Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The human iron-proteome†
Claudia
Andreini
ab,
Valeria
Putignano
a,
Antonio
Rosato
 ab and
Lucia
Banci
ab and
Lucia
Banci
 *ab
*ab
aMagnetic Resonance Center, University of Florence, Via Luigi Sacconi 6, Sesto Fiorentino 50019, Italy. E-mail: banci@cerm.unifi.it; Fax: +39 055 4574253; Tel: +39 055 4574273
bDepartment of Chemistry, University of Florence, Sesto Fiorentino 50019, Italy
First published on 10th August 2018
Abstract
Organisms from all kingdoms of life use iron-proteins in a multitude of functional processes. We applied a bioinformatics approach to investigate the human portfolio of iron-proteins. We separated iron-proteins based on the chemical nature of their metal-containing cofactors: individual iron ions, heme cofactors and iron–sulfur clusters. We found that about 2% of human genes encode an iron-protein. Of these, 35% are proteins binding individual iron ions, 48% are heme-binding proteins and 17% are iron–sulfur proteins. More than half of the human iron-proteins have a catalytic function. Indeed, we predict that 6.5% of all human enzymes are iron-dependent. This percentage is quite different for the various enzyme classes. Human oxidoreductases feature the largest fraction of iron-dependent family members (about 37%). The distribution of iron proteins in the various cellular compartments is uneven. In particular, the mitochondrion and the endoplasmic reticulum are enriched in iron-proteins with respect to the average content of the cell. Finally, we observed that genes encoding iron-proteins are more frequently associated to pathologies than the all other human genes on average. The present research provides an extensive overview of iron usage by the human proteome, and highlights several specific features of the physiological role of iron ions in human cells.
Significance to metallomicsIron is one of the most ancient and abundant metal ions in living organisms: it participates in fundamental biological processes, such as photosynthesis, and respiration. It is an essential metal ion for humans. Here, we applied a bioinformatics approach to predict the entire set of human proteins that use iron as cofactor. We found that about 2% of human genes encode an iron-protein. In particular, 35% are proteins binding individual iron ions, 48% are heme-binding proteins and 17% are iron–sulfur proteins. Most of these proteins are enzymes: 37% of the human oxidoreductases need an iron ion to perform their catalytic mechanisms. The analysis of the subcellular location highlighted that some organelles are enriched in iron-proteins, in particular about 7% of the proteins localized in the endoplasmic reticulum and in the mitochondrion bind iron. Finally, our data show that mutations in genes encoding iron-binding proteins are more likely to be associated with pathology than all human genes on average. |
Introduction
During evolution, organisms have selected some of the available elements from the environment to catalyze physiological reactions. Consequently, some metal ions became essential to life. Iron is one of the most ancient and abundant transition metal ions in living organisms,1,2 as it was highly available as ferrous ion in the early days of terrestrial life.3 Iron is essential to all forms of life and participates in fundamental biological processes, such as photosynthesis, respiration and nitrogen fixation.4,5 In cells, it is normally found in the +2 (ferrous) and/or +3 (ferric) oxidation states. Higher oxidation states may be generated transiently in the course of the catalytic cycle of enzymatic reactions. Besides individual iron ions, proteins can bind also iron-containing cofactors, such as heme or iron–sulfur clusters.6–8 Heme is one of the most versatile prosthetic groups in metalloproteins. The porphyrin constituting the heme group can be of several types, including e.g. heme a, heme b, and heme c. The heme proteins that transfer electrons mainly belong to the cytochromes class, and may contain one or several heme groups; globins are heme-containing proteins involved in dioxygen binding and/or transport; other heme proteins serve as biological sensors for oxidative stress. The broad range of possible reactions occurring at the heme center is mainly based on the ability of the heme iron to coordinate small molecules like CO, NO, and O2. The protein matrix can modulate the affinity towards the different exogenous ligands. Iron–sulfur clusters contain two or more iron ions bridged by sulfide ions. Each iron ion is tetracoordinated, with its coordination sphere typically completed by the sulfur or nitrogen atoms of cysteine and histidine side chains, respectively.9 The metal site of rubredoxin, which contains a single iron ion coordinated by four cysteines, is generally classified as the simplest unit of iron–sulfur clusters. Iron–sulfur clusters are among the most versatile inorganic cofactors.5 They are involved in a plethora of functional processes, including aerobic as well as anaerobic respiration, regulation of gene expression, amino acid and nucleotide metabolism, DNA modification and repair and tRNA modification.Heme and iron–sulfur clusters are cofactors featuring a high chemical complexity. Therefore, their biosynthesis as well as the biosynthesis of the final holo-proteins containing these cofactors involve a significant number of different protein components, some of which are iron-binding proteins. In the human cell, these biosynthetic processes have multiple pathways, related also to cellular compartmentalization. Nevertheless, some components may move across different compartments; furthermore, the various pathways can communicate with one another via the exchange of biosynthetic intermediates.
While iron is essential for life, it can catalyze the formation of potentially toxic reactive oxygen species (ROS). This process is unavoidable in the present oxygen-rich environment, and iron and ROS are increasingly recognized as important initiators and mediators of cell death in various organisms as well as in pathological conditions in humans.10 Therefore, biological systems must control iron metabolism by providing the adequate amount of iron for proper cellular function while limiting iron toxicity.11,12 Iron has also a role in pathogen virulence. The growth of microbial pathogens within the host usually requires iron as an essential nutrient.13,14 Heme-containing proteins, such as hemoglobin, and transferrin are the preferential iron sources for human pathogens.15,16 Therefore, another crucial reason for the cell to maintain a strict control on iron homeostasis is to restrict its access by pathogens.
