Open Access Article
Open Access ArticleUsing thiol–ene click chemistry to engineer 3D printed plasmonic hydrogel scaffolds for SERS biosensing†
Lara
Troncoso-Afonso
 abc,
Yolany M.
Henríquez-Banegas
ad,
Gail A.
Vinnacombe-Willson
abc,
Yolany M.
Henríquez-Banegas
ad,
Gail A.
Vinnacombe-Willson
 ac,
Junkal
Gutierrez
ac,
Junkal
Gutierrez
 e,
Gorka
Gallastegui
e,
Gorka
Gallastegui
 d,
Luis M.
Liz-Marzán
d,
Luis M.
Liz-Marzán
 acf and
Clara
García-Astrain
acf and
Clara
García-Astrain
 *acfg
*acfg
aCIC biomaGUNE, Basque Research and Technology Alliance (BRTA), 20014 Donostia-San Sebastián, Spain. E-mail: clara.garciaastrain@polymat.eu; clara.garcia@ehu.eus
bDepartment of Applied Chemistry, University of the Basque Country (UPV/EHU), 20018 Donostia-San Sebastian, Spain
cCentro de Investigación Biomédica en Red de Bioingeniería Biomateriales, y Nanomedicina (CIBER-BBN), 20014 Donostia-San Sebastián, Spain
dChemical and Environmental Engineering Department, University of the Basque Country (UPV/EHU), 01006 Vitoria-Gasteiz, Spain
eChemical and Environmental Engineering Department, Univeristy of the Basque Country (UPV/EHU), 20018 Donostia-San Sebastián, Spain
fIkerbasque Basque Foundation for Science, 48009 Bilbao, Spain
gPOLYMAT, Applied Chemistry Department, Faculty of Chemistry, University of the Basque Country (UPV/EHU), Paseo Manuel Lardizábal 3, 20018 Donostia–San Sebastián, Spain
First published on 16th April 2025
Abstract
3D cell culture models allow the study of the biomolecular processes underlying pathophysiological conditions by mimicking tissues and organs. Despite significant progress in creating such 3D architectures, studying cell behaviour in these systems still poses some challenges due to their heterogeneity and complex geometry. In this context, surface-enhanced Raman spectroscopy (SERS) can be implemented for molecular detection in biological settings with high sensitivity. The incorporation of SERS sensors in 3D models can thus lead to powerful platforms to study cellular response to therapeutics, metabolic pathways, signaling, and cell–cell communication events. Here, we introduce a library of plasmonic hydrogels that can be orthogonally photo-crosslinked via thiol–ene click chemistry and identify the main physicochemical factors accounting for their SERS performance. Using hydrogel-forming polymers such as gelatin, alginate, and carboxymethylcellulose modified with complementary thiol and norbornene groups, we created hydrogels with tailored chemical backbones. We identified swelling, porosity, and chemical composition as crucial factors determining their potential to detect different molecules by SERS. We additionally assessed their biocompatibility and printability, to ensure that these hydrogels meet the requirements for their use as 3D cellular scaffolds, showing their potential for real-time and in situ detection of biorelevant metabolites.
1. Introduction
In vitro cell models are essential tools for unravelling the fundamental biophysical and biomolecular mechanisms underlying different pathological processes.1 Although two-dimensional (2D) cell cultures have been widely accepted for their use in preclinical trials and have contributed to broadening our understanding of cell behavior, they can deeply deviate from in vivo systems.2 Therefore, to narrow the translational gap between 2D cellular models and the human body, efforts have been made towards developing reliable three-dimensional (3D) cell models.3 In such systems, cells grow in an organ- or tissue-like fashion, together with relevant components of the extracellular matrix, to mimic native cell–cell and cell–matrix interactions, as well as their physicochemical and mechanical properties. In this regard, hydrogels are the most relevant tissue-like matrices to allow cell growth in 3D.4 These hydrophilic networks enable the flux of oxygen and nutrients and support cell growth and adhesion, and their mechanical properties can be tailored to meet those of tissues.5To gain a better understanding of the metabolic pathways implied under pathophysiological conditions or to assess the response to drugs, such 3D models must be combined with robust analytical tools. On one hand, monitoring techniques might range from conventional optical microscopy to the integration of thermal, piezoelectric or electrochemical sensors.6–11 However, many of these techniques are invasive and may result in sample destruction, with loss of relevant biochemical information and hindering time-dependent studies. On the other hand, standard techniques to assess cell response to pharmaceuticals include the determination of metabolic activity or cell death.12 However, these tests are difficult to apply in multi-layered matrices where cells are organized in three dimensions.13 In this context, surface-enhanced Raman spectroscopy (SERS) has become a popular technique for the detection of molecular biomarkers in complex biological environments.14 The reasons behind are its excellent sensitivity, specificity and biocompatibility: (i) SERS enhancement enables the detection of biomolecules at milli- and micro-molar concentrations;15 (ii) the SERS vibrational spectrum of an analyte provides its molecular fingerprint;16 (iii) the source of radiation can be selected to lie in the near-infrared region (NIR), known as the biological transparency window, with enhanced penetration through tissues without compromising the integrity of the biological systems and allowing deep measurements in 3D;17 and (iv) the background signal in aqueous media is minimal because the inelastic Raman scattering cross section for water is very low.18 Considering the characteristics described above, this technique is appealing toward monitoring and analysing cellular behaviour with high spatial and temporal resolution in 3D cellular models.
SERS is based on the amplification of the Raman vibrational modes of molecules when they are adsorbed onto (or nearby) the surface of a metallic nanostructure. This amplification arises from the enhancement of the electromagnetic field when the incident beam couples with a localized surface plasmon resonance (LSPR) in the nanostructure.19 The incorporation of such nanostructures into hydrogels has been shown to allow the in situ detection of metabolites and biomarkers that are secreted by cells laden in the hydrogel network.20 Moreover, hydrogel-based plasmonic bioinks designed for 3D printing enable the construction of multiscale architectures with precise definition in the x-, y-, and z-directions. So far, few examples of plasmonic-hydrogel hybrid scaffolds for cell culture in 3D can be found in the literature. For instance, the combination of alginate and gelatin has been reported for the detection of methylene blue whereas methacrylated hyaluronic acid and κ-carrageenan were used for adenosine detection.21,22 In the design of these materials, it is important to tailor the physicochemical factors (presence of certain chemical moieties, hydrophobicity/hydrophilicity, biocompatibility, etc.) that account for selective molecular biosensing. To broaden the scope of the metabolites that can be detected using plasmonic hydrogels in biological systems, we first need to understand the interplay between the main factors that define their SERS performance.
