 Open Access Article
Open Access ArticleGuiding vascular infiltration through architected GelMA/PEGDA hydrogels: an in vivo study of channel diameter, length, and complexity†
Martha
Fowler‡
a,
Alvaro
Moreno Lozano‡
a,
Julian
Krause
a,
Patrick
Bednarz
a,
Shalini
Pandey
 a,
Mina
Ghayour
a,
Qixu
Zhang
b and
Omid
Veiseh
a,
Mina
Ghayour
a,
Qixu
Zhang
b and
Omid
Veiseh
 *a
*a
aDepartment of Bioengineering, Rice University, Houston, TX 77030, USA. E-mail: omid.veiseh@rice.edu
bDepartment of Plastic Surgery, MD Anderson, Houston, TX 77030, USA
First published on 25th April 2025
Abstract
Organ shortages for transplantation in the United States impact over 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 patients, with 17 dying daily due to the lack of available organs. This growing need is exacerbated by the limited functionality and disease risk of donated organs. Tissue-engineered organs present a promising alternative, requiring optimized scaffold architecture and cell integration. Vascular networks within organs are essential for supplying oxygen and nutrients to cells, with a critical distance between blood vessels and surrounding tissue to allow effective diffusion. Various microfabrication techniques, such as electrospinning, freeze-drying, and gas foaming, have been employed to develop engineered organs. However, these techniques often lack the complexity needed to support vascularization. 3D bioprinting, particularly digital light projection (DLP)-based stereolithography, offers a solution by enabling high-resolution control of both external and internal architectures. Gelatin methacrylate (GelMA) and polyethylene glycol diacrylate (PEGDA) hydrogels have shown potential for tissue integration in simple structures but require further optimization for vascularization in more complex constructs. This study utilizes DLP to 3D bioprint GelMA/PEGDA hydrogels, exploring various channel designs to enhance tissue infiltration and vascularization in rodent models, providing a potential platform for cell and tissue transplantation. We demonstrate that GelMA/PEGDA hydrogels are mechanically robust, biocompatible, and support in vivo vascular infiltration. Channel diameter significantly influenced vascularization, with 1 mm channels yielding the highest infiltration, while channel length had minimal impact. Among five tested architectures, one design (GEO3) promoted the greatest vascular ingrowth, establishing a tunable hydrogel platform for prevascularized tissue engineering applications.
000 patients, with 17 dying daily due to the lack of available organs. This growing need is exacerbated by the limited functionality and disease risk of donated organs. Tissue-engineered organs present a promising alternative, requiring optimized scaffold architecture and cell integration. Vascular networks within organs are essential for supplying oxygen and nutrients to cells, with a critical distance between blood vessels and surrounding tissue to allow effective diffusion. Various microfabrication techniques, such as electrospinning, freeze-drying, and gas foaming, have been employed to develop engineered organs. However, these techniques often lack the complexity needed to support vascularization. 3D bioprinting, particularly digital light projection (DLP)-based stereolithography, offers a solution by enabling high-resolution control of both external and internal architectures. Gelatin methacrylate (GelMA) and polyethylene glycol diacrylate (PEGDA) hydrogels have shown potential for tissue integration in simple structures but require further optimization for vascularization in more complex constructs. This study utilizes DLP to 3D bioprint GelMA/PEGDA hydrogels, exploring various channel designs to enhance tissue infiltration and vascularization in rodent models, providing a potential platform for cell and tissue transplantation. We demonstrate that GelMA/PEGDA hydrogels are mechanically robust, biocompatible, and support in vivo vascular infiltration. Channel diameter significantly influenced vascularization, with 1 mm channels yielding the highest infiltration, while channel length had minimal impact. Among five tested architectures, one design (GEO3) promoted the greatest vascular ingrowth, establishing a tunable hydrogel platform for prevascularized tissue engineering applications.