In this paper, we carried out a systematic prediction of iron-binding proteins encoded in the human genome, extending our previous analysis on iron–sulfur proteins.17 By integrating this prediction with information on heme and individual iron ions, we achieved a complete landscape of the iron handling by proteins in human, thus providing a framework for the understanding of physiological iron metabolism and of its dysfunction in diseases.
Results
Iron binding by human proteins and their coordination spheres
We analysed iron usage by human proteome via three different possible modes of binding: as individual iron ions, as iron-containing heme cofactors and as iron–sulfur clusters. In total, we identified 398 human genes whose protein products interact with iron (iron-proteins hereafter), i.e. about 2% of the human genes. Of these, 139 genes express proteins binding individual iron ions (Table S1, ESI†), 192 express proteins binding heme (Table S2, ESI†) and 7017 express proteins binding iron–sulfur clusters (Table S3, ESI†).The coordination spheres of the three different iron-containing cofactors are quite diverse; we refer to the pattern of the protein residues coordinating the iron ion(s) of the cofactor as the iron-binding pattern (IBP). The IBP is a regular expression defined by the identity of the amino acids coordinating the metal and by their spacing along the protein sequence (e.g. CX4CX25C). Thus, the coordination sphere of each iron ion corresponds to a single IBP.
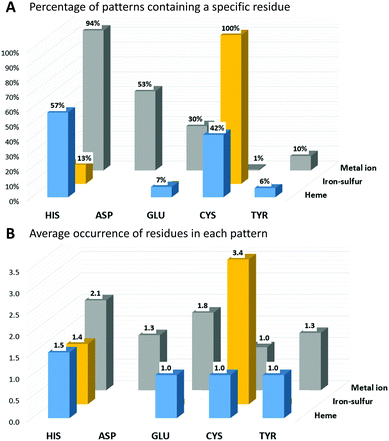
In IBPs of human iron-proteins binding individual iron ions, histidine is by far the most common residue. His is present in 94% of these IBPs, each of which contains on average two His (Fig. 1). Aspartate, glutamate and tyrosine are found in 53%, 30% and 10% of the identified patterns, respectively. On average, only one Asp and one Tyr are found in each IBP, whereas there can be one (such as in most iron-dependent enzymes) or two (such as in ferritins) Glu residues. All iron–sulfur binding proteins use on average three-four cysteines to coordinate the cluster. Cys is absolutely required in the IBPs of these proteins. In particular, in human iron–sulfur proteins the coordination sphere of the Fe4S4 clusters is always and only composed by cysteines whereas the IBPs of Fe2S2 clusters sometimes (37% of Fe2S2 IBPs) include one or two His residues. In human heme-binding proteins, IBPs commonly contain one or two His with the exception of catalytic heme sites (such as in cytochrome P450) where Cys is more common (83% of IBPs).
The function of the metal cofactor within the protein is correlated also to the number of coordinating residues provided by the protein (i.e. the number of residues in the IBP). Indeed, the coordination sphere of the metal ion is not always completed by atoms of the protein. 64% of the sites that bind individual iron ions contain three protein residues in the IBP, whereas the others contain four protein residues. Similarly, most of the iron ions in heme cofactors have only one ligand provided by the protein (about 58%), which allows the substrate to occupy the second heme axial position. The remaining 42% heme sites have two coordinating residues provided by the protein. In iron–sulfur proteins, the most common number of protein ligands is 4; however, all the iron–sulfur clusters that perform a catalytic function have only three Cys ligands in the IBP. It is thus evident that there is a trend for human iron-proteins to have a lower number of residues in their IBPs when the metal-binding site performs a catalytic function, in order to allow the iron ion to coordinate directly to the substrate as already observed for other metal containing proteins.18
Subcellular localization of human iron-proteins
We then analysed the subcellular localization of the human iron-proteins identified through our search (Tables S4–S6, ESI†). This information is not available for 94 proteins (37 binding individual iron ions, 10 binding iron–sulfur clusters, and 47 binding hemes), which were thus ignored for this analysis. Various proteins are present in more than one compartment, and thus were included in the statistics of each relevant organelle. Fig. 2 summarizes the distribution of the different types of iron-proteins within each cellular compartment and reports the fraction of iron-proteins with respect to the total number of proteins localized in each compartment (percentages within parenthesis). It appears that two subcellular locations stand out for their enrichment in iron-proteins: the mitochondrion and the endoplasmic reticulum.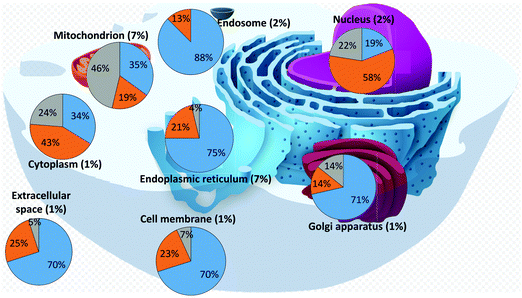 | ||
| Fig. 2 Distribution of iron-proteins in different cellular organelles of the human cell (heme-proteins: blue; iron–sulfur proteins: grey; individual iron ions: orange). | ||
Our dataset (iron-proteins for which cellular localization is known) is composed by 45% heme-binding proteins, 34% proteins binding individual iron ions, and 21% proteins binding iron–sulfur clusters. From Fig. 2, we can readily identify compartments that differ appreciably in the distribution of the types of iron-proteins. The nucleus is highly depleted of heme-binding proteins, whereas it features a relatively high number of proteins binding individual iron ions. On the other hand, the mitochondrion is the compartment most enriched in iron–sulfur proteins, with respect to both the two other types, whereas the endosome is mostly enriched in heme-binding proteins and does not contain any iron–sulfur protein. In addition, the endoplasmic reticulum is enriched in heme-binding proteins and depleted in iron–sulfur proteins. The distribution of the three types of iron-proteins in the cytoplasm closely resembles that of the overall dataset. It should be noted that in this respect, we are referring to the number of proteins and not to their relative quantity, which depends on their expression levels. We did not analyze such levels in this work.