To explore the physicochemical characteristics influencing SERS performance for diverse metabolites, we developed a library of plasmonic hydrogels by combining alginate, gelatin, and carboxymethyl cellulose (CMC), three widely used hydrogel-forming biopolymers for bioprinting. Although these biopolymers can form physically cross-linked hydrogels, chemical cross-linking is often employed to enhance their stability under physiological conditions. Therefore, these biopolymers were modified with complementary functional groups that enable thiol-ene click cross-linking reactions through the incorporation of norbornene groups or thiols, facilitating orthogonal photo-crosslinking.23 Our objective was to create a versatile range of chemically stable hydrogels that combine different polymers and incorporate gold nanorods (AuNRs) as SERS substrates for the detection of biologically relevant metabolites. We evaluated the hydrogels across a range of properties, including rheology, swelling, degradation, microstructure, and biocompatibility, selecting promising candidates for use as plasmonic inks in 3D-printed scaffolds. Finally, we assessed the impact of hydrogel composition on SERS sensitivity for detecting metabolites with diverse chemical structures, such as chemotherapeutics (doxorubicin or pemetrexed) or cancer-related biomarkers (adenine). Through these analyses, we identified key hydrogel characteristics linked to SERS performance and selectivity, providing insights for optimizing plasmonic hydrogels in metabolite detection applications.
2. Results and discussion
2.1. Functionalization of polymers
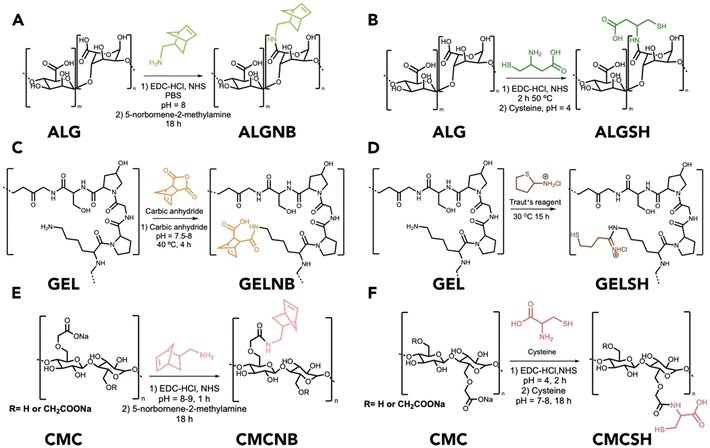
The preparation of the desired hydrogels required functionalization of three different polymers, namely alginate (ALG), gelatin (GEL), and carboxymethyl cellulose (CMC), with alkene and thiol groups to take part in thiol-ene cross-linking reactions. In particular, guluronic (G) units of alginate (where carboxylic groups are more available for modification) were modified with norbornene motifs via coupling reaction between a carboxylate group and 5-norbornene-2-methylamine (ALGNB) (Fig. 1A).24 Similarly, cysteine functionalized alginate (ALGSH) was obtained by coupling cysteine (through its amine group) with the carboxylate moiety in the G units (Fig. 1B). Gelatin was chemically modified through the lysine residues, by a coupling reaction with carbic anhydride, to obtain norbornene bearing gelatin (GELNB) (Fig. 1C), as well as by thiolation with 2-iminothiolane (GELSH), the so-called Traut's reagent (Fig. 1D). This chemical is a thiolation agent that reacts with primary amine groups (as in lysine) to yield sulfhydryl groups. Similar to alginate, norbornene bearing carboxymethyl cellulose (CMCNB) and thiolated carboxymethyl cellulose (CMCSH) were obtained by coupling the carboxymethylated group with 5-norbornene-2-methylamine (Fig. 1E) and cysteine (Fig. 1F), respectively. All synthetic procedures were optimized to obtain the maximum substitution degree, as detailed in the Experimental section.2.2. Physicochemical characterization and evaluation of the degree of functionalization
The presence of the targeted chemical groups was confirmed by 1H-NMR and FTIR spectroscopy. The success of these chemical modifications was evaluated by determining the degree of substitution in each case, by means of 1H-NMR for norbornene bearing polymers, or Ellman's assay for the thiolated polymers (further details can be found in the ESI, section 1†). The resulting degrees of functionalization are summarized in Table 1.The functionalization of alginate with norbornenes (ALGNB) and thiols (ALGSH) was confirmed by the appearance of a new vibrational band around 1730 cm−1 in the FTIR spectrum (Fig. S1A†), which can be attributed to the new amide carbonyl group stretching.23–25 Similarly, the FTIR spectra of GELNB and GELSH shows that the characteristic bands for amide I and amide II vibrational modes shifted from 1521 cm−1 to 1535 cm−1 and from 1627 cm−1 to 1639 cm−1, compared to non-functionalized gelatin (Fig. S2A†).25,26 Finally, the presence of norbornene units in CMCNB and cysteine motifs for CMCSH was confirmed by a new vibrational band around 1730 cm−1 in the FTIR spectrum (Fig. S3A†).25,25–27 Similarly, the FTIR spectra of GELNB and GELSH showed that the characteristic bands for amide I and amide II vibrational modes shifted from 1521 cm−1 to 1535 cm−1 and 1627 cm−1 to 1639 cm−1, respectively, in comparison with non-functionalized gelatin (Fig. S2A†).26,27 Finally, the presence of norbornene units in CMCNB and cysteine motifs for CMCSH was confirmed by a new vibrational band around 1730 cm−1 in the FTIR spectrum (Fig. S3A†).26,28
For norbornene functionalized polymers, the 1H-NMR spectra revealed new peaks in the range of 5.90-6.30 ppm, which can be attributed to the norbornene ring and used to assess the degree of functionalization (Fig. S1–3B†).24,26,27 In the case of the thiolated polymers, the 1H-NMR spectra did not include any particular signal for the thiol group, likely because it can exchange a proton with the solvent (Fig. S1B–3B†). Therefore, Ellman's assay was used instead (see further details in the ESI, section 2†), with the exception of gelatin. In this case, as the reaction takes place at the lysine residues, both Ellman's assay and 1H-NMR can be used for quantification (ESI, section 1†).