1 Introduction
Shortage of organs for transplantation affects over 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 patients in the United States leading to 17 of those dying every day due to the lack of organs. For all organs and especially the liver the shortage of organs keeps growing with time as more patients need them every year.1,2 In addition, the functionality and the risk for disease and/or infection of the donated organs is another issue that limits the final number of available and functional organs that can be donated and transplanted.3,4 Therefore, there is an important need to develop tissue-engineered organs that can be used instead of donor organs for organ transplantation. There is a wide presence of blood vessel networks within solid organs that allow for the supply of oxygen, and nutrients to the embedded cells and the removal of waste from the organ. The maximum distance between blood vessels and the implanted cells needs to be within 200–300 μm to allow for effective diffusion of oxygen and nutrients that can keep the corresponding cells alive long-term.5 However other factors such as the metabolic rate of the tissue might also influence diffusion distance.6,7
000 patients in the United States leading to 17 of those dying every day due to the lack of organs. For all organs and especially the liver the shortage of organs keeps growing with time as more patients need them every year.1,2 In addition, the functionality and the risk for disease and/or infection of the donated organs is another issue that limits the final number of available and functional organs that can be donated and transplanted.3,4 Therefore, there is an important need to develop tissue-engineered organs that can be used instead of donor organs for organ transplantation. There is a wide presence of blood vessel networks within solid organs that allow for the supply of oxygen, and nutrients to the embedded cells and the removal of waste from the organ. The maximum distance between blood vessels and the implanted cells needs to be within 200–300 μm to allow for effective diffusion of oxygen and nutrients that can keep the corresponding cells alive long-term.5 However other factors such as the metabolic rate of the tissue might also influence diffusion distance.6,7
Different microfabrication techniques have been used to develop tissue engineered organs with specific architectures that can facilitate blood vessel formation in close proximity to the nearby cells. Some of these techniques include electrospinning,8 freeze-drying,9 and gas foaming10 which are mainly focusing on the bulk properties of the tissue-engineered device instead of developing complex internal architectures and topologies within the device that can promote vascular formation within the scaffold.11 Organs within the body follow a hierarchical structure that is guided by large arteries leading to arterioles and capillaries. These hierarchical structures are based on vascular patterning which plays an important role in tissue engraftment and functionality of the tissue.12–14 Some of these patterned architectures are specific to different organs but can be conserved among multiple species.15–17
3D bioprinting is a technique that allows for layer-by-layer fabrication of complex models that are designed using computer-aided design software. 3D bioprinting leads to high resolution, replicability, and reproducibility of highly complex architecture constructures allowing to control not only the external geometry of the construct but also the internal architecture to mimic the structure and function of tissues and organs.18–21 There are several 3D bioprinting techniques which include extrusion bioprinting, inkjet bioprinting, laser-assisted bioprinting, dual head printing, and light-mediated stereolithography.22,23 Stereolithography is a 3D bioprinting technique that allows for fabrication of perfusable architectures using biocompatible material via digital light projection (DLP).24–27 In addition, the biomaterial or bioink used for 3D printing can also affect tissue integration, interconnectivity of different components within the organ, and diffusion dynamics of oxygen and nutrients within the 3D printed device.20,28,29 Gelatin based biomaterials for 3D printing hydrogels have shown potential for integration of the host tissue within simple architectures inside implanted 3D printed gelatin methacrylate (GelMA) hydrogels in mice models.26 However, rapid vascularization within more complex architectures is still required to mimic the structure and function of organs and tissues.
To isolate the specific effects of hydrogel architecture on vascularization, we use a cell-free hydrogel platform in this study. Several recent findings support the use of cell-free hydrogel to elucidate how structural parameters such as channel diameter, length, and topography influence vascular infiltration and tissue integration whilst removing cofounding factors from embedded cells. Researchers have demonstrated that introducing patterned channels into acellular constructs can enhance vascular ingrowth and host cell migration.30 Additionally, cell-free hydrogels have been utilized to study how microarchitecture alone can promote neovascularization and integration with the host tissue.31,32 These strategies mirror the body's natural reliance on the extracellular matrix (ECM) and topological cues to guide vessel formation and remodeling.33
We and others have previously utilized 3D projection stereolithography bioprinting for patterned vascularization within GelMA and polyethylene glycol diacrylate (PEGDA) hydrogels in vivo.24,26 In this study, we further evaluate 3D bio-printed GelMA/PEGDA hydrogels to screen different channel diameters, channel lengths, and channel architectures that can improve tissue infiltration and vascularization within the hydrogels in rodent models (Fig. 1). Our use of an acellular hydrogel platform uniquely enables this high-throughput architectural screening while maintaining a physiologically relevant environment for host-driven vascularization. This 3D printing screening for enhanced vascularization uniquely offers a vascular hydrogel architectural platform for cell and tissue transplantation.
 | ||
| Fig. 1 Schematic diagram of the development and screening for hydrogel channel morphology optimized for enhanced vascular infiltration. | ||
2 Materials and methods
2.1 GelMA/PEGDA synthesis
The synthesis of photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), gelatin methacrylate (GelMA), and polyethylene glycol diacrylate (PEGDA) were prepared following a previously published method.24 Briefly, PEGDA (3.4 kDa) was synthesized by reacting poly(ethylene glycol) with acryloyl chloride and triethylamine overnight. GelMA was prepared by adding methacrylic anhydride to gelatin in carbonate/bicarbonate buffer (pH 9.5) at 50 °C for 4 hours.24,252.2 3D projection stereolithography bio-printing of GelMA/PEGDA hydrogels
Monolithic matrices were fabricated using projection stereolithography (pSLA) method, that builds 3D structures by sequentially crosslinking photosensitive polymers layer by layer through projected photomasks.34 Pre-hydrogel mixtures were prepared to contain 3.25 wt% 3.4 kDa PEGDA, 10 wt% GelMA, 17 mM LAP, 10% glycerol, and 2.255 mM tartrazine (FD&C Yellow 5, E102) as a photo absorber previously established.24 The pre-hydrogel mixture is loaded into the PDMS-coated dish, and the build platform is lowered to the initial layer position. Since GelMA is thermally sensitive, the vat is kept at 37 °C to maintain the aqueous pre-hydrogel mixture until crosslinked. Each layer height was set to 50 μm with an exposure time of 6 seconds per layer at 19.5 mW cm−2 power output. Once printing is complete, the 3D hydrogel is removed from the glass slide using a razor and allowed to swell overnight in PBS with multiple washes.2.3 Mechanical testing
Mechanical testing of 3D printed GelMA/PEGDA hydrogels and GelMA only hydrogels was performed with a disc hybrid rheometer (Discovery HR-1, TA Instruments). Hydrogels were printed into 8 mm discs and compressed using an 8 mm probe by applying a constant crosshead linear rate of 10 μm s−1. Young's modulus was calculated by plotting stress over strain and calculating the slope over the linear region using a custom MATLAB code developed in our laboratory.2.4 Swelling ratio and equilibrium water content (EWC)
Swelling ratio and equilibrium water content of 3D printed GelMA/PEGDA and GelMA only hydrogels were performed by obtaining the wet weight of the 3D printed hydrogels and then placing them overnight in the oven at 65 °C and measuring the dry wet weight. The equation used for the swelling ratio was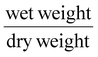 and the equation used for equilibrium water content was
and the equation used for equilibrium water content was  .