The mitochondrion and the endoplasmic reticulum are the compartments with the largest percentage of iron-proteins. As mentioned, the mitochondrion is significantly enriched in iron–sulfur proteins (about 2.5 times the average fraction for the whole cell), whereas the endoplasmic reticulum is enriched in heme-binding proteins (1.6 times the cell average). The nucleus is the only compartment where proteins binding individual iron ions are the majority of iron-proteins (1.7 times the cell average).
Functional roles
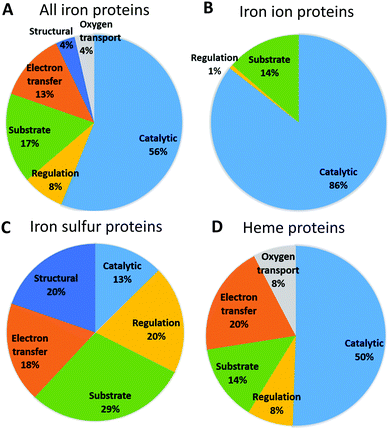
Fig. 3 shows the functional roles of sites binding iron and iron-containing cofactors in human proteins (Tables S4–S6, ESI†). This information is not available for 24 proteins (14 binding iron–sulfur clusters, and 10 binding heme), which were thus ignored for this analysis. It appears that sites binding heme or individual iron ions most commonly have a catalytic role, i.e. are directly involved in enzymatic mechanisms. This is also the most common role for the entire set of iron-proteins, partly due to the low number of iron–sulfur proteins. For sites binding individual iron ions the only other relevant function is its use as a substrate, i.e. in storage and transport processes (this classification of sites is taken from the MetalPDB database9). Heme-binding sites have the largest variety of functional roles, among which electron transfer is the second most common. As it is well known, human heme-binding proteins also play a crucial role in the transport of molecular dioxygen and in sensing, particularly of small gaseous molecules such as NO, leading to a regulatory function. Heme-binding proteins associated with a substrate function (i.e. when the heme cofactor is the target/substrate of the protein) are involved in the biosynthesis, transport and degradation of the heme cofactor. This may be linked also to the fact that there are as many as seven different types of heme cofactors in human heme-binding proteins (heme a, b, c, d, i, o, m). While the most common type is heme b, occurring in 90% of the heme-proteins, the synthesis of all the other heme types requires the action of specific enzymes that modify the cofactor and/or the protein binding it (e.g. cytochrome c19).20,21 The most common role for iron–sulfur proteins is transport, biosynthesis and insertion into the final target proteins of the clusters themselves (tagged as substrate).22–26 This is the result of both the chemical complexity of the iron-containing clusters, thus requiring elaborate biosynthetic and degradation pathways, and the potential toxicity of free iron ions. The second most common roles for iron–sulfur proteins are structural and regulatory. The role of iron–sulfur clusters in several DNA- and RNA-binding proteins is not completely understood, in particular for the many systems involved in DNA repair, where the presence of the cluster could be instrumental to detect lesions. Curiously, sites performing electron transfer are less common.We then checked whether there is a relationship between cellular localization and protein function in order to rationalize the patterns reported in Fig. 2. To do this we examined the lists of the iron-proteins localized to the various compartments and identified all the processes, as defined by the Gene Ontology (GO27,28), associated with the corresponding genes. Seven processes involve 81% of the genes coding for iron-proteins localized to the endoplasmic reticulum (Table 1). The process involving more iron-proteins is lipid metabolism, which is a key cellular role played by cytochromes P450; only one tenth of the genes involved in lipid metabolism codes for proteins binding individual iron ions. Xenobiotic metabolic process and drug metabolism are common processes which involve exclusively heme-binding proteins and are essentially associated to cytochromes P450, which are involved in the modification of exogenous molecules, from drugs to pollutants. Proteins binding individual iron ions are involved in different pathways, such as peptidyl amino acid hydroxylation. These pathways do not involve any heme-binding protein. Overall, 92% of the iron-proteins localized to the endoplasmic reticulum are oxidoreductases, as directly observed from their Enzyme Commission (EC) numbers, and these are either members of the cytochrome P450 family (heme-containing enzymes) or iron-dependent hydroxylases (typically harboring two iron ions in their active site). The functional role of the iron-proteins in the endoplasmic reticulum is thus tightly linked to their catalytic activity, most commonly in biosynthetic or metabolic processes.
| All | iron_ion | iron_heme | iron_sulfur | |
|---|---|---|---|---|
| Endoplasmic reticulum | ||||
| Drug metabolism | 14 | 0 | 14 | 0 |
| Peptidyl amino acid hydroxylation | 6 | 6 | 0 | 0 |
| Lipid metabolic process | 43 | 5 | 38 | 0 |
| Cell proliferation | 12 | 4 | 8 | 0 |
| Response_to_stress | 9 | 0 | 9 | 0 |
| Vitamin metabolism | 8 | 0 | 8 | 0 |
| Xenobiotic metabolic process | 20 | 0 | 20 | 0 |
| Nucleus | ||||
| Cell death/apoptotic process | 20 | 10 | 5 | 5 |
| Gene expression | 46 | 33 | 9 | 4 |
| Cell proliferation | 20 | 11 | 5 | 4 |
| Peptidyl amino acid hydroxylation | 8 | 8 | 0 | 0 |
| Response to stress | 25 | 9 | 6 | 10 |
| Mitochondrion | ||||
| Cell death/apoptotic process | 13 | 4 | 5 | 4 |
| Iron ion homeostasis | 11 | 4 | 4 | 3 |
| Iron sulfur cluster biosynthesis | 6 | 0 | 0 | 6 |
| Cellular respiration | 18 | 1 | 7 | 10 |
| Response to drug | 9 | 1 | 5 | 3 |
| Response to stress | 16 | 3 | 5 | 8 |
In the nucleus, 5 processes involve about 89% of the iron-proteins present in this cell compartment. Gene expression is the process associated to most of these proteins, because several genes encode iron-proteins involved in the regulation of transcription e.g. through DNA binding or histone modification. Many iron-proteins in the nucleus are also involved in response to stress, for instance by repairing damaged DNA, in apoptosis17 and in cell proliferation. About half of the nuclear iron-enzymes are oxydoreductases; transferases and hydrolases are relatively common.