2.3. Formulation of thiol–ene photo-crosslinked hydrogels
To formulate thiol-ene cross-linked hydrogels, norbornene functionalized polymers (ALGNB, GELNB and CMCNB) were combined with the corresponding thiolated counterparts (ALGSH, GELSH and CMCSH). An initial screening of hydrogel formation was performed by preparing 3%, 5%, and 10% weight/volume (w/v) aqueous solutions of the polymers. Concentrations higher than 5% (w/v) produced more robust, white-opaque hydrogels, which would likely hinder SERS detection within gels due to increased light scattering. Therefore, our study focused on those formulations containing 3% (w/v) concentration of the polymer with a higher degree of substitution and the stoichiometric amount of its counterpart. It is also worth noting that hydrogels with lower polymer content often lead to softer mechanical properties and, therefore, are more likely to promote cell proliferation and enhance cellular viability in 3D.2 As a result, using 3% (w/v) hydrogel concentrations would enhance cell viability in these formulations. All key polymers, ALG, CMC, and GEL, providing different chemical environments were represented in the study, thereby providing a diverse range of functional hydrogels for evaluation of their use as bioinks and SERS substrates (Fig. 2A). For application in SERS detection, AuNRs were incorporated into the polymeric matrices (Fig. S4†). The resulting hydrogels featured a homogeneous dark red colour after UV-promoted crosslinking (Fig. 2B) and the yield of the thiol–ene reaction was estimated through the determination of free thiols in the matrix (additional details can be found in the Experimental section and in the ESI, section 3†). The yield ranged between 80% and 100% for all formulations, except for the ALGSH-ALGNB combination, which achieved only 12% yield, suggesting a less efficient cross-linking for alginate-based hydrogels (Table 2).2.4. Rheological characterization of plasmonic inks and hydrogels
Real-time photo-rheology measurements were performed to monitor changes in the storage modulus (G′) and loss modulus (G′′) of the composite inks upon irradiation with UV light as the thiol-ene reaction proceeded. Even though the inks were already in a gel state before irradiation (G′ > G′′), both moduli increased upon irradiation, proving that the photoreaction induced a more rigid and stable network within the hydrogels. The newly formed covalent bonds between polymer chains (R–S–R′) strengthened the overall structure in the composite inks (Fig. 3A). The reaction time was approximately 10 s for all formulations, and after this period all systems reached a steady state. We also found that both storage and loss moduli were lower when AuNRs were not included in the ink (Fig. S6A†), meaning that the presence of AuNRs enhances the mechanical properties of the hydrogels, as previously reported. This effect is likely due to electrostatic interactions between polymers and AuNRs, resulting in a reinforcement of the hydrogel network.28–30 Additionally, control experiments carried out without the photoinitiator, lithium phenyl (2,4,6-trimethylbenzoyl) (LAP), did not lead to any increase in the moduli, supporting the chemical photo-crosslinking of the ink of the hydrogels (Fig. S6B†).We additionally carried out frequency sweep tests on the different formulations to evaluate the viscoelastic behaviour of the hydrogels (Fig. 3B). Considering that hydrogels are designed for biomedical purposes, the tests were conducted at 37 °C, mimicking physiological conditions to align with the intended applications of the hydrogels.31 In all cases, G′ was consistently higher than G′′ across the different frequencies, meaning that they behave as gels (G′ > G′′). Those gels exhibiting more elastic behaviour typically show a value of G′ largely independent of frequency, meaning that the gel is more likely to maintain its structure and is more stable. In contrast, weak hydrogels show frequency dependence, with G′′ increasing as frequency rises due to their viscous nature.32 In the case of these hydrogels, different viscoelastic behaviours were observed depending on the hydrogel composition. For instance, ALGNB-ALGSH showed an increase in both moduli, indicating the poor stability of the hydrogel. This can be related to a poor yield of the thiol-ene photo-crosslinking for this formulation, meaning that many polymeric chains are not involved in the reaction and remain embedded in the gel, hindering its stability.
Regarding CMCNB-CMCSH, it was found that G′ remained constant while G′′ decreased, suggesting alterations in the hydrogel network that result in a lower energy loss as heat during deformation and an alteration in the viscous behaviour. Finally, ALGNB-GELSH, CMCNB-ALGSH, CMCNB-GELSH, and GELNB-GELSH showed frequency-independent G′ (elastic behaviour) and increasing G′′ (viscous behaviour), i.e. a typical behaviour for viscoelastic materials. We conclude that the composition of the hydrogels can be tailored to match the viscoelastic properties required for a particular application where either less stable/soft (degradable) or more stable/stiffer hydrogels may be required.
2.5. Characterization of hydrogels: SEM, swelling, degradability, and biocompatibility
The microstructure of hydrogels influences molecule diffusion within the 3D network and, consequently, affects the distribution and release rate of metabolites or drugs. Parameters such as porosity (i.e. pore size and interconnectivity) or network density (i.e. more compact or open networks) determine how easily molecules can diffuse through the hydrogel matrix.33,34 To study such parameters, scanning electron microscopy (SEM) was used to capture the microstructural differences between the different hydrogels (Fig. 4A). Regarding their microstructure, while ALGNB-ALGSH and ALGNB-GELSH exhibited low porosity, CMCNB-CMCSH, CMCNB-ALGSH and CMCNB-GELSH showed highly porous microstructures with large voids. GELNB-GELSH exhibited a distinct porous structure, with small, dense pores in comparison with CMC-bearing hydrogels. Higher magnification images were used to identify AuNRs on the surface of these hydrogels (Fig. S7†). In such images, it can be observed that AuNRs retain their shape after hydrogel processing.On the other hand, hydrophilicity of the polymers and network swelling also affect molecular diffusion through the hydrogels. The swelling ratio (SR) is the fractional increase in the weight of a hydrogel caused by water absorption. Hydrogel swelling introduces additional space within the microstructure, facilitating molecular diffusion. The extent of swelling is influenced by several factors: the cross-linking density (where more tightly cross-linked networks restrict swelling), the porosity (with more porous networks allowing greater swelling), the hydrophilicity of the polymers, and the hydrogel responsiveness to external stimuli such as pH or temperature changes. To mimic the real swelling conditions that hydrogels would face in the desired application, we determined swelling in complete cell media (cDMEM), used for biomedical applications. The gravimetric method was applied to determine the swelling and degradation behaviour of the six obtained hydrogels. All combinations exhibited high SR without significant degradation over the course of one week (Fig. 4B), CMCNB-CMCSH swelled the most and GELNB-GELSH the least. The high and low swelling, respectively, of these gels could be partly due to the relatively large and small pores observed with SEM. However, the swelling behaviour of the gels also depends on other factors beyond porosity, such as the chemical moieties of the hydrogel. In general, GEL-containing hydrogels showed lower SR whereas CMC- and ALG-based hydrogels showed higher SR, with CMCNB-GELSH and ALGNB-GELSH having medium values. Actually, CMCNB-ALGSH had a higher SR than the incorporation of ALGNB-ALGSH, meaning that the presence of CMCNB increases the swelling capacity further, compared to ALGNB. Additionally, the swelling experiment was repeated without AuNRs in the formulations, and it was observed that the presence of AuNRs enhanced the SR (Fig. S8†).22 All these differences in porosity and density, together with their swelling ability are all intimately interconnected and also related to the SERS performance of these gels, as discussed in the following section.
