorf® Thermomixer comfort.Acknowledgements
The authors would like to thank J. Eijgenstein for ICP-MS analysis and P. J. H. M. Adams for synthesis of the tripeptide. S. A. Meeuwissen and M. F. Debets are acknowledged for providing the diazotransfer reagent and the aza-dibenzocyclooctyne functionalized PEG, respectively. This work was financially supported by the Netherlands Organization for Scientific Research NWO and the Dutch Polymer Institute.Notes and references
- (a) C. W. Tornøe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057 CrossRef CAS; (b) V. V. Rostovtsev, L. G. Green, V. V. Fokin and B. K. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596 CrossRef CAS.
- (a) J.-F. Lutz, Angew. Chem., Int. Ed., 2008, 47, 2182 CrossRef CAS; (b) M. F. Debets, C. W. J. van der Doelen, F. P. J. T. Rutjes and F. L. van Delft, ChemBioChem, 2010, 11, 1168 CrossRef CAS.
- (a) A. Deiters, T. A. Cropp, D. Summerer, M. Mukherji and P. G. Schultz, Bioorg. Med. Chem. Lett., 2004, 14, 5743 CrossRef CAS; (b) K. L. Kiick, E. Saxon, D. A. Tirrell and C. R. S. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 19 CrossRef CAS; (c) Schoffelen, M. H. L. Lambermon, M. B. van Eldijk and J. C. M. van Hest, Bioconjugate Chem., 2008, 19, 1127 CrossRef CAS.
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007 CrossRef CAS.
- M. Fernández-Suárez, H. Baruah, L. Martínez-Hernández, K. T. Xie, J. M. Baskin, C. R. Bertozzi and A. Y. Ting, Nat. Biotechnol., 2007, 25, 1483 CrossRef CAS.
- J. Xiao and T. J. Tolbert, Org. Lett., 2009, 11, 4144 CrossRef CAS.
- S. F. M. Van Dongen, R. L. M. Teeuwen, M. Nallani, S. S. van Berkel, J. J. L. M. Cornelissen, R. J. M. Nolte and J. C. M. van Hest, Bioconjugate Chem., 2009, 20, 20 CrossRef CAS.
- E. D. Goddard-Borger and R. V. Stick, Org. Lett., 2007, 9, 3797 CrossRef CAS.
- Y. H. Hui, in Handbook of Food Science, Technology, and Engineering, CRC Press, 2006, vol. 1, ch. 6, pp. 10–11 Search PubMed.
- R. M. Sterritt and J. N. Lester, Sci. Total Environ., 1980, 14, 5 CrossRef CAS.
- (a) C. J. Cavender and V. J. Shiner, J. Org. Chem., 1972, 37, 3567 CrossRef CAS; (b) J. Zaloom and D. C. Roberts, J. Org. Chem., 1981, 46, 5173 CrossRef CAS.
- P. B. Alper, S.-C. Hung and C.-H. Wong, Tetrahedron Lett., 1996, 37, 6029 CrossRef CAS.
- (a) H. S. G. Beckmann and V. Wittmann, Org. Lett., 2007, 9, 1 CrossRef CAS; (b) N. M. Smith, M. J. Greaves, R. Jewell, M. W. D. Perry, M. J. Stocks and J. P. Stonehouse, Synlett, 2009, 1391 CrossRef CAS; (c) S. Maisonneuve and J. Xie, Synlett, 2009, 18, 2977.
- P. T. Nyffeler, C.-H. Liang, K. M. Koeller and C.-H. Wong, J. Am. Chem. Soc., 2002, 124, 10773 CrossRef CAS.
- R. L. M. Teeuwen, F. A. de Wolf, H. Zuilhof and J. C. M. van Hest, Soft Matter, 2009, 5, 4305 RSC.
- V. Z. Spassov, A. D. Karshikov and B. P. Atanasov, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 1989, 999, 1 Search PubMed.
- G. T. Hermanson, in Bioconjugate Techniques, Academic Press, London, 2nd edn, 2008, ch. 1, p. 9 Search PubMed.
- D. J. Vocadlo, G. J. Davies, R. Laine and S. G. Withers, Nature, 2001, 412, 835 CrossRef CAS.
- M. F. Debets, S. S. van Berkel, S. Schoffelen, F. P. J. T. Rutjes, J. C. M. van Hest and F. L. van Delft, Chem. Commun., 2010, 46, 97 RSC.
- (a) T. C. Logan, D. S. Clark, T. B. Stachowiak, F. Svec and J. M. J. Frechet, Anal. Chem., 2007, 79, 6592 CrossRef CAS; (b) S. F. M. van Dongen, M. Nallani, J. L. L. M. Cornelissen, R. J. M. Nolte and J. C. M. van Hest, Chem.–Eur. J., 2009, 15, 1107 CAS; (c) O. I. Wilner, Y. Weizmann, R. Gill, O. Lioubashevski, R. Freeman and I. Willner, Nat. Nanotechnol., 2009, 4, 249 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: CalB and ELP production, synthesis of imidazole-1-sulfonyl azide hydrochloride, proteolytic digestion, mass spectrometry, LC-MS/MS, synthesis and diazotransfer to tripeptide Lys-Phe-Phe, click reaction, SDS PAGE analysis, and activity assays. See DOI: 10.1039/c0sc00562b |
| ‡ It should be noted that diazotransfer reagent 1 is shelf stable, but should not be kept as a concentrated aqueous solution for prolonged time due to risk of decomposition. |
| This journal is © The Royal Society of Chemistry 2011 |