In the mitochondrion, 6 processes involve about 63% of all iron-proteins within this cellular compartment. The process involving the largest number of iron-proteins is cellular respiration, which leverages both heme-binding and iron–sulfur proteins (6 vs. 10 genes, respectively). Other processes involving more than 10 genes are cell death, iron ion homeostasis and response to stress (which is mainly response to oxidative stress), half of which are iron–sulfur proteins. The biosynthesis of iron–sulfur clusters comprises genes encoding require iron–sulfur proteins. At the functional level, the observed enrichment of the mitochondrion in iron–sulfur proteins (Fig. 2) is largely accounted for by the involvement of these proteins in the respiratory chain, in stress response and in the assembly of iron–sulfur clusters themselves. For the latter, the clusters are transiently bound by various proteins along the biosynthetic pathway, also depending upon the final target for cluster insertion.25,26,29 The electron transfer capabilities of iron–sulfur proteins are important but not the only determinant of the higher abundance in the mitochondrion of iron–sulfur proteins with respect to all iron-proteins.
Uncharacterized putative human iron-proteins
Our analysis identified several proteins that had not been described in the literature as binding iron or iron-containing cofactors. In particular, Retinoid-related Orphan Receptors-alpha, beta and gamma (RORα, RORβ, and RORγ, hereafter) were predicted to have a heme-binding site similar to that found in REV-ERBα and REV-ERBβ. The REV-ERB family binds heme with two axial ligands: one His and one Cys.30 The sequence alignment of these two families (Fig. S1, ESI†) clearly shows that the His ligand is strictly conserved also in the ROR family whereas the Cys ligand is not. However, the superimposition of the heme-containing 3D structure of REV-ERBβ (PDB code 3CQV30) with the experimental structures of RORα, RORβ and RORγ (PDB codes 1N83,311NQ7,324WLB,33 respectively) shows that the latter contain a Cys (Cys323, Cys262 and Cys320, respectively) that is essentially in the same position as the heme-binding Cys384 of REV-ERBβ (Fig. 4A). A small rearrangement of the side chains of the Cys residues would bring their Sγ atoms at a distance from the iron ion compatible with the formation of a coordination bond. This Cys corresponds to a strictly conserved position in the multiple sequence alignment of the ROR family (Fig. S1, ESI†). Furthermore, the cavities of the 3D structures of ROR are sterically compatible with the binding of a heme molecule and the regions in contact with the cofactor have a high sequence similarity with the REV-ERB family. Another new putative heme-binding protein is the extracellular matrix protein FRAS1. This protein is in the plasma membrane: it has a very long region exposed in the extracellular matrix and a short cytoplasmatic tail. We identified three putative heme-binding sites in the extracellular part. We predicted the occurrence of a site with two potential axial ligands (His2080 and His3301) whereas for the other two sites, we predicted only one ligand, i.e. His1799 and His1945, respectively. The structure of this protein is not available and we were not able to build a 3D structural model, which would have allowed us to evaluate the possible geometrical features of the three predicted sites. The HSPB1-associated protein 1 is another potential iron-binding protein which could bind a single iron ion via its residues His175, Asp177 and His257; all these three residues are highly conserved in the protein family. For this protein we could identify a suitable template in the PDB for 3D structural prediction by homology modeling: the Hypoxia-inducible factor 1-alpha inhibitor which has a sequence identity to human HSPB1-associated protein 1 as high as 26%, and contains a site binding a single iron ion. The structural model in Fig. 4B, shows that the predicted ligands of HSPB1-associated protein 1 have the proper spatial configuration to bind an iron ion. Finally, we predicted as putative heme-binding protein the phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2. A structure as well as a suitable 3D template for the putative heme-binding region of this protein are not available. This prediction, however, appears less reliable than the previous ones.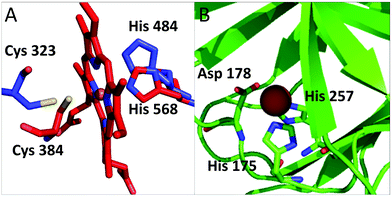 | ||
| Fig. 4 (A) Superposition of RORα (pdb code: 1n83, in blue) and REV-ERB (pdb code: 3cqv, in red). Only the relative positions of the putative ligands of RORα and the iron ligands of REV-ERB are reported. The side chain of Cys 323 is rotated to bring it closer to the heme iron. In this configuration the distance between the potential sulfur donor and the iron ion is 3.4 Å. (B) Putative iron-binding site in the structural model of HSPB1-associated protein 1. | ||
Pathogenic alterations associated to human iron-proteins
To assess the impact of the iron-proteome on the human health, we investigated how often defects or mutations affecting genes encoding iron-proteins are associated to pathologies (Tables S4–S6, ESI†). We analysed only proteins in the Swiss-Prot database (Reviewed proteins)34 and excluded those from the trEMBL database, which are just predicted and do not have mutational studies associated. Thus, we took into account 385 proteins (137 binding individual iron ions, 178 binding heme, and 70 binding iron–sulfur clusters). Of these, 148 are related to one or more pathogenic mutations or alterations, corresponding to about 38% of the total. Interestingly, if we consider the different types of iron sites, we found that more than half of the identified iron–sulfur proteins are involved in pathologies (37/70 corresponding to 53%). For proteins binding individual iron ions or heme cofactors, the percentage of proteins associated to pathologies is 31% (i.e. 43/137) and 38% (i.e. 68/178), respectively. As of January 2018, the total number of human proteins in the Swiss-Prot database was 20259. Of these, 4014 are associated to pathogenic mutations, corresponding to about 20% of the dataset. It thus appears that on average defects or mutations affecting genes encoding iron-proteins are more commonly associated to pathologies than all the other genes.In Table 2 we broke down the cumulative data reported in the previous paragraph for the whole human cell by looking at specific compartments. In particular, we took into consideration the compartments with the highest number of iron-proteins. In the mitochondrion, 36% of all proteins are associated to pathologies, whereas as many as 60% of mitochondrial iron-proteins are disease-related, with the main contribution of heme-proteins and iron–sulfur proteins. Similarly, in the cytoplasm and in the nucleus, heme-proteins and iron–sulfur proteins are more commonly associated to pathologies than all other human genes (Table 2).