Finally, in the interest of obtaining 3D printed constructs for biomedical applications, the cytotoxicity of the hydrogels was assessed. We evaluated the indirect cytotoxicity of the hydrogels according to ISO 10993 standards for the in vitro testing of biomaterials. Briefly, human breast cancer cells (MDA-MB-231) were cultured with media extracted from the hydrogels over a period of 72 h. After that period, a material is considered non-cytotoxic if cell viability is greater than 70%, compared to the control sample incubated with conventional cell media. The study results showed that all hydrogels were non-cytotoxic at 3% (w/v) concentrations and therefore suitable for biomedical applications (Fig. 4C).
2.6. SERS performance
The main objective of the present study was to build a library of hydrogels with different physicochemical properties such that the main parameters influencing their SERS performance can be elucidated. Hence, once the hydrogels were fully characterized, their SERS performance was initially evaluated by incubation with 4-mercaptobenzoic acid (4-MBA) (Fig. 5A). This molecule features a large Raman cross-section and high affinity for AuNRs (via strong Au–thiol binding), so it can be used as a model Raman molecule. The SERS spectrum of 4-MBA features two main bands at 1080 cm−1 and 1590 cm−1, which are assigned to aromatic ring vibrations (Fig. 5B).34,35 All six different hydrogels were incubated with 1 mM 4-MBA aqueous solution and SERS spectra were recorded across the xy plane along the hydrogels’ surface. The SERS intensity maps for the peak at 1080 cm−1 are shown in Fig. 5C. The background of the hydrogels shows three peaks at around 1200-1400 cm−1, which decrease in intensity after incubation with 4-MBA because this molecule exhibits a greater affinity for the gold surface than the polymeric moieties (Fig. S9†). The measurements were carried out in triplicate to assess the reproducibility of the SERS measurements on each hydrogel. The observed differences in SERS intensity range from 5% to 35% for the different combinations (Fig. S10A†). Notably, the intensity of the SERS signal for 4-MBA could be correlated with the SR of the hydrogels, with CMCNB-CMCSH exhibiting the highest and GELNB-GELSH having the lowest 4-MBA SERS signal intensity. Higher SR enhanced the uptake of 4-MBA, leading to stronger SERS signals resulting from greater interaction with the AuNRs embedded in the hydrogel matrix. Overall, there is a monotonic relationship between the intensity of the 1080 cm−1 peak of 4-MBA and the swelling ratio of each hydrogel, with the exception of ALGNB-ALGSH (Fig. S10B†). The reason behind this outlier might be the low yield of the thiol–ene reaction leading to a less stable hydrogel that shows a poor performance as a SERS substrate. Nonetheless, we envision that there is a limit on this relationship, so that high swelling of the hydrogel would separate the Au nanoparticles, resulting in lower density and a corresponding reduction in the intensity of SERS signals.However, we hypothesized that swelling was not the only parameter that influenced SERS performance, but the size of the molecule, polarity, and main chemical groups also are key to explaining differences in the sensing performance of the hydrogels. Therefore, the hydrogels were also incubated with doxorubicin (DOXO), an anthracycline derivative used as a DNA intercalant in the treatment of cancer (Fig. 6A). The molecular structure of doxorubicin includes four ortho-fused aromatic rings coupled to a pyranose moiety through an o-glycosidic bond. Doxorubicin has a well-described SERS spectrum with main bands at 1413, 1434, and 1576 cm−1, related to skeletal ring stretching modes (Fig. 6B).36 We envisioned that such a large, rigid and strained structure could give more insights into the different SERS performance of the gels. Accordingly, all six combinations were incubated with DOXO and SERS spectra were recorded across the xz plane perpendicular to the surface of each gel.
The intensity maps for the peak at 1413 cm−1 are plotted in Fig. 6C, showing that the signal from DOXO was clearly detected in three of the formulations. From these, two of them contain alginate in their compositions, suggesting that the carboxylic groups of ALG might interact with the amine of the pyranose moiety, causing doxorubicin molecules to get closer to the AuNR surface. Meanwhile, the CMCNB-CMCSH combination was also capable of sensing doxorubicin and gave a more uniform signal, possibly due to its highly porous structure. However, the difference in intensities between the four compositions was not as notable as in the case of 4-MBA. Overall, these results indicated that supramolecular interactions (i.e. carboxylate–amine hydrogen bonding) between chemical groups of both the polymer and the analyte of interest can overcome deficiencies in SERS sensing capabilities that result from poor swelling or porosity when targeting difficult-to-diffuse molecules.
Next, we explored the effect of lowering the concentration of the polymers on molecule diffusion, using 1.5% (w/v) hydrogels. For instance, we incubated CMCNB-CMCSH with adenine at 3% and 1.5% (w/v). Adenine is one of the four nucleobases in the nucleic acids of DNA (Fig. 7A). Its chemical structure encompasses an amino-substituted purine, and the SERS spectrum shows a characteristic intensity band at 734 cm−1, corresponding with the purine ring vibrational mode (Fig. 7C). The intensity maps across the xz plane showed greater intensity of this band in 3% (w/v) hydrogels but the depth of the signal was greater for 1.5% (w/v) (Fig. 7B). This effect might be caused by the enhanced transparency of the 1.5% (w/v) gels because the polymeric matrix is highly diluted. Therefore, we focused on less concentrated CMCNB-ALGSH and CMCNB-CMCSH, as both combinations gave greater intensities, to study the effect of ALG and CMC on the compositions. These gels were incubated with two similar molecules: adenine and pemetrexed. While adenine is a nucleobase, pemetrexed is a nucleotide derivative used as a folate antimetabolite in the treatment of cancer (Fig. 7A). This molecule is sensitive to pH and exhibits at neutral pH a characteristic band at 1615 cm−1 related to the scissoring vibrational mode of NH2 groups (Fig. 7D). Regarding polarity, adenine is less polar than pemetrexed. After incubating CMCNB-ALGSH and CMCNB-CMCSH with these two molecules, SERS spectra were collected along the xz directions, so that the distribution of the molecules within the whole hydrogel could be observed. When the intensity of the 734 cm−1 band was tracked for the gels incubated with adenine, the highest intensity was obtained for the CMCNB-CMCSH combination (Fig. 7E). In contrast, the intensity of the 1615 cm−1 band was followed for the gels incubated with pemetrexed, obtaining the highest intensity for the CMCNB-ALGSH combination (Fig. 7F). Therefore, we see that molecules with higher polarity are able to get closer to the AuNRs in CMCNB-ALGSH gel. This trend may relate to the higher availability of polar carboxylate groups in the CMCNB-ALGSH hydrogel, which should bear an overall negative charge.