.
2.5 Intraperitoneal screening of channel diameter and length
All work involving animals was performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Rice University and was approved by the Institutional Animal Care and Use Committee (IACUC) under protocol IACUC-24-069-RU. Groups with n = 3 male C57BL/6 mice (6–9 weeks old, Charles River, strain code 027) were implanted with GelMA/PEGDA 3D bio-printed hydrogels in between the fad pads in the intraperitoneal (IP) space. Implantation of 3D bio-printed hydrogels was performed by first anesthetizing the mice using 2–4% isoflurane, followed by shaving and sterilizing the abdomen region with betadine and isopropanol. Prior to making the initial incision mice were administered with a pain reliever, 1 mg kg−1 buprenorphine SR lab (Zoopharm), and 0.9% saline to prevent dehydration. During the surgical procedure, a scalpel and scissors were used to create an incision at the linea alba, and Adson forceps were used to externalize the left and right gonadal fat pads. Hydrogels were gently rinsed with PBS before implantation and sterile 0.9% saline was used to maintain gel hydration intraoperatively. For the channel diameter screening two gels were implanted in each mouse, one containing 0.5 mm and 1 mm channels and the other containing 1.5 mm and 2 mm channels as depicted in Fig. 3. Each gel was wrapped around and sutured to the fat pads and then inserted into the abdominal cavity. Finally, both muscle and skin layers were sutured. For the length screening, only one gel was implanted per mice and were implanted as described above. Mice were monitored post-operatively. Hydrogels were explanted 2 weeks post-surgery.2.6 Intraperitoneal implantation of 3D bio-printed hydrogel geometries
Groups with n = 3–5 male C57BL/6 mice (6–9 weeks old, Charles River, strain code 027) were implanted with GelMA/PEGDA 3D bio-printed hydrogels between the fad pads in the intraperitoneal (IP) space. Implantation of 3D bio-printed hydrogels was performed following the same procedure as described in section 2.5. One hydrogel was implanted per mouse by wrapping and suturing the gel to both gonadal fat pads before inserting the gel into the abdominal region and suturing the muscle and skin layers. Mice were monitored post-operatively and provided care and husbandry as needed. Implants were retrieved and evaluated at 4-, or 8-weeks post-surgery to obtain time-dependent data.2.7 Dextran injection, hydrogel retrieval, and confocal microscopy of 3D hydrogel explants
CF680-conjugated fixable dextran, 70 kDa (Biotium, catalog #80129) was prepared at 2.5 mg mL−1 in PBS. A total of 100 μL of 680 nm fixable dextran solution was injected in the tail vein of C57BL/6 mice 15 minutes before euthanasia. After euthanasia, hydrogels were retrieved by opening the abdominal cavity and separating them from the fat pads. Retrieved gels were then imaged using a Sony A7R3 camera (Sony) and a Canon macro lens EF 100 mm 1:2.8 L IS USM (Canon). Hydrogels were placed in 10% formalin at 4 °C after explant procedure. 3D bio-printed gels were images with a Nikon A1 rsi confocal microscope using a 10× objective and 633.3 μm laser source. Laser power between 50–100% and PMT voltage between 50–150 kV was used, with a pinhole size of 122 μm.2.8 Matlab code development and quantification for vascularization
Vascularization was quantified by measuring the mean fluorescent intensity of the region of interest. Because of the complexity of the architectures, a MATLAB app was coded to be able to quantify the mean fluorescent intensity efficiently and accurately. Once uploaded into the app, the architecture was traced and identified on the confocal image. From there, the image was binarized to remove noise using MATLAB's built-in Otsu algorithm. The mean fluorescent intensity in the selected region was calculated from the binarized image and reported by the app. All confocal images were quantified three times to limit any effect of random error in the architecture selection.2.9 Imaris imaging of hydrogel infiltrated vessels
Z-Stacked Nikon nd2 files were converted in Imaris software and analyzed in 2D. Manual selection of the boundaries of the hydrogels and their channels was followed by an analysis of the infiltrated blood vessels in these channel areas using intensity thresholding. Here, only pixels with higher intensity than the background were included. This background intensity was estimated by measuring the maximum intensity in a larger non-channel hydrogel area. Additionally, all depicted confocal scans were standardized by setting their maximum intensity to ten times the background intensity. Then, the vessel-covered area was plotted in GraphPad Prism as the percentage of the total channel area. Secondly, the total intensity measurements of neovascularization signals were adjusted by the infiltration percentage to prevent designs with larger channel areas from appearing superior solely due to their size.2.10 Histology
For histological analysis, animals were euthanized by first anesthetizing them using 1.0 L min−1 O2 and 2% isoflurane. After animals were anesthetized, O2 and isoflurane were stopped, and the gas line switched to CO2 at 2 L min−1. The CO2 was left on until the animals stopped showing signs of breathing. The animals were then removed from the induction chamber, cervically dislocated to ensure euthanasia, and hydrogels were explanted and fixed in paraffin before being submitted for hematoxylin and eosin (H&E) histology staining at the Human Tissue Acquisition and Pathology Core at MD Anderson.2.11 Statistical analysis