| Heme | Individual iron-ions | Iron–sulfur clusters | Total iron-proteins | All human proteins | |
|---|---|---|---|---|---|
| Cytoplasm | 13/27 (48%) | 10/34 (29%) | 8/19 (42%) | 31/80 (39%) | 1413/5569 (25%) |
| Endoplasmic reticulum | 15/60 (25%) | 9/17 (53%) | 0/3 (0%) | 24/80 (30%) | 362/1163 (31%) |
| Mitochondrion | 20/28 (72%) | 5/15 (33%) | 23/37 (62%) | 48/80 (60%) | 420/1174 (36%) |
| Nucleus | 7/17 (41%) | 10/52 (19%) | 11/20 (55%) | 28/89 (31%) | 1180/5389 (22%) |
Discussion
398 human genes encode iron-proteins, which correspond to about 2% of all human genes. This number should be regarded as a lower limit because within our approach to the identification of iron-proteins false positives (i.e. proteins that do not bind iron but are predicted to do so) are quite unlikely to occur. This is due to the fact that we rely significantly on the known 3D structures of iron-proteins, while in the absence of structural data we scan the literature for supporting evidence. On the other, it is possible that we did not detect completely uncharacterized iron-proteins, especially if they are membrane-associated. Therefore, this number (398) should be taken as a lower limit even if we foresee that the actual number should not be much different.Of the 398 human iron-proteins, 48% are heme-binding proteins, 35% are proteins binding individual iron ions and 17% are iron–sulfur proteins. The intracellular distribution of these proteins is uneven, with some organelles containing a larger share of iron-proteins than others do. In particular, 7% of all the proteins localized in the endoplasmic reticulum and in the mitochondrion are iron-proteins. Thus these two organelles are significantly enriched (in comparative terms) in iron-proteins with respect to the average of the entire human cell (2%, as mentioned above). Within heme-binding proteins, 90% bind heme b and 61% are membrane-associated.
The three types of iron-proteins feature highly diverse preferences in the coordination sphere of the bound iron ions (i.e. IBPs). Cys is always present in the IBPs of iron–sulfur proteins, whereas it is practically absent from the coordination sphere of individual iron ions. Conversely, His, which is nearly always present in the IBPs of proteins binding individual iron ions, is observed rarely in the IBPs of iron–sulfur proteins. Asp is the second most common ligand in proteins binding individual iron ions. Heme-proteins have a similar preference for His and Cys in their IBPs. Cys is particularly common in the IBPs of heme-proteins that have catalytic function. This is presumably linked to the role of Cys in promoting the heterolytic breakage of the O–O bond of the iron-bound peroxide intermediate that forms along the catalytic cycle of cytochromes P450 or of nitric oxide synthase.35–37 This feature is independent of the overall protein fold, and is defined by the coordination chemistry properties of the sites.
6.5% of the human enzymes are iron-proteins. Unsurprisingly, this percentage is not the same for all enzyme classes. In particular, 37% of human oxidoreductases use a catalytic iron ion. 56% of all human iron-proteins have a catalytic function (Fig. 3). Proteins that bind individual iron ions mainly represent them: 86% of these proteins (119 out of 139) are iron-dependent enzymes. The large majority of these enzymes are oxidoreductases, in particular dioxygenases, where the iron ion is directly involved in the transfer of electron from/to the substrate. Also, about half of the heme-sites in the human proteins have a catalytic function. These enzymes are primarily members of the human cytochrome P450 family, whose isoforms are significantly differentiated in terms of expression but have typically broad and overlapping substrate specificities.
Iron-binding enzymes are commonly located in the nucleus and cytoplasm, followed by the mitochondrion and endoplasmic reticulum. The latter features the highest number of heme-binding proteins as it is the most common localization for cytochromes P450. Consistently with this, we observed that processes such as drug metabolism, lipid metabolism or xenobiotic stimulus are the most common processes associated with iron-proteins localized to the endoplasmic reticulum (Table 1). In the mitochondrion, 63% of all iron-proteins are involved in only 6 processes; the process involving the largest number of iron-proteins is respiration, which leverages both heme-binding and iron–sulfur proteins. The mitochondrion is the most likely localization for iron–sulfur proteins (Fig. 2), whose primary processes within this compartment are, besides respiration, the biosynthesis of iron–sulfur clusters and the response to oxidative stress. The biosynthesis of iron–sulfur clusters is among the most common functional roles of iron–sulfur proteins at the level of the whole cell,17,38 owing to the chemical complexity of this group of cofactors. Within the nucleus, iron-proteins are largely involved in various aspects of the regulation of protein expression, such as histone modification. In addition, also DNA binding, DNA biosynthesis and DNA replication involve several iron-proteins, especially iron–sulfur proteins.