2.7. 3D printing of composite bioinks
To evaluate the use of the prepared plasmonic hydrogels to build scaffolds by 3D printing, we first performed printability tests by measuring flow curves for the different formulations. Such rheological tests are widely used in bioprinting to investigate the dynamic viscosity of the ink and, thus, to understand flow properties during extrusion. Polymer solutions are typically non-Newtonian and exhibit either shear thickening (viscosity increases with stress) or shear thinning behaviour (viscosity decreases beyond a critical shear rate). This shear thinning behaviour is crucial for the bioinks to be extruded through the narrow orifices of the needles used for bioprinting. All our hydrogel precursors (inks) exhibited the expected decrease in viscosity when increasing the shear rate (Fig. 8A). However, even though all ink compositions could be printed, other factors such as shape fidelity and recovery had to be considered for optimizing printability of hydrogel inks into scaffolds. For instance, ALGNB-ALGSH did not show any shape fidelity, so it cannot be used to print well-defined structures. Replacing ALGSH with GELSH increased the shape fidelity of the bioink, but it was still difficult to achieve high-resolution printing. Similarly, CMCNB-CMCSH and CMCNB-ALGSH did not show sufficient resolution whereas it was possible to obtain resolved scaffolds with CMCNB-GELSH and even better with GELNB-GELSH (Fig. 8B).At this point, the highest SERS performance was obtained with those inks that showed poor printability. In other words, high SERS signals could primarily be obtained at the expense of the printing resolution. Therefore, we selected those gels with balanced SERS and printing properties: CMCNB-CMCSH, CMCNB-ALGSH, and CMCNB-GELSH. To improve the printing of CMCNB-CMCSH, we used a supporting bath of citric acid, which can partially induce CMC crosslinking.37 By printing in a citric acid bath, we achieved crosslinking of the ink, prior to photo-crosslinking with UV light. In this manner, it was possible to obtain cubes and scaffolds using 3% (w/v) CMCNB-CMCSH. Unfortunately, it was not possible to further decrease the concentration of the bioink (Fig. 8C). Similarly, as alginate can be physically crosslinked using Ca2+ ions, by using a calcium chloride (CaCl2) bath, it was possible to obtain cubes and scaffolds with both bioinks containing 3% (w/v) and 1.5% (w/v) CMCNB-ALGSH (Fig. 8D). Finally, with CMCNB-GELSH, we decided to print at low temperatures (5 °C) because both polymers are thermo-responsive, with their viscosity increasing at low temperature, yielding an improvement in the resolution of the printed structure. By doing so, we obtained both cubes and scaffolds without using any assisting bath (Fig. 8E). In all cases, the pressure, printing speed and needle type were optimized for each particular ink (Experimental section 4.9). In general, the inks containing gelatin required a higher pressure at similar printing speeds and UV-crosslinking was promoted in all cases after deposition of each layer because it improved the formation of stable constructs. Additionally, the biocompatibility of the hydrogels at 1.5% (w/v) concentration was evaluated, resulting in highly biocompatible materials (Fig. S11†).
3. Conclusions
The functionalization of different polymers – alginate, gelatin and carboxymethyl cellulose – with norbornene and thiol groups has been optimized to obtain the highest degree of substitution possible. These modifications significantly enhanced the reactivity of the polymers and enabled the preparation of different (6 in this work) orthogonally photo-crosslinked hydrogels via a thiol–ene click reaction. The equimolar combination of such polymers with AuNR colloids was shown to yield suitable inks for advanced bioprinting, cell culture, and biosensing applications. The mechanical properties, biocompatibility, microstructural features, swelling, printing capabilities, and the SERS performance of the resulting plasmonic hydrogels were carefully assessed. We found that the incorporation of carboxymethylcellulose in the formulations enhanced both swelling and porosity, while hindering the printability of the hydrogels. In contrast, including gelatin resulted in to the opposite effect, offering the highest printing resolution but yielding structures with low porosity and poor swelling. Alternatively, hydrogels containing alginate produced the most balanced properties for SERS sensing and bioprinting. Moreover, even though the swelling and porosity of the gels did influence their SERS performance, these may not be the main factors. Other parameters such as the polymer concentration or the availability of chemical groups to establish supramolecular interactions with the analyte (hydrogen bonding, dipole–dipole interactions, etc.) are crucial for sensing certain analytes, as is the case of pemetrexed, which could only be detected by SERS in CMCNB-ALGSH despite its lesser swelling and porosity compared to CMCNB-CMCSH. The limit of detection for relevant molecules and the effect of hydrogel composition at lower concentrations remain to be addressed, so additional clues can be gathered on the SERS performance of these materials. In conclusion, with this study, we have determined relevant parameters that account for the SERS performance of plasmonic composite hydrogels. Nevertheless, building hydrogels combining more than two polymers, controlling the distribution of nanoparticles in the hydrogel, or adding sacrificial materials to the bioinks might unlock even greater potential for these materials, toward biomedical applications.4. Experimental section
4.1. Materials
Acetone (≥99.5%), alginic acid sodium salt (alginate, MW 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), L-ascorbic acid (AA, ≥99%), 5-bromosalicylic acid (5-BrSA, technical grade, 90%), hexadecyltrimethylammonium bromide (CTAB, ≥96%), L-cysteine hydrochloride monohydrate (cysteine, ≥98%), hydrogen tetrachloroaurate trihydrate (HAuCl4·3H2O, ≥99.9%), deuterium oxide (D2O, ≥99), dithiobis 2-nitrobenzoic acid (DTNB/Ellman's reagent, ≥98%), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC–HCl, ≥98%), ethanol (≥99.5%), 2-iminothiolane hydrochloride (Traut's reagent, ≥98%), lithium phenyl-2,4-6-trimethylbenzoylphosphinate (LAP, ≥95%), 4-mercaptobenzoic acid (4-MBA, 99%), cis-5-norbornene-endo-2-3-dicarboxylic anhydride (CA, 99%), 5-5′-,5-norbornene-2-methylamine (NMA, ≥98%), N-hydroxysuccinimide (NHS, 98%), sodium borohydride (NaBH4, 99%), potassium chloride (KCl, ≥99%), potassium phosphate monobasic (KH2PO4, ≥99%), sodium carboxymethyl cellulose (CMC, MW 90