One-way ANOVA statistical analyses were performed using GraphPad Prism 8 software (GraphPad Software); *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.3 Results and discussion
3.1 Characterization of GelMA/PEGDA 3D bio-printed hydrogels
Nuclear magnetic resonance (NMR) spectroscopy demonstrated sufficient methacrylation of gelatin and polyethylene glycol (Fig. 2a). GelMA was prepared by substituting the hydroxyl and lysine residues on the gelatin backbone with acrylate groups from methacrylic anhydride. For GelMA (left panel), the distinct peaks at δ = 5.7–5.3 ppm corresponds to the vinyl protons while the peak at δ = 1.9 ppm corresponds to the methyl protons of the methacrylate group introduced via methacrylic anhydride modification. Further the peak around δ ∼2.9 ppm which corresponds to the free lysine residues of the gelatin also got diminished. This is due to the methacrylation of the lysine residues on gelatin backbone.For PEGDA (right panel), the spectrum also shows clear vinyl proton peaks around δ = 6.4–5.9 ppm, consistent with the acrylate end groups of the diacrylate. The peaks at 4.2 and 3.6 ppm belong to the (–OCH2CH2) and (–CH2OCO–) protons of the PEG backbone. The absence of unexpected peaks and the presence of defined vinyl signals confirm that PEGDA is pure and functionalized with acrylate groups at both termini.
Taken together, these spectra validate the successful functionalization of gelatin with methacrylate groups (GelMA) and confirm the presence of acrylate groups on PEGDA, both of which are critical for subsequent photo-crosslinking via radical polymerization during 3D printing. This chemical verification is essential for ensuring crosslinking efficiency and mechanical stability of the final hydrogel constructs. The biomaterial properties of our 3D-bioprinted GelMA/PEGDA hydrogels were assessed by measuring their mechanical strength, swelling ratio, and water content. As a control, we compared them to 10% w/v GelMA hydrogels without PEGDA. Both GelMA/PEGDA and GelMA-only hydrogels, with an 8 mm diameter, were 3D-printed using a Lumen Bioprinter. After printing, the hydrogels were weighed and incubated at 60 °C overnight. Once dried, they were reweighed, and the equilibrium water content and swelling ratio were calculated. GelMA/PEGDA hydrogels exhibited significantly higher water content and swelling ratios compared to GelMA-only controls, which we attribute to the presence of PEGDA as demonstrated by others with the incorporation of PEGDA increasing swelling due to the stability of the poly(ether) backbone and therefore promoting a lack of degradation that natural proteins and polymers are subjected to.34 In addition, the incorporation PEGDA to the hydrogels enhances the mechanical properties through physicochemical interactions such as the formation of covalent crosslinking as PEGDA contains acrylate groups that participate in photopolymerization with the methacrylate groups on GelMA. Additionally the introduction of PEGDA can influence the hydrogen bonding and ionic interactions presence of ions can lead to ion–dipole interactions between water molecules and polymer functional groups, reinforcing the tightly bound water fraction and impacting the hydrogel's swelling properties.35,36 Overall, the incorporation of PEGDA improves hydrogel print stability and increases initial weights, resulting in greater equilibrium water content and swelling ratios (Fig. 2b and c). Mechanical strength was assessed through compression testing of the 8 mm cylindrical GelMA/PEGDA and 10% GelMA hydrogels using a hybrid rheometer (Fig. 2d). The GelMA/PEGDA 3D printed hydrogels showed significantly higher young modulus compared to GelMA-only hydrogels, confirming the notion that PEGDA enhances mechanical stability, resulting in stronger, more durable hydrogels capable of withstanding stress without breaking, making them a better candidate for in vivo applications.
Fibrotic tissue formation is a common hinderance for the therapeutic success and translation of implantable 3D printed biomaterial constructs. Therefore, mitigating the fibrotic response is essential for long-term or replacement therapy. The biomaterial and surface properties determine the immune response and overall implant fate.37 The 3D printed GelMA/PEGDA hydrogels were implanted between the fat pads in the intraperitoneal space of mice for 4 and 8 weeks to evaluate the presence of a fibrotic layer. After retrieval, the hydrogels were fixed and sectioned for H&E staining. The resulting images demonstrated minimal to no fibrotic response and biocompatibility as evident by the absence of a fibrotic layer in the gel surface both at 4 and 8 weeks (Fig. 2e and f). While GelMA is of naturally derived gelatin, it is thus subjected to material properties that promote degradation and enhanced cell recruitment, such as fibroblasts that encourage collagen deposition and matrix remodeling as well as vascularization.38,39 On the other hand, PEGDA is thought to be antiadhesive and when combined with other biomaterials, such as GelMA, can reduce fibrosis formation.40,41 Therefore making the GelMA/PEGDA biomaterial formulation a candidate for further in vivo 3D printing applications.