We identified three human members of the retinoid-related orphan receptor (ROR) family as potentially harbouring a heme-binding site similar to those observed in proteins of the REV-ERB family. In the absence of experimental evidence in the literature, our hypothesis is supported by the strict conservation of the two potential heme ligands. The experimental structures of RORα, RORβ, and RORγ, feature a His and a Cys residue in a spatial position corresponding to His and Cys ligands of iron in REV-ERBβ. Another putative human iron-binding protein is the HSPB1-associated protein 1. A structural model of this proteins shows that the reciprocal position in 3D space of the putative ligands is completely consistent with our prediction (Fig. 4).
As an important aspect of the present study, we analysed how many pathologies are associated to human genes encoding iron-proteins, based on the occurrence of disease-associated mutations reported in the Swiss-Prot database. The percentage of pathologies associated to genes encoding iron-proteins is almost 40%, which is higher than the percentage of pathologies associated to all human genes (about 20%). In practice, two genes out of 10 are associated with pathogenic mutations in the human genome, whereas this percentage is essentially doubled if we take into account specifically the genes encoding iron-proteins. Interestingly, this percentage peaks at 72% for all heme-binding proteins in the mitochondrion.
In summary, this work provided an extensive overview of iron usage by human proteins, spanning from iron coordination properties to biochemical/cellular function and compartmentalization, and addressing the interplay between these aspects. We observed that the distribution of the type of iron cofactors and of their catalytic properties is quite uneven, with some organelles such as the mitochondrion or the nucleus displaying higher occurrence than the others. The main localization of iron-dependent enzymes, which constitute 6.5% of all human enzymes, is the endoplasmic reticulum, where they catalyze the modification of both endo- and exogenous molecules and metabolites. Human iron-enzymes have a lower number of protein residues in their IBPs, in order to allow the iron ion to coordinate directly to the substrate.
Materials and methods
Proteins are generally composed of one or more functional regions, commonly termed domains. The identification of domains that occur within proteins can therefore provide insights into their function. Pfam is a database of protein domains, defined on the basis of the comparison of ensembles of protein regions that share a significant degree of sequence similarity, thereby suggesting homology. Each domain is represented by a multiple sequence alignment and by a more complex mathematical representation called a hidden Markov model (HMM). HMMs can be used for analyzing proteomes to search for occurrences of the corresponding domain (see below). Each domain entry in the Pfam database has an annotation, which may include the ability to bind metal cofactors.Using the approach described in ref. 39 as implemented in the RDGB program,40 we predicted all iron-binding proteins (IBPs) encoded by the human genome. RDGB is a computational tool written in Python. The approach of RDGB exploits the protein domains of the Pfam database to identify putative homologues of the proteins of interest in any desired genome or list of genomes. Thus, the input to RDGB is a list of Pfam domains of interest (in our case, domains associated with iron-binding capability) and a list of genomes to be analyzed (in our case only the human genome).
The input list of Pfam domains is created by merging two lists: first, the list of all Pfam domains annotated as iron-binding, retrieved by mining the text of the annotations in the database; second, from the analysis of the sequence of iron-binding proteins with known 3D structure that are available from the Protein Data Bank (PDB). In the latter case, we extract from the PDB database also the pattern of amino acids that are responsible for metal binding (i.e. the metal binding pattern, MBP) and its position within the domain sequence. The MBP is defined by the identity and spacing of the amino acids, e.g., CX4CX20H, where X is any amino acid. This pattern provides a way to filter the initial results in order to reduce the number of false positives39 (i.e., of the proteins containing a Pfam domain annotated as iron-binding but which in reality are unable to bind it) by rejecting the proteins that lack the MBP or that have the MBP in the wrong position within the domain. The MBP filter cannot be applied in the absence of a relevant 3D structure available from the PDB. The MetalPDB database contains information on all the MBPs and the Pfam domains found in structurally characterized metalloproteins.9 Our search started from 352 Pfam domains: 261 with an associated iron-containing 3D structure (102 binding individual iron ions, 80 binding iron–sulfur clusters, and 79 binding heme) and 91 annotated as iron-binding domains.
This search was integrated by locally searching from MBPs within all human protein sequences. This is done by extracting from the HMM representing the Pfam domain that contains the binding site of interest only the regions around the MBP. This “trimmed domain” provides a convenient way to search for a MBP regardless of the agreement with the whole Pfam domain, thus affording a better sensitivity in the detection of MBPs in divergent sequences.41
In total we retrieved 363 human iron-proteins. As a qualitative indicator of reliability of our dataset, we checked whether one of the following conditions applied (in decreasing order of reliability):
(1) A 3D structure of the human protein in the iron-bound form is available (105 proteins).
(2) A 3D structure of a close homolog (sequence identity ≥50%) of the human protein in the iron-bound form is available (76 proteins).
(3) The predicted protein contains an iron-binding Pfam domain with a conserved MBP (147 proteins).
(4) The predicted protein contains a conserved MBP (based on local search) (22 proteins).
(5) The predicted protein contains an iron-binding Pfam domain, but the occurrence of the MBP cannot be verified due to the lack of a 3D structure for that domain family (13 proteins).
We integrated these predictions by adding the proteins annotated in the Uniprot database, a public comprehensive resource of protein sequence and functional information, as “iron-binding”, “iron–sulfur-binding”, or “heme-binding”. This contributed 35 additional iron-proteins.