000 g mol−1), L-ascorbic acid (AA, ≥99%), 5-bromosalicylic acid (5-BrSA, technical grade, 90%), hexadecyltrimethylammonium bromide (CTAB, ≥96%), L-cysteine hydrochloride monohydrate (cysteine, ≥98%), hydrogen tetrachloroaurate trihydrate (HAuCl4·3H2O, ≥99.9%), deuterium oxide (D2O, ≥99), dithiobis 2-nitrobenzoic acid (DTNB/Ellman's reagent, ≥98%), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC–HCl, ≥98%), ethanol (≥99.5%), 2-iminothiolane hydrochloride (Traut's reagent, ≥98%), lithium phenyl-2,4-6-trimethylbenzoylphosphinate (LAP, ≥95%), 4-mercaptobenzoic acid (4-MBA, 99%), cis-5-norbornene-endo-2-3-dicarboxylic anhydride (CA, 99%), 5-5′-,5-norbornene-2-methylamine (NMA, ≥98%), N-hydroxysuccinimide (NHS, 98%), sodium borohydride (NaBH4, 99%), potassium chloride (KCl, ≥99%), potassium phosphate monobasic (KH2PO4, ≥99%), sodium carboxymethyl cellulose (CMC, MW 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, 70% carboxylation), sodium chloride (NaCl, ≥99%), sodium hydroxide (NaOH, ≥97%), sodium phosphate dibasic (Na2HPO4, ≥99%), sodium phosphate monobasic (NaH2PO4, ≥99%), silver nitrate (AgNO3, ≥99.9%), thioglycolic acid (TGA, ≥99%), triethylamine (TEA, ≥99.5%) and type A gelatin from porcine skin (Bloom 300 g) were purchased from Sigma-Aldrich. alamarBlue HS cell viability reagent, Invitrogen ethylenediaminetetraacetic acid disodium salt 0.5 M, pH = 8.9, RNase-free (EDTA, ≥99%) and Gibco Dulbecco's modified Eagle's medium (DMEM) were purchased from Thermo Fisher. Hydrochloric acid (HCl, 37%) was purchased from Scharlau. Fetal bovine serum (FBS) and penicillin–streptomycin (PS) were purchased from Invitrogen. FBS and PS were used at 10% and 1% to prepare complete DMEM (cDMEM). The human breast MDA-MB-231 cell line was obtained from American Type Culture Collection (ATCC HTB-26). All chemicals were used as received. All glassware used for the synthesis of AuNRs was washed with aqua regia, rinsed with water and dried before use.
000 g mol−1, 70% carboxylation), sodium chloride (NaCl, ≥99%), sodium hydroxide (NaOH, ≥97%), sodium phosphate dibasic (Na2HPO4, ≥99%), sodium phosphate monobasic (NaH2PO4, ≥99%), silver nitrate (AgNO3, ≥99.9%), thioglycolic acid (TGA, ≥99%), triethylamine (TEA, ≥99.5%) and type A gelatin from porcine skin (Bloom 300 g) were purchased from Sigma-Aldrich. alamarBlue HS cell viability reagent, Invitrogen ethylenediaminetetraacetic acid disodium salt 0.5 M, pH = 8.9, RNase-free (EDTA, ≥99%) and Gibco Dulbecco's modified Eagle's medium (DMEM) were purchased from Thermo Fisher. Hydrochloric acid (HCl, 37%) was purchased from Scharlau. Fetal bovine serum (FBS) and penicillin–streptomycin (PS) were purchased from Invitrogen. FBS and PS were used at 10% and 1% to prepare complete DMEM (cDMEM). The human breast MDA-MB-231 cell line was obtained from American Type Culture Collection (ATCC HTB-26). All chemicals were used as received. All glassware used for the synthesis of AuNRs was washed with aqua regia, rinsed with water and dried before use.
4.2. Synthesis of gold nanorods
AuNRs were prepared following well-established procedures.38,39 First, gold seeds were prepared by adding 25 μL of 50 mM HAuCl4 to 4.7 mL of 0.1 M CTAB solution at 30 °C and the resulting mixture was stirred for 5 min. Then, 300 μL of freshly prepared 10 mM NaBH4 was injected under vigorous stirring. The solution remained under moderate stirring for 30 min more at room temperature in order to consume the excess of borohydride. For the growth solution, 45 mg of 5-BrSA was dissolved into 25 mL of 0.1 M CTAB before the addition of 480 μL of 100 mM AgNO3. The mixture was stirred for 15 min at room temperature. Then, 500 μL of HAuCl4 50 mM was added to cause prereduction of Au(III) to Au(I), causing the solution to change its colour from orange to clear yellow. This process was monitored via UV-vis spectroscopy until the absorbance value at 396 nm was in between 0.80–0.85, causing AuNRs to show a longitudinal plasmon band around 780 nm. Then, 130 μL of 100 mM AA was added, and the stirring was vigorously increased for the addition of 80 μL of the seed solution previously prepared. The resulting solution remained undisturbed overnight to allow AuNRs to grow. Finally, AuNRs were purified by centrifuging at 8000 rpm for 30 min and redispersing the precipitate first in water and, after a second centrifugation, in 1 mM CTAB solution.4.3. Synthesis of functionalized polymers
4.4. Preparation of inks and hydrogels
The preparation of inks and hydrogels required further purification of AuNRs that were centrifuged at 8000 rpm and redispersed in Milli-Q water to remove the excess of CTAB. Then, the corresponding norbornene partner (ALGNB, CMCNB or GELNB) was dissolved in 1 mL of Milli-Q water. Meanwhile, the thiolated partner (ALGSH, CMCSH or GELSH) was dissolved in 1 mL of Milli-Q water. Both solutions were stirred at 40 °C overnight before being mixed together. Then, the solution was allowed to cool down to room temperature and 1 mL of 4.5 mM dispersion of AuNRs was added to achieve a final concentration of 1.5 mM AuNRs. The concentration of nanoparticles in the bioink can be confirmed by measuring the absorbance at 400 nm and taking into account that a value of 1.2 in absorbance corresponds to 0.5 mM Au0.38 Finally, 0.1% (w/v) LAP (lithium phenyl-(2,4,5-trimethylbenzoyl) phosphonate) was dissolved in the previous mixture at room temperature in order to promote the thiol–ene click reaction with a UV lamp (405 nm) for 120 s. Inks, in turn, were prevented from cross-linking for 3D printing. Quantities of polymers were adjusted to have 3% (w/v) concentration of the highest functionalized polymer and to ensure the equimolar presence of norbornene and thiols in the final mixture.4.5. Characterization of AuNRs
4.6. Characterization of polymers
4.7. Characterization of hydrogels
Amplitude sweeps were performed on both inks and hydrogels to identify the linear viscoelastic (LVE) region over a shear strain range of 0.01% to 100%. In order to study gelation kinetics, ink samples were subjected to a shear strain of 0.1% within the LVE range and a frequency of 1 Hz. Measurements were taken over 170 s, with gelation initiated by exposure to a UV curing lamp at 365 nm. For hydrogels, frequency sweeps were conducted in triplicate to determine the storage modulus (G′) and loss modulus (G′′) over an angular frequency (ω) range of 0.1 rad s−1 to 100 rad s−1, using a fixed shear strain (γ) of 0.1% within the LVE region, identified from the amplitude sweep at 37 °C. The flow curve test for ink samples was conducted over a shear rate ( ) range of 1–1000 s−1.