3.2 In vivo channel diameter and channel length screening within 3D bio-printed GelMA/PEGDA hydrogels
Blood vessel growth into an implanted construct is required for tissue regeneration, functionality, transplanted cell survival, and integration of the implant.42 Current tissue engineering approaches such as porous scaffolds are limited and are not sufficient for robust in-growth of blood vessels from the host, leading to poor delivery of nutrients and oxygen, followed by implant morbidity.43 Therefore, we and others have employed 3D printing strategies to fabricate macro- and microchannels into biomaterial construct.44 However, the majority of this work is focused on the development of microchannels that can be perfused with endothelial cells to fabricate in vitro tissues, rather than investigating how these open channels influence blood vessel growth and guidance in vivo. Therefore, studies have focused on microchannels between 0.2–1 mm in diameter.45–47 The significance of these channel diameters on blood vessel infiltration has yet to be determined. Here, GelMA/PEGDA hydrogels were 3D printed with channel diameters of 0.5 mm, 1 mm, 1.5 mm and 2 mm. Both hydrogels were implanted between each fat pad in the IP space of C57BL6 mice for 2 weeks. Prior to retrieval, 680 nm fixable dextran was injected into the mice via the tail vein to visualize infiltration of blood vessels into 3D printed constructs as previously established (Fig. 3a).26 Confocal analysis revealed increased blood vessel infiltration within the 1 mm channels compared to the other diameters. To quantify the dextran signal within each channel, two complementary methods were employed: a custom MATLAB code developed in our lab and Imaris imaging software. Both analyses indicated that the mean fluorescence intensity within the 1 mm channels was significantly higher compared to the rest of the channels (Fig. 3b d and ESI Fig. 2a†). These results demonstrated that 1 mm channels have a higher potential to allow for blood vessel infiltration in vivo as they are large enough to let vessels and pro-angiogenic molecules invade through the channels. Therefore, there may be a limited channel diameter that alone encourages vessel integration in vivo.Following this, we aimed to evaluate whether channel length would influence the degree of blood vessel infiltration within the 3D-printed GelMA/PEGDA hydrogels. Individual hydrogels with a channel diameter of 1 mm and varying lengths of 6 mm, 10 mm, and 16 mm were printed. Each hydrogel was implanted between the fat pads in the intraperitoneal space of C57BL6 mice for 2 weeks and injected with 680 nm dextran as previously done. Confocal microscopy showed no clear differences in pro-angiogenic signal between the three-channel length conditions (Fig. 3a). MATLAB and Imaris software were used to analyze the pro-angiogenic signal within the channels in each of the hydrogels. While the 6 mm channels exhibited a higher signal, the differences between the 6 mm, 10 mm, and 16 mm channels were not statistically significant (Fig. 3c, e and ESI Fig. 2b†). Gross imaging and H&E staining confirmed the presence of blood vessels, with the most notable infiltration observed in the 6 mm and 10 mm channels ( Fig. 3f and ESI Fig. 2c–e†). However, there appeared to be infiltration at the opening and end of the inlet or outlet after 2 weeks of implantation.
Together, these results suggest the diameter of the inlet and outlet of the 3D bio-printed channels significantly influence blood vessel infiltration within the hydrogel, while hydrogel length appears to have a lesser impact. These findings were then incorporated into the design of the remaining studies, with a consistent 1 mm opening channel diameter maintained throughout.