For each predicted iron-protein, we retrieved the following annotations from UniProt:42 intracellular location, EC number, biological processes as reported in the Gene Ontology database,43 involvement in diseases. Further annotation such as the cofactor role and type were manually added by inspecting the literature. We used the Swiss-Prot database (at February 2018 contained 20259 entries)34 to compare the iron-protein dataset with all human proteins. For the latter dataset, annotations were retrieved from Uniprot in the same way as for the iron-protein dataset.
The 3D structural model of the HSPB1-associated protein 1 was built using MODELER v.9.244 and energy-refined using the AMBER45 web server provided by the WeNMR platform.46
Abbreviations
| IBP | Iron-binding pattern |
| ROS | Reactive oxygen species |
| ROR | Retinoid-related orphan receptor |
Conflicts of interest
There are no conflicts to declare.References
- C. Andreini, L. Banci, I. Bertini, S. Elmi and A. Rosato, Non-heme iron through the three domains of life, Proteins: Struct., Funct., Bioinf., 2007, 67, 317–324 CrossRef PubMed.
- C. Andreini, A. Rosato and L. Banci, The relationship between environmental dioxygen and iron–sulfur proteins explored at the genome level, PLoS One, 2017, 12, e0171279 CrossRef PubMed.
- A. Camacho, X. A. Walter, A. Picazo and J. Zopfi, Photoferrotrophy: Remains of an Ancient Photosynthesis in Modern Environments, Front. Microbiol., 2017, 8, 323 CrossRef PubMed.
- J. J. R. Frausto da Silva and R. J. P. Williams, The Biological Chemistry of the Elements, Oxford, Oxford, 1991 Search PubMed.
- I. Bertini, A. Sigel and H. Sigel, Handbook on Metalloproteins, New York, Marcel Dekker, 2001, p. 1800 Search PubMed.
- N. Maio and T. A. Rouault, Iron–sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery, Biochim. Biophys. Acta, 2015, 1853, 1493–1512 CrossRef PubMed.
- T. A. Rouault, Mammalian iron-sulphur proteins: novel insights into biogenesis and function, Nat. Rev. Mol. Cell Biol., 2015, 16, 45–55 CrossRef PubMed.
- S. Ciofi-Baffoni, V. Nasta and L. Banci, Protein networks in the maturation of human iron–sulfur proteins, Metallomics, 2018, 10, 49–72 RSC.
- V. Putignano, A. Rosato, L. Banci and C. Andreini, PDB in 2018: a database of metal sites in biological macromolecular structures, Nucleic Acids Res., 2018, 46, D459–D464 CrossRef PubMed.
- S. J. Dixon and B. R. Stockwell, The role of iron and reactive oxygen species in cell death, Nat. Chem. Biol., 2014, 10, 9–17 CrossRef PubMed.
- M. D. Knutson, Iron transport proteins: gateways of cellular and systemic iron homeostasis, J. Biol. Chem., 2017, 292, 12735–12743 CrossRef PubMed.
- R. Coffey and T. Ganz, Iron homeostasis: an anthropocentric perspective, J. Biol. Chem., 2017, 292, 12727–12734 CrossRef PubMed.
- H. Contreras, N. Chim, A. Credali and C. W. Goulding, Heme uptake in bacterial pathogens, Curr. Opin. Chem. Biol., 2014, 19, 34–41 CrossRef PubMed.
- C. Ratledge and L. G. Dover, Iron metabolism in pathogenic bacteria, Annu. Rev. Microbiol., 2000, 54, 881–941 CrossRef PubMed.
- T. A. Rouault and W. H. Tong, Iron–sulfur cluster biogenesis and human disease, Trends Genet., 2008, 24, 398–407 CrossRef PubMed.
- M. Caza and J. W. Kronstad, Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans, Front. Cell. Infect. Microbiol., 2013, 3, 80 Search PubMed.
- C. Andreini, L. Banci and A. Rosato, Exploiting bacterial operons to illuminate human iron–sulfur proteins, J. Proteome Res., 2016, 15, 1308–1322 CrossRef PubMed.
- C. Andreini and I. Bertini, A bioinformatics view of zinc enzymes, J. Inorg. Biochem., 2012, 111, 150–156 CrossRef PubMed.
- R. G. Kranz, R. Lill, B. Goldman, G. Bonnard and S. Merchant, Molecular mechanisms of cytochrome c biogenesis: three distinct systems, Mol. Microbiol., 1998, 29, 383–396 CrossRef PubMed.
- F. Fontanesi, I. C. Soto, D. Horn and A. Barrientos, Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process, Am. J. Physiol.: Cell Physiol., 2006, 291, C1129–C1147 CrossRef PubMed.
- F. Fontanesi, I. C. Soto and A. Barrientos, Cytochrome c oxidase biogenesis: new levels of regulation, IUBMB Life, 2008, 60, 557–568 CrossRef PubMed.
- R. Lill, R. Dutkiewicz, S. A. Freibert, T. Heidenreich, J. Mascarenhas, D. J. Netz, V. D. Paul, A. J. Pierik, N. Richter, M. Stumpfig, V. Srinivasan, O. Stehling and U. Muhlenhoff, The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron–sulfur proteins, Eur. J. Cell Biol., 2015, 94, 280–291 CrossRef PubMed.
- O. Stehling, C. Wilbrecht and R. Lill, Mitochondrial iron–sulfur protein biogenesis and human disease, Biochimie, 2014, 100, 61–77 CrossRef PubMed.
- O. Stehling and R. Lill, The role of mitochondria in cellular iron–sulfur protein biogenesis: mechanisms, connected processes, and diseases, Cold Spring Harbor Perspect. Biol., 2013, 5, a011312 Search PubMed.