) range of 1–1000 s−1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 EtOH/H2O during 5 min; (b) 50
75 EtOH/H2O during 5 min; (b) 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 EtOH/H2O during 5 min; (c) 75
50 EtOH/H2O during 5 min; (c) 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 EtOH/H2O during 5 min; (d) EtOH during 10 min; (e) cDMEM during 10 min; and (f) cDMEM during 30 min. Then, the discs were incubated with cDMEM overnight and the resulting extractive media were used for the incubation of MDA.MB.231 breast cancer cells over a period of 72 h. Once completed, the cytotoxicity was evaluated using the alamarBlue assay. In the case of CMCNB-CMCSH, due to its higher swelling, the disc should be washed with cDMEM for 6 h, changing the media every 2 h before incubation overnight to obtain the extractive media.
25 EtOH/H2O during 5 min; (d) EtOH during 10 min; (e) cDMEM during 10 min; and (f) cDMEM during 30 min. Then, the discs were incubated with cDMEM overnight and the resulting extractive media were used for the incubation of MDA.MB.231 breast cancer cells over a period of 72 h. Once completed, the cytotoxicity was evaluated using the alamarBlue assay. In the case of CMCNB-CMCSH, due to its higher swelling, the disc should be washed with cDMEM for 6 h, changing the media every 2 h before incubation overnight to obtain the extractive media.
4.8. Surface-enhanced Raman spectroscopy (SERS)
Prior to SERS measurements, hydrogels were incubated overnight with 1 mM solutions of the corresponding analyte. Then, the gels were placed in homemade holders and immersed in the solution of the corresponding molecule for measurement through a Leica 40× immersion objective, with a working distance of 3 mm and a numerical aperture of 1.4. The spectra were collected using a 785 nm laser at 100% power (205 mW μm−2) over 1 s for 4-MBA and over 3 s for DOXO, pemetrexed, and adenine. The measurements were taken using a backscattering configuration of the instrument and a Renishaw 1024 CCD Camera-1024 × 256 detector with a grating of 1200 l mm−1 and a slit opening of 65 μm. All the collected spectra were baseline corrected and smoothed, and cosmic rays were removed before obtaining the intensity maps using WiRE 4.4. Renishaw Raman microscope software.4.9. Bioprinting
A multiheaded 3D Discovery bioprinter (RegenHU, Switzerland) was used to print the scaffolds. The printing parameters for each ink composition are listed in Table 3. The supportive bath used for printing 3% (w/v) CMCNB-CMCSH based ink contained 100 mM citric acid aqueous solution. The supportive bath used for printing 3% (w/v) and 1.5% (w/v) CMCNB-ALGSH based inks contained 100 Mm CaCl2 aqueous solution. 3% (w/v) and 1.5% (w/v) CMC-GELSH inks were kept at 5 °C by using a cartridge cooler. The G-code for square scaffolds (2 mm spacing) and cubes was produced using BioCAD software (RegenHU, Switzerland). In situ UV-crosslinking was performed after deposition of each layer, using a light curing cartridge at 405 nm wavelength, with a power density of 25.3 mW cm−2.| Sample | Pressure (MPa) | Printing speed (mm s−1) | Needle (internal diameter = ID) |
|---|---|---|---|
| CMCNB–CMCSH | 0.13 | 10 | Stainless steel needle |
| 3% (w/v) | ID = 0.4 mm | ||
| CMCNB–CMCSH | — | — | — |
| 1.5% (w/v) | |||
| CMCNB–ALGSH | 0.39 | 10 | Stainless steel needle |
| 3% (w/v) | ID = 0.4 mm | ||
| CMCNB–ALGSH | 0.07 | 15 | Stainless steel needle |
| 1.5% (w/v) | ID = 0.4 mm | ||
| CMCNB–GELSH | 0.63 | 15 | Plastic conic needle |
| 3% (w/v) | ID = 0.6 mm | ||
| CMCNB–GELSH | 0.55 | 10 | Plastic conic needle |
| 1.5% (w/v) | ID = 0.4 mm |
Data availability
The data underlying this study are available in the published article and its ESI.† If required, after acceptance of the paper, raw data will be made publicly available at an open repository such as Zenodo.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
Financial support was provided by the European Research Council (ERC-AdG-2017 #787510 4DBIOSERS). C. G. A. thanks the Spanish State Research Agency for a Juan de la Cierva Incorporación Fellowship (FJCI-2016-28887). L. T. A. thanks the Spanish Ministry of Universities for a Formación de Profesorado Universitario fellowship (FPU-05867). Y. M. H. B. thanks the University of the Basque Country for an i3 GAZtE fellowship. G. A. V.-W. acknowledges support from the European Union's Horizon Europe research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 101105300 (PLASMOSTEMFATE). We acknowledge support from the María de Maeztu Excellence Unit CEX2023-001303-M funded by MCIN/AEI/10.13039/501100011033.References
- V. Foglizzo, E. Cocco and S. Marchiò, Cancers, 2022, 14, 3692 CrossRef CAS PubMed.
- F. Ruedinger, A. Lavrentieva, C. Blume, I. Pepelanova and T. Scheper, Appl. Microbiol. Biotechnol., 2015, 99, 623–636 CrossRef CAS PubMed.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed.
- D. Antoni, H. Burckel, E. Josset and G. Noel, Int. J. Mol. Sci., 2015, 16, 5517–5527 CrossRef CAS PubMed.
- S. R. Caliari and J. A. Burdick, Nat. Methods, 2016, 13, 405–414 CrossRef CAS PubMed.
- H. Li and A. B. Hummon, Anal. Chem., 2011, 83, 8794–8801 CrossRef CAS PubMed.
- J. P. Celli, I. Rizvi, A. R. Blanden, I. Massodi, M. D. Glidden, B. W. Pogue and T. Hasan, Sci. Rep., 2014, 4, 3751 CrossRef CAS PubMed.
- M. Oliveira, P. Conceição, K. Kant, A. Ainla and L. Diéguez, Cancers, 2021, 13, 1381 CrossRef CAS PubMed.
- J. Dornhof, J. Kieninger, H. Muralidharan, J. Maurer, G. A. Urban and A. Weltin, Lab Chip, 2022, 22, 225–239 RSC.