3.3 Fabrication and characterization of intrinsic architectures within 3D bio-printed GelMA/PEGDA hydrogels
Current bioprinting approaches for micro and macro-channel blood vessels only use a single layer of endothelial cells, similar to that of capillaries within channel dimension comparable to arteries or arterioles. This often leads to poor translation as it does not perform the necessary physiological functions of native blood vessels.48,49 Self-assembled vessel networks provide a more promising approach; however, these can lead to irregular vascular network geometries.42,49 After investigating the optimal channel diameter and length for GelMA/PEGDA hydrogels and guidance of the native vasculature, an alternative to current engineering of vascular networks could be providing the host vasculature with guiding channels to dictate and pattern vascular networks to similar geometries found within the native environment. We designed hydrogels with various channel architectures using Blender software to explore how variations in channel design, such as altering channel complexity influence blood vessel infiltration within open channels. Each of the architectures featured channels with an initial opening of 1 mm, as this diameter previously demonstrated enhanced blood vessel infiltration. The dimensions of the hydrogels were 9.3 mm × 4 mm × 16.1 mm based on the dimensions used for a previously published channel architecture labeled as serpentine, which we used as a control in these studies.26,27 To evaluate the impact of channel complexity on vascularization, we developed four new architectures inspired by capillary and alveoli features – geometry 1 (GEO1), geometry 2 (GEO2), geometry 3 (GEO3), and geometry 4 (GEO4) (Fig. 4a). To verify that the channel architectures designed within the hydrogels were feasible to be infiltrated by blood vessels and pro-angiogenic molecules, we studied computational fluid dynamics simulations using COMSOL, a mathematical modeling software that was used to analyze the flow of fluids through the channels. The settings used for the COMSOL simulation were laminar flow with pressure of 1 atm and a temperature of 293.15 degrees kelvin following a previously published system.24 All the designed channel architectures showed different fluid flow patterns. The control architecture, serpentine, exhibited the fastest fluid velocity symbolized by a red-colored pattern in the flow of the fluid. We hypothesized that the lack of retention of fluid inside the gel could decrease the opportunity for vascularizing cells and pro-angiogenic agents to infiltrate and form blood vessel structures. In contrast, GEO4, GEO1, GEO2, and GEO3 architectures all showed areas within the channel architecture with slower fluid flow, (symbolized by a blue-colored pattern) which could promote blood vessel formation within the channels (Fig. 4b). Hydrogels were printed and then perfused with red ink dye to validate flow patterns. All the hydrogels were able to be perfused without the channels bursting or breaking. In addition, the perfusion of the 3D bio-printed hydrogels via manual injection of red dye with a 1 ml syringe confirmed the simulation findings, demonstrating that all the architectures have the potential to be perfused with pro-angiogenic fluids and allow for blood vessel infiltration (Fig. 4c).3.4 In vivo architecture screening of intrinsic architectures within 3D bio-printed GelMA/PEGDA hydrogels
Following the in vitro characterization of the five distinct hydrogel architectures, these were implanted between the fat pads in the intraperitoneal space of C57BL6 mice for 4 and 8 weeks (Fig. 5a). To assess blood vessel infiltration, 680 nm fixable dextran was injected via the tail vein as described above. Gross images of explanted hydrogels were taken, GEO3 hydrogels exhibited an increase in blood vessel and tissue infiltration compared to the rest of the architectures (ESI Fig. 3c†). Retrieved samples were evaluated for dextran positive signal. Initial visual observations of confocal microscopy indicated insufficient blood vessel infiltration at the 4-week mark (Fig. 5b and c). Quantification using MATLAB and Imaris software confirmed this, showing minimal signal that made it difficult to distinguish which architecture promoted enhanced vascularization (Fig. 5c and ESI Fig. 3a†). We hypothesized that the limited infiltration at this point was due to the larger dimensions of the hydrogels (16 mm in length) that were necessary to accommodate the different architectures and to make it comparable to the previously published control serpentine architecture.However, by week 8 confocal images suggest that GEO3 architecture exhibited enhanced blood vessel infiltration (Fig. 5b and d). This was further validated by both quantification methods, which demonstrated that GEO3 significantly outperformed the other architectures in promoting vascularization within the 3D-printed hydrogels (Fig. 5d and ESI Fig. 3b†). We hypothesize that this improved performance is due to the larger internal volume and unique patterning of GEO3, which allowed greater infiltration of pro-angiogenic molecules and vascularizing cells, facilitating the formation of more robust blood vessel networks. In addition, H&E-stained images confirmed the presence of blood vessels within the lead hydrogel, GEO3, 8 weeks post implantation (Fig. 5e). One possible rationale for the GEO3 architecture demonstrating superior host vascular infiltration, could be due to the ability for macro-channels to recruit other cells, such as smooth muscle cells from the host environment rather than solely endothelial cells alone as seen with microchannel constructs, to form complete and robust vascular networks.42 Furthermore, others have shown that the incorporation of topographical cues such as channel branching, curvature, and multiple channels can accelerate vascularization from the host into a biomaterial implant.50 Our leading architecture was designed with macro-channels to maximize the volume of infiltrating blood vessels and possess branching for sufficient guidance through multiple channels from the inlet to the outlet.
With these findings, we demonstrated not only that we were able to screen 3D-printing parameters in vivo, such as channel length, diameter and architecture, to enhance vascularization but also that we have developed a 3D printing hydrogel that can guide blood vessel infiltration for future applications in tissue engineering, as well as organ transplantation.
Overall, these results demonstrate potential as an in vivo vascularizing platform, which could further be improved by perfusion through the channels or incorporation in the bulk of the hydrogel of pro-angiogenic factors, peptides or cells to accelerate blood vessel formation and growth for organ transplant as well as tissue engineering applications. Additionally, this work presents an opportunity for an up-and-coming approach; that prevascularized networks as the most ideal vascularized engineered tissue or organ must already possess robust and functional blood vessels within the biomaterial implant.50 Therefore, this leading construct could be implanted within a host for 8 weeks to form blood vessel networks and then retrieved and transplanted to a host.