- F. Camponeschi, S. Ciofi-Baffoni and L. Banci, Anamorsin/Ndor1 Complex Reduces [2Fe–2S]-MitoNEET via a Transient Protein–Protein Interaction, J. Am. Chem. Soc., 2017, 139, 9479–9482 CrossRef PubMed.
- V. Nasta, A. Giachetti, S. Ciofi-Baffoni and L. Banci, Structural insights into the molecular function of human (2Fe–2S) BOLA1–GRX5 and (2Fe–2S) BOLA3–GRX5 complexes, Biochim. Biophys. Acta, 2017, 1861, 2119–2131 CrossRef PubMed.
- M. Ashburner, C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin and G. Sherlock, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nat. Genet., 2000, 25, 25–29 CrossRef PubMed.
- The Gene Ontology Consortium, The Gene Ontology project in 2008, Nucleic Acids Res., 2008, 36, D440–D444 CrossRef PubMed.
- L. Banci, S. Ciofi-Baffoni, K. Gajda, R. Muzzioli, R. Peruzzini and J. Winkelmann, N-terminal domains mediate [2Fe–2S] cluster transfer from glutaredoxin-3 to anamorsin, Nat. Chem. Biol., 2015, 11, 772–778 CrossRef PubMed.
- K. I. Pardee, X. Xu, J. Reinking, A. Schuetz, A. Dong, S. Liu, R. Zhang, J. Tiefenbach, G. Lajoie, A. N. Plotnikov, A. Botchkarev, H. M. Krause and A. Edwards, The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta, PLoS Biol., 2009, 7, e43 CrossRef PubMed.
- J. A. Kallen, J. M. Schlaeppi, F. Bitsch, S. Geisse, M. Geiser, I. Delhon and B. Fournier, X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha, Structure, 2002, 10, 1697–1707 CrossRef PubMed.
- C. Stehlin-Gaon, D. Willmann, D. Zeyer, S. Sanglier, D. A. Van, J. P. Renaud, D. Moras and R. Schule, All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta, Nat. Struct. Biol., 2003, 10, 820–825 CrossRef PubMed.
- M. B. van Niel, B. P. Fauber, M. Cartwright, S. Gaines, J. C. Killen, O. Rene, S. I. Ward, B. G. de Leon, Y. Deng, C. Eidenschenk, C. Everett, E. Gancia, A. Ganguli, A. Gobbi, J. Hawkins, A. R. Johnson, J. R. Kiefer, H. La, P. Lockey, M. Norman, W. Ouyang, A. Qin, N. Wakes, B. Waszkowycz and H. Wong, A reversed sulfonamide series of selective RORc inverse agonists, Bioorg. Med. Chem. Lett., 2014, 24, 5769–5776 CrossRef PubMed.
- A. Bairoch and R. Apweiler, The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000, Nucleic Acids Res., 2000, 28, 45–48 CrossRef PubMed.
- R. Davydov, S. Im, M. Shanmugam, W. A. Gunderson, N. M. Pearl, B. M. Hoffman and L. Waskell, Role of the Proximal Cysteine Hydrogen Bonding Interaction in Cytochrome P450 2B4 Studied by Cryoreduction, Electron Paramagnetic Resonance, and Electron-Nuclear Double Resonance Spectroscopy, Biochemistry, 2016, 55, 869–883 CrossRef PubMed.
- J. H. Dawson and K. S. Eble, Cytochrome P450: heme iron coordination structure and mechanism of action, in Advances in Inorganic and Bioinorganic Mechanism, ed. Sykes J., London, Academic Press, 1986, pp. 2–64 Search PubMed.
- H. Li and T. L. Poulos, Structural variation in heme enzymes: a comparative analysis of peroxidase and P450 crystal structures, Structure, 1994, 2, 461–464 CrossRef PubMed.
- R. Lill and U. Muhlenhoff, Iron–sulfur protein biogenesis in eukaryotes: components and mechanisms, Annu. Rev. Cell Dev. Biol., 2006, 22, 457–486 CrossRef PubMed.
- C. Andreini, I. Bertini and A. Rosato, Metalloproteomes: a bioinformatic approach, Acc. Chem. Res., 2009, 42, 1471–1479 CrossRef PubMed.
- C. Andreini, I. Bertini, G. Cavallaro, L. Decaria and A. Rosato, A simple protocol for the comparative analysis of the structure and occurrence of biochemical pathways across superkingdoms, J. Chem. Inf. Model., 2011, 51, 730–738 CrossRef PubMed.
- Y. Valasatava, A. Rosato, L. Banci and C. Andreini, Metalpredator: a web server to predict iron–sulfur cluster binding proteomes, Bioinformatics, 2016, btw 238 Search PubMed.
- The Uniprot Consortium, UniProt: the universal protein knowledgebase, Nucleic Acids Res., 2017, 45, D158–D169 CrossRef PubMed.
- E. Camon, M. Magrane, D. Barrell, V. Lee, E. Dimmer, J. Maslen, D. Binns, N. Harte, R. Lopez and R. Apweiler, The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology, Nucleic Acids Res., 2004, 32, D262–D266 CrossRef PubMed.
- B. Webb and A. Sali, Comparative Protein Structure Modeling Using MODELLER, Curr. Protoc. Bioinformatics, 2014, 47, 5 Search PubMed.
- D. A. Case, T. A. Darden, T. E. Cheatham, III, C. L. Simmerling and J. Wang, et al., AMBER 12, version 12, University of California, San Francisco, CA, 2012 Search PubMed.
- I. Bertini, D. A. Case, L. Ferella, A. Giachetti and A. Rosato, A grid-enable web portal for NMR structure refinement with AMBER, Bioinformatics, 2011, 27, 2384–2390 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8mt00146d |
| This journal is © The Royal Society of Chemistry 2018 |