- S.-M. Lee, N. Han, R. Lee, I.-H. Choi, Y.-B. Park, J.-S. Shin and K.-H. Yoo, Biosens. Bioelectron., 2016, 77, 56–61 CrossRef CAS PubMed.
- A. R. Kherlopian, T. Song, Q. Duan, M. A. Neimark, M. J. Po, J. K. Gohagan and A. F. Laine, BMC Syst. Biol., 2008, 2, 74 CrossRef PubMed.
- S. Avnet, G. D. Pompo, G. Borciani, T. Fischetti, G. Graziani and N. Baldini, Biomed. Mater., 2024, 19, 025033 CrossRef CAS PubMed.
- G. Häusler and S. Ettl, in Optical Measurement of Surface Topography, ed. R. Leach, Springer Berlin Heidelberg, Berlin, Heidelberg, 2011, pp. 23–48 Search PubMed.
- J. Plou, P. S. Valera, I. García, C. D. L. De Albuquerque, A. Carracedo and L. M. Liz-Marzán, ACS Photonics, 2022, 9, 333–350 CrossRef CAS PubMed.
- Q. Zhao, H. Hilal, J. Kim, W. Park, M. Haddadnezhad, J. Lee, W. Park, J.-W. Lee, S. Lee, I. Jung and S. Park, J. Am. Chem. Soc., 2022, 144, 13285–13293 CrossRef CAS PubMed.
- K. Kneipp, H. Kneipp and H. G. Bohr, Surface-Enhanced Raman Scattering. Topics in Applied Physics, in Single-Molecule SERS Spectroscopy, ed. K. Kneipp, M. Moskovits and H. Kneipp, Springer, Berlin, Heidelberg, 2006, vol. 103, pp. 261–277 Search PubMed.
- D. Jimenez De Aberasturi, M. Henriksen-Lacey, L. Litti, J. Langer and L. M. Liz-Marzán, Adv. Funct. Mater., 2020, 30, 1909655 CrossRef CAS.
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner, P. L. Martin-Hirsch, M. J. Walsh, M. R. McAinsh, N. Stone and F. L. Martin, Nat. Protoc., 2016, 11, 664–687 CrossRef CAS PubMed.
- S. M. Morton and L. Jensen, J. Am. Chem. Soc., 2009, 131, 4090–4098 CrossRef CAS PubMed.
- C. García-Astrain, E. Lenzi, D. Jimenez De Aberasturi, M. Henriksen-Lacey, M. R. Binelli and L. M. Liz-Marzán, Adv. Funct. Mater., 2020, 30, 2005407 CrossRef.
- J. Plou, B. Molina-Martínez, C. García-Astrain, J. Langer, I. García, A. Ercilla, G. Perumal, A. Carracedo and L. M. Liz-Marzán, Nano Lett., 2021, 21, 8785–8793 CrossRef CAS PubMed.
- C. García-Astrain, E. Lenzi, D. Jimenez De Aberasturi, M. Henriksen-Lacey, M. R. Binelli and L. M. Liz-Marzán, Adv. Funct. Mater., 2020, 30, 2005407 CrossRef.
- C. E. Hoyle and C. N. Bowman, Angew. Chem., Int. Ed., 2010, 49, 1540–1573 CrossRef CAS PubMed.
- P. Gacesa and N. J. Russell, Pseudomonas Infection and Alginates: Biochemistry, genetics and pathology, Springer Netherlands, Dordrecht, 1990 Search PubMed.
- H. W. Ooi, C. Mota, A. T. Ten Cate, A. Calore, L. Moroni and M. B. Baker, Biomacromolecules, 2018, 19, 3390–3400 CrossRef CAS PubMed.
- G. Socrates, Infrared and Raman Characteristic Group Frequencies: Tables and Charts, John Wiley & Sons Inc, New York, 3rd edn, 2003, vol. 1 Search PubMed.
- Z. Mũnoz, H. Shih and C.-C. Lin, Biomater. Sci., 2014, 2, 1063–1072 RSC.
- T. X. Morrison and W. M. Gramlich, Carbohydr. Polym., 2023, 319, 121173 CrossRef CAS PubMed.
- K. Zhu, S. R. Shin, T. Van Kempen, Y. Li, V. Ponraj, A. Nasajpour, S. Mandla, N. Hu, X. Liu, J. Leijten, Y. Lin, M. A. Hussain, Y. S. Zhang, A. Tamayol and A. Khademhosseini, Adv. Funct. Mater., 2017, 27, 1605352 CrossRef PubMed.
- B. W. Walker, R. Portillo Lara, E. Mogadam, C. Hsiang Yu, W. Kimball and N. Annabi, Prog. Polym. Sci., 2019, 92, 135–157 CrossRef CAS PubMed.
- R. Lapasin, in Polysaccharide hydrogels, Jenny Stanford Publishing, 1st edn, 2015, vol. 1, p. 56 Search PubMed.
- G. Stojkov, Z. Niyazov, F. Picchioni and R. K. Bose, Gels, 2021, 7, 255 CrossRef CAS PubMed.
- J. Li and D. J. Mooney, Nat. Rev. Mater., 2016, 1, 16071 CrossRef CAS PubMed.
- J.-T. Zhang, S. Petersen, M. Thunga, E. Leipold, R. Weidisch, X. Liu, A. Fahr and K. D. Jandt, Acta Biomater., 2010, 6, 1297–1306 CrossRef CAS PubMed.
- A. Michota and J. Bukowska, J. Raman Spectrosc., 2003, 34, 21–25 CrossRef CAS.
- J. Gautier, E. Munnier, L. Douziech-Eyrolles, A. Paillard, P. Dubois and I. Chourpa, Analyst, 2013, 138, 7354 RSC.
- D. J. Sarkar and A. Singh, Carbohydr. Polym., 2017, 156, 303–311 CrossRef CAS PubMed.
- L. Scarabelli, A. Sánchez-Iglesias, J. Pérez-Juste and L. M. Liz-Marzán, J. Phys. Chem. Lett., 2015, 6, 4270–4279 CrossRef CAS PubMed.
- L. Scarabelli, M. Grzelczak and L. M. Liz-Marzán, Chem. Mater., 2013, 25, 4232–4238 CrossRef CAS.
- H. Yao, M. Zhu, P. Wang, Y. Liu and J. Wei, Nanomaterials, 2021, 11, 2191 CrossRef CAS PubMed.
- S. Deng, X. Li, W. Yang, K. He and X. Ye, J. Biomater. Sci., Polym. Ed., 2018, 29, 1643–1655 CrossRef CAS PubMed.
- S. Kommareddy and M. Amiji, Bioconjugate Chem., 2005, 16, 1423–1432 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01529k |
| This journal is © The Royal Society of Chemistry 2025 |