4 Conclusions
In summary, this work has demonstrated the potential of our GelMA/PEGDA platform to screen various 3D printing parameters to enhance blood vessel infiltration and vascularization in vivo. Additionally, key parameters were identified, showing improved blood vessel presence and guidance within the 3D printed hydrogels in a mouse model. Specifically, these studies highlight that when 3D printing guiding hydrogels the diameter of the open channels significantly influences blood vessel infiltration, with 1 mm channels leading to the greatest infiltration. In contrast, channel length had a lesser effect on vascularization compared to channel diameter. Additionally, this platform demonstrates the ability to screen various channel architectures, which were shown to influence and guide blood vessel infiltration within the open channels. These findings highlight that the lead hydrogel design after optimization of channel diameter, length and architecture can significantly impact the extent of blood vessels infiltration. Whether used alone, or in combination with other pro-angiogenic factors, peptides or cells, the optimized platform presented in this work holds great potential as a tissue engineering and organ transplant platform, facilitating blood vessel formation in close proximity to the implant.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Breakthrough T1D.References
- U.S. Department of Health & Human Services, Organ Donation Statistics, 2023, OrganDonor.gov.
- United Network for Organ Sharing, Organ Transplantation Statistics, 2009, UNOS.org; U.S. Department of Health & Human Services, Organ Procurement and Transplantation Network Data, 2023, OPTN.
- J. Neuberger and C. Callaghan, Transplant Int., 2020, 33(12), 1597–1609 CrossRef PubMed.
- S. Shah, M. Ibrahim, M. Delaney, S. Schey, C. Bygrave, M. Streetly and R. Benjamin, Clin. Kidney J., 2019, 216–223 CrossRef CAS PubMed.
- R. Cohen, E. S. Baruch, I. Cabilly, A. Shapira and T. Dvir, Gels, 2023, 9(10), 792 CrossRef CAS PubMed.
- D. M. Hoganson, H. I. Pryor II, I. D. Spool, O. H. Burns, J. Randall Gilmore and J. P. Vacanti, Tissue Eng., Part A, 2010, 16, 1469–1477 CrossRef CAS PubMed.
- W. Song, A. Chiu and L. H. Wang, et al., Engineering transferrable microvascular meshes for subcutaneous islet transplantation, Nat. Commun., 2019, 10, 4602 CrossRef PubMed.
- A. A. Chaudhari, K. Vig, D. R. Baganizi, R. Sahu, S. Dixit, V. Dennis, S. R. Singh and S. R. Pillai, Int. J. Mol. Sci., 2016, 17(12), 1974 CrossRef PubMed.
- M. K. Lee, M. H. Rich, J. Lee and H. Kong, Biomaterials, 2015, 58, 26–34 CrossRef CAS PubMed , ISSN 0142-9612.
- H. J. Kim, I. K. Park and J. H. Kim, et al. , Tissue Eng. Regener. Med., 2012, 9, 63–68 CrossRef CAS.
- J. Zhu, Y. Wang, L. Zhong, F. Pan and J. Wang, Dev. Dyn., 2021, 250(12), 1717–1738 CrossRef PubMed.
- J. D. Baranski, R. R. Chaturvedi, K. R. Stevens, J. Eyckmans, B. Carvalho, R. D. Solorzano, M. T. Yang, J. S. Miller, S. N. Bhatia and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(19), 7586–7591 CrossRef CAS PubMed.
- T. Mirabella, J. W. MacArthur, D. Cheng, C. K. Ozaki, Y. J. Woo, M. Yang and C. S. Chen, Nat. Biomed. Eng., 2017, 1, 0083 CrossRef CAS PubMed.
- W. Song, A. Chiu, L. H. Wang, R. E. Schwartz, B. Li, N. Bouklas, D. T. Bowers, D. An, S. H. Cheong, J. A. Flanders, Y. Pardo, Q. Liu, X. Wang, V. K. Lee, G. Dai and M. Ma, Nat. Commun., 2019, 10(1), 4602 CrossRef PubMed.
- C. D. Murray, Proc. Natl. Acad. Sci. U. S. A., 1926, 12(5), 299–304 CrossRef CAS PubMed.
- T. F. Sherman, J. Gen. Physiol., 1981, 78(4), 431–453 CrossRef CAS PubMed.
- K. A. McCulloh, J. S. Sperry and F. R. Adler, Nature, 2003, 421(6926), 939–942 CrossRef CAS PubMed.
- J. M. Lee and W. Y. Yeong, Adv. Healthcare Mater., 2016, 5(22), 2856–2865 CrossRef CAS PubMed.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32(8), 773–785 CrossRef CAS PubMed.
- W. Zhu, X. Ma, M. Gou, D. Mei, K. Zhang and S. Chen, Curr. Opin. Biotechnol., 2016, 40, 103–112 CrossRef CAS PubMed.
- E. S. Bishop, S. Mostafa, M. Pakvasa, H. H. Luu, M. J. Lee, J. M. Wolf, G. A. Ameer, T. C. He and R. R. Reid, Genes Dis., 2017, 4(4), 185–195 CrossRef CAS PubMed.
- C. Mandrycky, Z. Wang, K. Kim and D. H. Kim, Biotechnol. Adv., 2016, 34(4), 422–434 CrossRef CAS PubMed.
- S. Derakhshanfar, R. Mbeleck, K. Xu, X. Zhang, W. Zhong and M. Xing, Bioact. Mater., 2018, 3, 144–156 Search PubMed.
- B. Grigoryan, S. J. Paulsen, D. C. Corbett, D. W. Sazer, C. L. Fortin, A. J. Zaita, P. T. Greenfield, N. J. Calafat, J. P. Gounley, A. H. Ta, F. Johansson, A. Randles, J. E. Rosenkrantz, J. D. Louis-Rosenberg, P. A. Galie, K. R. Stevens and J. S. Miller, Science, 2019, 364(6439), 458–464 CrossRef CAS PubMed.
- I. S. Kinstlinger, G. A. Calderon, M. K. Royse, A. K. Means, B. Grigoryan and J. S. Miller, Nat. Protoc., 2021, 16(6), 3089–3113 CrossRef CAS PubMed.
- S. Parkhideh, G. A. Calderon, K. D. Janson, S. Mukherjee, A. K. Mai, M. D. Doerfert, Z. Yao, D. W. Sazer and O. Veiseh, Biomater. Sci., 2023, 11(2), 461–471 RSC.
- M. K. Royse, M. Fowler, A. K. Mai, Y. He, M. R. Durante, N. Buist, A. Procopio, J. Xu and O. Veiseh, Biomater. Sci., 2024, 12(17), 4363–4375 RSC.
- Y. Zhang, F. Zhang and Z. Yan, et al. , Nat. Rev. Mater., 2017, 2, 17019 CrossRef CAS.
- P. S. Gungor-Ozkerim, I. Inci, Y. S. Zhang, A. Khademhosseini and M. R. Dokmeci, Biomater. Sci., 2018, 6(5), 915–946 RSC.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32(8), 773–785 CrossRef CAS PubMed.
- S. J. Yoon, Y. S. Park, J. S. Lee, D. W. Cho and J. Y. Kim, Front. Bioeng. Biotechnol., 2018, 6, 153 CrossRef PubMed.
- B. J. Klotz, D. Gawlitta, A. J. Rosenberg, J. Malda and F. P. W. Melchels, Trends Biotechnol., 2016, 34(5), 394–407 CrossRef CAS PubMed.
- D. G. Seifu, A. Purnama, K. Mequanint and D. Mantovani, Biomed. Mater., 2013, 8(1), 014106 CrossRef PubMed.
- K. Arcaute, B. K. Mann and R. B. Wicker, Ann. Biomed. Eng., 2006, 34, 1429–1441 CrossRef PubMed.
- M. Vigata, C. Meinert, N. Bock, B. L. Dargaville and D. W. Hutmacher, Biomedicines, 2021, 19(5), 574 CrossRef PubMed.
- Y. Wang, M. Ma, J. Wang, W. Zhang, W. Lu, Y. Gao, B. Zhang and Y. Guo, Materials, 2018, 11(8), 1345 CrossRef PubMed.
- S. Capuani, G. Malgir, C. Y. X. Chua and A. Grattoni, Bioeng. Transl. Med., 2022, 7(3), e10300 CrossRef PubMed.
- A. E. Chalard, A. W. Dixon, A. J. Taberner and J. Malmström, Front. Cell Dev. Biol., 2022, 10, 946754 CrossRef PubMed.
- R. I. R. Ibañez, R. J. F. C. do Amaral, R. L. Reis, A. P. Marques, C. M. Murphy and F. J. O'Brien, Polymers, 2021, 13(15), 2510 CrossRef PubMed.
- J. J. Y. Tan, D. V. Nguyen, J. E. Common, C. Wu, P. C. L. Ho and L. Kang, Int. J. Mol. Sci., 2021, 22(4), 2143 CrossRef CAS PubMed.
- J. A. Belgodere, D. Son, B. Jeon, J. Choe, A. C. Guidry, A. X. Bao, S. A. Zamin, U. M. Parikh, S. Balaji, M. Kim and J. P. Jung, ACS Biomater. Sci. Eng., 2021, 7(6), 2212–2218 CrossRef CAS PubMed.
- J. B. Lee, D. H. Kim and J. K. Yoon, et al. , Nat. Commun., 2020, 11, 615 CrossRef CAS PubMed.
- K. Zheng, M. Chai, B. Luo, K. Cheng, Z. Wang, N. Li and X. Shi, Smart Mater. Med., 2024, 5, 183–195 CrossRef.
- T. Simon-Yarza, M. N. Labour, R. Aid and D. Letourneur, Mater. Sci. Eng., C, 2021, 118, 111369 CrossRef CAS PubMed.
- H. J. Kim, H. C. Rundfeldt, I. Lee and S. Lee, Biomech. Model. Mechanobiol., 2023, 22(3), 1095–1112 CrossRef PubMed.
- A. W. Justin, R. A. Brooks and A. E. Markaki, J. R. Soc. Interface, 2016, 13(125), 20160768 CrossRef PubMed.
- J. Schöneberg, et al. , Sci. Rep., 2018, 8, 10430 CrossRef PubMed.
- C. O’Connor, E. Brady and Y. Zheng, et al. , Nat. Rev. Mater., 2022, 7, 702–716 CrossRef PubMed.
- S. M. White, et al. , Tissue Eng., Part A, 2014, 20, 2316 CrossRef CAS PubMed.
- Y. Wang, M. Liu, W. Zhang, H. Liu, F. Jin, S. Mao, C. Han and X. Wang, Burns Trauma, 2024, 12, tkae39 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5bm00193e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |




