Open Access Article
Open Access ArticleGold labelling of a green fluorescent protein (GFP)-tag inside cells using recombinant nanobodies conjugated to 2.4 nm thiolate-coated gold nanoparticles†
Nadja
Groysbeck
 a,
Mariel
Donzeau
a,
Mariel
Donzeau
 a,
Audrey
Stoessel
a,
Anne-Marie
Haeberle
b,
Stéphane
Ory
a,
Audrey
Stoessel
a,
Anne-Marie
Haeberle
b,
Stéphane
Ory
 b,
Danièle
Spehner
c,
Patrick
Schultz
b,
Danièle
Spehner
c,
Patrick
Schultz
 c,
Ovidiu
Ersen
c,
Ovidiu
Ersen
 d,
Mounib
Bahri
d,
Mounib
Bahri
 d,
Dris
Ihiawakrim
d,
Dris
Ihiawakrim
 d and
Guy
Zuber
d and
Guy
Zuber
 *a
*a
aUniversité de Strasbourg – CNRS, UMR 7242 Laboratoire de Biotechnologie et Signalisation Cellulaire, Boulevard Sébastien Brant, 67400 Illkirch, France. E-mail: zuber@unistra.fr
bCentre National de la Recherche Scientifique, Université de Strasbourg, Institut des Neurosciences Cellulaires et Intégratives, F-67000 Strasbourg, France
cUniversité de Strasbourg – Department of Integrated Structural Biology, Institut de Génétique et de Biologie Moléculaire et Cellulaire, 67400 Illkirch, France
dUniversité de Strasbourg – CNRS, UMR 7504, Institut de Physique et Chimie des Matériaux de Strasbourg (IPCMS), 23 rue de Loess, 67034 Strasbourg, France
First published on 24th September 2021
Abstract
Advances in microscopy technology have prompted efforts to improve the reagents required to recognize specific molecules within the intracellular environment. For high-resolution electron microscopy, conjugation of selective binders originating from the immune response arsenal to gold nanoparticles (AuNPs) as contrasting agents is the method of choice to obtain labeling tools. However, conjugation of the minimal sized 15 kDa nanobody (Nb) to AuNPs remains challenging in comparison to the conjugation of 150 kDa IgG to AuNPs. Herein, effective Nb-AuNP assemblies are built using the selective and almost irreversible non-covalent associations between two peptide sequences deriving from a p53 heterotetramer domain variant. The 15 kDa GFP-binding Nb is fused to one dimerizing motif to obtain a recombinant Nb dimer with improved avidity for GFP while the other complementing dimerizing motif is equipped with thiols and grafted to a 2.4 nm substituted thiobenzoate-coordinated AuNP via thiolate exchange. After pegylation, the modified AuNPs are able to non-covalently anchor Nb dimers and the subsequent complexes demonstrate the ability to form immunogold label GFP-protein fusions within various subcellular locations. These tools open an avenue for precise localization of targets at high resolution by electron microscopy.
1. Introduction
The investigation of protein trafficking and localization is fundamental to understand cell functioning. Fluorescence microscopy (FM) and electron microscopy (EM) are two valuable tools in this endeavour.1 Continuous technological advances in both FM and EM are steadily pushing the resolution boundaries and providing new insights into how cellular constituents precisely organize together and participate in the life and wellbeing of biological organisms. Pinpointing selected elements within the subcellular organization is unfortunately not always feasible due to the lack of differential contrast between the various cellular constituents. Specific labelling methodologies must therefore be developed and optimized to accompany the progress of microscopy technology. The Aequorea victoria green fluorescent protein (GFP) and fluorescent derivatives are popular tags to label chosen proteins through genetically made fusion proteins and to localize them with FM even in living cells and organisms. For EM, gold nanoparticles (AuNPs) are the contrasting agents of choice.2 The traditional AuNP-antibody conjugates reported for the first time by Faulk and Taylor,3 consisting of 150 kDa antibodies that are physically adsorbed onto colloidal AuNPs of 10–15 nm, are still in use today. Yet, the large size of AuNPs and IgG molecules, as well as the difficulty to define bonds between the AuNPs and the IgG hamper the efficiency of labelling as well as accurate target localization.4 Reduction of the AuNP diameter to less than 1.4 nm led to improved labelling agents with favourable diffusion and penetration abilities into tissues.5 Replacement of the full 150 kDa IgG with the smaller 50 kDa fragment antigen-binding domain (Fab) was also beneficial.6,7 In a step toward further optimization, 12–15 kDa single-domain camelid antibody fragments (VHH) or nanobodies (Nbs) appear highly appealing.8–10 They are interesting not only because of their compactness (molecular weight (MW) of ca. 15 kDa for a size of 4 nm × 2.5 nm), but also due to their bioengineering properties.11 Point mutations of selected amino-acids to cysteines12,13 afford precise attachment sites for thiol-reactive fluorophores. The resulting fluorescently-labelled Nbs have known spacing distances between the targeted epitope and the fluorescence signal,14 affording probes with enhanced target localization accuracy suited for super-resolution microscopy imaging of biological targets.15,16 Nanobodies have also been applied to EM immunolabelling.Kijanka and colleagues developed a protocol for the labelling of HER2 using an HER2 binding Nb followed by a secondary anti-Nb antibody and then a protein A-gold particle conjugate.21 Ariotti and colleagues developed fusion proteins between GFP-binding Nbs and a soybean ascorbate peroxidase to locally produce a stain upon the addition of diaminobenzidine (DAB) and H2O2.22,23 The brownish stain visible by optical microscopy (OM) could be further converted into an electron dense product through treatment with OsO4, permitting correlative light and electron microscopy observation. Investigation of the direct conjugation of Nbs to 10–15 nm AuNPs by adsorption has been reported by Goossens and colleagues.24 Functional conjugates were obtained but maintaining the colloidal stability of the nanobody-AuNP conjugates was clearly extremely challenging because of the huge difference in bulkiness between the Nb and the AuNP. Decreasing the difference can be achieved by decreasing the diameter of the AuNPs.25 However, decreasing the diameters weakens the strength of protein adsorption onto the AuNP surface, leading to highly dynamic protein coronas.26,27 If the gold-bound protein layer is mandatory for target recognition, linkage of the protein to the AuNP should hence be performed using stronger coordination links such as the Au–S bonds28,29 and/or functionalization strategies involving covalent bonds.30 To design Nb-AuNP conjugates with usefulness for gold immunolabelling application of cellular specimens, we explored methods for strongly linking thionitrobenzoate-coated AuNPs (AuG)31,32 to bioengineered GFP binding Nbs.8,20 We discovered that the non-covalent conjugation approach based on the complementary associating dimers originating from a p53 heterotetramer variant17,18,33 led to Nb:AuNP assemblies with excellent immunogold labelling abilities (Fig. 1).
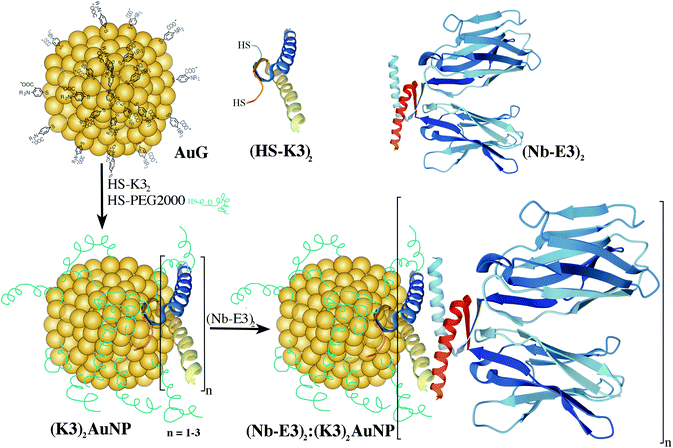 | ||
| Fig. 1 Schematic route for the synthesis of Nb2-AuNP conjugates using the strong and mutual associating ability of E3 and K3 dimers originating from a p53 heterotetramer.17,18 According to the structure of the p53 tetramerization domain (pdb1OLG),19 the dimers mainly fold into α-helices. The GFP binding nanobody (Nb) mainly folds into β-sheets (in blue pdb3OGO).20 | ||
2. Results and discussion
2.1 Synthesis of GFP nanobody-AuNP conjugates
To generate immunogold probes based on Nbs, we selected the GFP binding Nb cAbGFP4 with a previously determined structure. It displays a dissociation constant (Kd) within the nanomolar range.20 For the gold domain, we used 2.4 nm AuG that was previously described and extensively characterized.32This AuG is water-soluble and contains a single layer of substituted thiobenzoates that can be readily replaced with other thiol-containing molecules via a thiolate-to-thiolate exchange. Functionalized AuNPs with robust Au–S coordination bonds might hence be conveniently synthesized.
In our first attempt, we investigated the preparation of nanobody-AuNP conjugates directly from Nbs that were point-mutated to contain an additional thiol.14 The GFP binding Nb sequence was bioengineered to possess either a C-terminal cysteine (C143-Nb) or a cysteine at position 7 (C7-Nb). The C143-Nb displayed severe proteolytic instability and was not further used (ESI, Fig. S1†). The C7-Nb was stable at 4 °C for more than six months, had GFP binding ability similar to that of the wild type Nb (ESI, Fig. S2†) and could be linked to AuG via Au–S coordination. The conjugates could be further purified and characterized by SDS-PAGE (ESI, Fig. S3†). Unfortunately, in contrast to the covalent appending of fluorescent molecules,14 the direct coordination of the C7-Nb onto the 2.4 nn AuNP dramatically altered its ability to bind to GFP and labelling potency (Fig. S3†). The direct Au–S coordination bond at this C7 position may have exposed the nanobody to the AuNP's corona, resulting in obliteration of the nearby GFP-Nb binding interface.
Such an alteration of the Nb specificity for its target after conjugation has already been pointed out in the literature.10 Even the functionalization of Nbs with fluorophores of M.W. smaller than 1000 Da fluorophores via random coupling to lysines can drastically reduce Nb binding.14
To obtain a Nb-AuNP conjugate without loss of binding affinity of the Nb for its target and loss of protein integrity, we played around by comprehensive modification of the peptide sequence at the C-terminus. Instead of carrying out this classical approach, we chose to investigate a non-covalent functionalization approach using the properties of 2 peptides derived from the p53 protein to specifically form a stable heterotetramer composed of two homodimers (Fig. 1). Relative to the native p53 tetramerization domain, one sequence was enriched in lysine (K) while the replacement of one K with glutamic acid (E) provided an E-enriched sequence (E3).17 These E- and K-enriched sequences, named E3 and K3, respectively, were recently demonstrated to conveniently favour the selective non-covalent union of 2 distinctly tagged functional proteins into highly stable heterotetramers even in complex biological fluids crowded with all sorts of proteins and diverse compounds.18,33
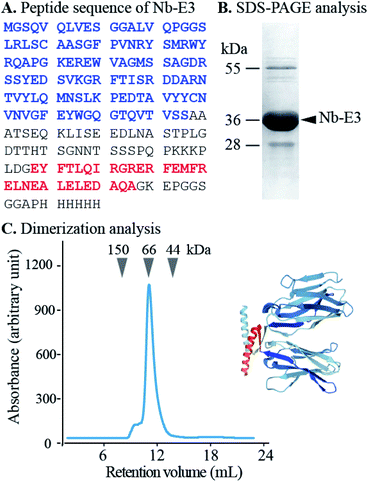
First, the Nb was genetically engineered at its C-terminal end with the aspartate-enriched variant of the p53 heterotetramer domain (E3) along with a polyhistidine sequence (Fig. 2).18 The recombinant Nb-E3 fusion protein was then produced in E. coli and purified by immobilized metal affinity chromatography followed by size exclusion chromatography (SEC) with high yield and purity (Fig. 2B). The SEC profile showed elution of a species with an apparent molecular weight (MW) close to 66 kDa corresponding to the MW of a dimer of Nb-E3 (Fig. 2C). The integrity of the Nb-E protein was fully maintained upon storage at −80 °C. Proteolytic cleavage was nonetheless observed upon storage >120 days at 4 °C (ESI, Fig. S4†).
The K-enriched sequence K3 (ref. 17 and 18) was fused to the CALNN motif.34 According to extrapolation from the structure of the p53 domain,19 the two cysteines at the N-terminal end are 2 nm apart with little chance to cross react. They can also point toward the same direction, offering possible bidentate coordination to AuG upon thiolate exchange reaction. Functionalization of the AuG with this dimer in 0.1 M HEPES buffer, pH 7.5, was then assayed at increasing dimer/AuG ratios (ESI, Fig. S5†). After a 3 h incubation time at 25 °C to allow thiolate-to-thiolate exchange of Au(I)-coordinated ligands on the AuG surface, the crude reactions were analysed by SDS-PAGE. Functionalization of the AuNP with the dimer via exchange of ligands proceeded smoothly with an increased number of dimer attachment per particle by increasing the dimer/AuG ratios. At the dimer/AuG ratio of 3, the mixture was devoid of unreacted AuG. Three distinctive AuNP species with different electrophoretic mobility were produced, likely corresponding to AuNPs equipped with 1, 2 and 3 dimers. It should be noted that, the masking of the gold surface of AuG by coordination of thiolated PEG 2000 Da was mandatory to minimize non-specific adsorption of the probe onto the cellular ultrastructure (ESI, Fig. S6†). Therefore, this reaction condition was scaled-up and immediately followed by the addition of excess thiolated PEG 2000 Da to ensure full PEG-mediated shielding of the AuNP surface (Fig. 3). The end product (K3)2AuNP, barely migrated into the polyacrylamide gel, likely because of the properties of PEG to shield the superficial charge of the nanomaterial.35,36
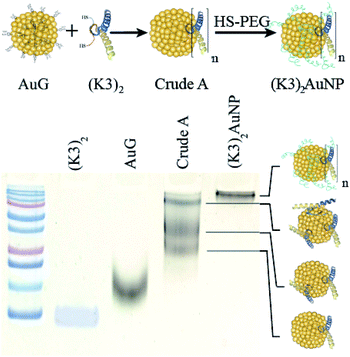 | ||
| Fig. 3 PAGE analysis of reaction products obtained by the reaction of AuG with 3 molar equivalent of the thiolated K3 dimer (Crude A) and then thiolated PEG 2000 Da ((K3)2AuNP). | ||
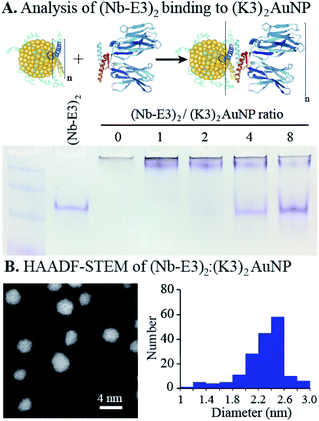
The non-covalent association of (Nb-E3)2 with the (K3)2AuNP was next evaluated by titrating the (K3)2AuNP PAGE with increasing amounts of (Nb-E3)2. A PAGE analysis under non-denaturing conditions allowed the assessment of binding by detecting free/unbound (Nb-E3)2 (Fig. 4). A small amount of free (Nb-E3)2 started to be detected at the (Nb-E3)2/(K3)2AuNP ratio of 2 indicating that each (K3)2AuNP can anchor slightly less than 2 (Nb-E3)2. In the next experiments, the non-covalent (Nb-E3)2:(K3)2AuNP assemblies were generated by mixing the modified AuNP with 2 molar equivalents of (Nb-E3)2.
The high-angle annular dark field scanning transmission electron microscopy (HAADF-STEM) observation of (Nb-E3)2:(K3)2AuNP assemblies revealed gold particles of roughly spherical shape (Fig. 4B). Analysis of a TEM image containing 180 gold particles showed that the sizes of the AuNP were distributed mostly between 2 and 2.6 nm (average diameter: 2.31 nm; standard deviation: 0.31 nm) (ESI, Fig. S7†). Moreover, the image was almost devoid of clustered particles (less than 2%) indicating that the used synthetic condition mainly favoured the coordination of the two thiols of the bidentate K3 dimer with gold atoms of one single AuNP, as seen in the illustration (Fig. 1).
2.2 Binding ability of the non-covalent (Nb-E3)2:(K3)2AuNP to GFP
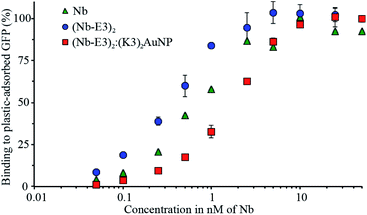
In order to use the complex as a probe for GFP in microscopy, we had to check whether the (Nb-E3)2:(K3)2AuNP complex maintained its ability to bind to GFP. To do so, we performed an indirect ELISA assay using purified GFP as the plastic-immobilized antigen and compared the binding profiles of the wild type Nb, (Nb-E3)2 and (Nb-E3)2:(K3)2AuNP complex (Fig. 5). The dose-dependent binding of the various Nbs tested showed similar sigmoidal profiles from which we assessed the apparent Kds. Accordingly, we found a Kd of 0.8 nM for the wild type Nb, 0.4 nM for each Nb of (Nb-E3)2 and 2 nM for each Nb of the (Nb-E3)2:(K3)2AuNP.The Kd values indicated that the E3-mediated dimerization of the Nb results in a gratifying avidity effect (four fold increase in apparent affinity when (Nb-E3)2 is compared to Nb).18,37 In contrast, the grafting of about 2 (Nb-E3)2 onto the (K3)2AuNP resulted in a 5-fold affinity loss relative to (Nb-E3)2.
The reason behind the apparent affinity loss of (Nb-E3)2 after anchoring to the (K3)2AuNP might be explained by the presence of PEG. PEG is required to minimize unspecific association of proteins with the AuNP surface. However, its bulkiness and vicinity to the Nb-anchored AuNP may entail a weakening of the apparent binding affinity of the AuNP-attached nanobodies to GFP.
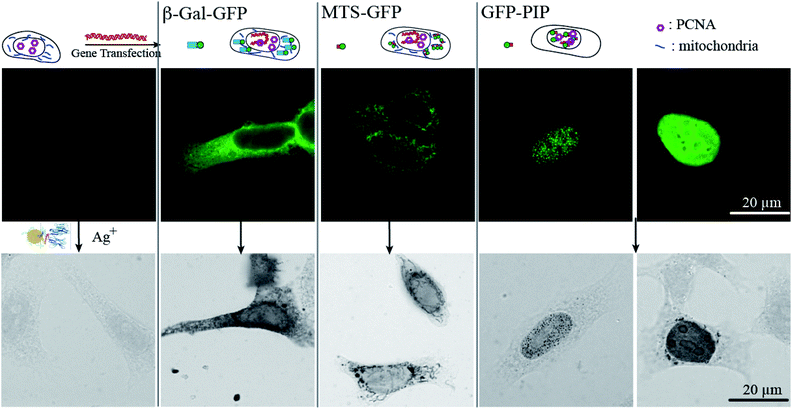
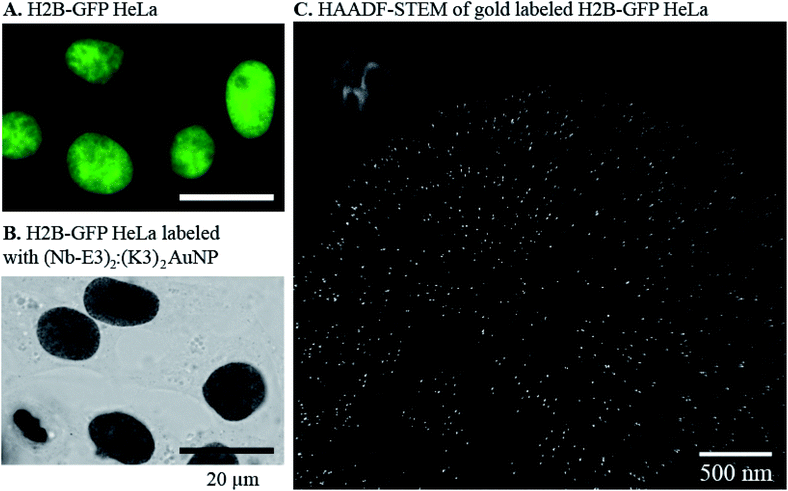
The efficiency of the (Nb-E3)2:(K3)2AuNP complex to mark GFP-protein fusions with AuNP inside cells was then evaluated using the immunocytochemistry method followed by bright field microscopy (BFM) (Fig. 6). HeLa cells were transiently transfected with DNA plasmids encoding a β-Galactosidase-GFP (β-Gal-GFP), a mitochondria-targeting signal tagged GFP (MTS-GFP) and a GFP tagged-PCNA-interacting peptide (PIP) (GFP-PIP). The cells expressing various levels of each exogenous protein were then observed by FM (Fig. 6, upper images). The subcellular localization of these GFP-protein fusions was then assayed by immunogold labelling with the (Nb-E3)2:(K3)2AuNP followed by silver enhancement to sizes observable by BFM (Fig. 6, lower images). Comparison between the FM and BFM images of the transfected HeLa cells showed an excellent correlation between the fluorescence signal and the silver-enhanced AuNPs. The 520 kDa β-galactosidase-GFP fusion protein was seen inside the cytosol but not inside nuclei in agreement with the function of the nuclear envelop to block the passive diffusion of macromolecules above 50 kDa. When the 520 kDa β-galactosidase-GFP fusion was further equipped with a nuclear localization signal (NLS), FM and immunogold labelling experiments also showed the expected transport of the β-Gal-GFP-NLS into the cell nuclei (ESI, Fig. S8†). The preferential accumulation of MTS-GFP in mitochondria (filamentous ultrastructure near surrounding the nucleus) was also observed by the immunogold labelling protocol. The GFP-PIP can be used as a marker for cell replication.38 Two patterns could be observed in transfected cells: GFP-PIP fluorescent foci in the nucleus corresponding to early DNA replication initiation sites, or the nucleus filled with GFP-PIP with accumulation near the nucleoli corresponding to a more advanced state of DNA replication. Both of these patterns were also observed using the (Nb-E3)2:(K3)2AuNP.
The ability of the (Nb-E3)2:(K3)2AuNP to detect GFP in cellular specimen fixed with a low percentage of glutaraldehyde39 was preserved although with a loss in sharpness (ESI, Fig. S9 and S10†). For comparison of labelling activity, a mouse anti-GFP antibody-gold particle conjugate made from the same TAB- and TNB- coated gold particles and a monoclonal 150 kDa IgG was prepared (ESI, Fig. S11†). Its gold labelling ability was similar to the one of the (Nb-E3)2:(K3)2AuNP (ESI, Fig. S12–S14†). A tendency of the anti-GFP IgG-AuNP conjugate to preferentially label the nuclear periphery of the H2B-GFP HeLa was noticed (Fig. S12 and S14†). If the (K3)2AuNP is preferably mixed with (Nb-E3)2 right before use, the (K3)2AuNP complex was seen to remain active for 7 days after storage at 4 °C (ESI, Fig. S15†). The selectivity of the E3 dimer for the K3 dimer could also be exploited by detecting the specimen-bound (GFP Nb-E3)2 with the (K3)2AuNP through sequential incubations (ESI, Fig. S15†). Altogether, the experiments clearly demonstrate the usefulness of the (Nb-E3)2:(K3)2AuNP as a probe to label GFP and fusion proteins with gold particles either inside the cytosol or inside the nucleus.
2.3 (Nb-E3)2:(K3)2AuNP assemblies for immunogold labelling of GFP-protein fusion inside cells by electron microscopy
We next evaluated whether (Nb-E3)2:(K3)2AuNP complexes were suitable for EM labelling of GFP-tagged proteins in cells using a stably transformed HeLa cell line that expressed the nuclear histone protein H2B fused to GFP.40 First, the stable H2B-GFP HeLa and wild type HeLa cell lines were fixed and permeabilized for immunocytochemistry labelling with extensive silver-mediated amplification of AuNPs for FM and BFM imaging (Fig. 7, images A and B). Since the silver staining correlated well with the GFP signal, the cells were processed for gold immunolabelling and moderate silver enhancement before resin embedding, sectioning and EM observation of the specimen. We also prepared an EM specimen of the immunolabelled HeLa H2B-GFP without the silver enhancement step. This non-silver enhanced specimen was analysed for gold content in the nucleus using energy dispersive X-ray (EDX) spectroscopy (ESI, Fig. S16†). The EDX spectrum clearly demonstrated the presence of gold atoms inside the cell nucleus (AuMa = 2.123 keV, AuMb = 2.203 keV, AuLa = 9.713 keV, and AuLb = 11.443 keV). Unfortunately, in our hands, the direct visualization of non-silver enhanced AuNPs in the cell sections was unfruitful due to instability of this low contrasted specimen during observation. We therefore performed a mild silver enhancement procedure of the specimen containing (Nb-E3)2:(K3)2AuNP-labeled H2B-GFP HeLa cells to grow the size of the particle to a size below 20 nm. After sectioning, the specimen was analysed by TEM. The HAADF-STEM (Fig. 7, image C) image clearly showed the presence of discrete particles inside the nucleus. EDX spectroscopy confirmed that these particles were composed of silver and gold (ESI, Fig. S17†). The density of AuNPs in the different subcellular compartments was then measured by compiling data from six images to obtain the following: 37 AuNPs per μm2 in the heterochromatin (HC) near the nuclear periphery; 14 AuNPs per μm2 within the perinucleolar compartment (PC); and 4 AuNPs per μm2 within the euchromatin (EC) regions within the nucleus. The nucleoli and the cytosol were almost devoid of probes (0.5 and 0.2 AuNPs per μm2 for the nucleoli and cytosol, respectively). A similar H2B-GFP nuclear labelling pattern was also obtained using conventional TEM whereas the wild type HeLa was devoid of any particles (ESI, Fig. S18†). A preferential localization of H2B-GFP in the heterochromatin near the nuclear or nucleolar periphery was in agreement with published studies.40,41 We can nonetheless note a discrepancy between nuclear staining seen by OM and inhomogeneous nuclear labelling observed by EM. This variation likely results from the silver staining procedure (sizes of AuNP-seeded silver particles for BFM imaging were made larger than the ones for EM imaging) and from the difference in the specimen thickness (BFM images were projections of light intensity variation throughout the whole cell; EM were performed on about 100 nm thick specimen). The density of gold labelling inside the nuclei was lower than the expected calculated density of nearly 104 H2B per μm2 (one human cell nucleus contains approximately 30 million nucleosomes, corresponding to 60 million H2B molecules).42 However, the immunogold labelling protocol often underestimates the real number because PFA might preclude recognition of a proportion of the target epitope. A high local concentration of targets might also play a role.43 In our specific case, the H2B-GFP fusion was moreover expressed in addition to the endogenous non-labelled H2B, likely at a low percentage.3. Materials and methods
3.1 Genetic engineering of the Nb-E3 construct
The GFP binding nanobody was engineered to contain the E3 peptide sequence18 and polyhistidine at the C terminal end. The GFP binding nanobody20 gene was amplified using PCR and primer sequences are as follows: CGTCAG CCATGG GGTCCC AGGTTCAGC and CCACAG GAATTC ACAATG GTGATG ATGGTG ATGTGCG. The DNA fragment was then inserted into a pETOM vector containing the E3 tag DNA sequence using NcoI and SpeI restriction sites.183.2 Expression and purification of (Nb-E3)2
The recombinant nanobody was expressed in E. coli BL21(DE3)pLys after induction with 1 mM IPTG in 100 mL LB medium at 20 °C for 24 h. The bacteria were lysed by ultrasonication. The his-tagged proteins were then purified by immobilized metal affinity chromatography using a HisTrap HP column (1 mL) charged with NiSO4 and then by gel filtration on a HiLoad Superdex 200 PG preparative column operating at a flow rate of 0.5 mL min−1. Protein fractions were analysed by SDS-PAGE. Selected fractions were pooled and protein solutions were concentrated with Amicon Ultra 4 mL centrifugal devices (MWCO 3 kDa).3.3 Synthesis of AuNP-nanobody conjugates
Typically, the AuG in 0.1 M HEPES, pH 7.5 solution (70 μL, 3 nmol) was added to the peptide sequence [CALNNGEYFTLQIRGRERFEMFRKLNKALELKDAQA] (176 μL of a 1 mM solution, 18 nmol) freshly prepared in 0.1 M HEPES, pH 7.5. After 3 h at 25 °C, the remaining exchangeable thiolates on the AuG surface were then exchanged with polyethyleneglycol 2000 Da by the addition of alpha-methoxy-omega-mercapto poly(ethylene glycol) 2000 Da (276 μL of a 1 mM solution, 276 nmol) followed by a 3 h incubation time at 25 °C. The crude mixture was then purified by ultrafiltration (30 kDa cut-off) (5 times with 0.5 mL of 0.1 M HEPES, pH 7.5) to yield 50 μL of the modified AuNP ((K3)2AuNP). The concentration of the AuNP was determined spectrophotometrically to be 18 μM.44,45 The (Nb-E3)2:(K3)2AuNP was finally obtained by the addition of (Nb-E3)2 (20 μL of a 5 μM solution, 100 pmol) to the (K3)2AuNP (5 μL of a 10 μM solution).3.4 Transfection experiments
The plasmids encoding the mitochondria targeting signal-GFP fusion (MTS-GFP), GFP-β-galactosidase fusion (GFP-β-gal), GFP-PCNA interacting peptide fusion (GFP-PIP) were gifts from Prof. Étienne Weiss and Dr Mariel Donzeau.44 They were transfected into HeLa cells using PEI derivatives.46,47 Cells were seeded onto glass coverslips in 24-well plates at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/well the day before the transfection experiment. The transgene expression was analysed after 24 h.
000 cells/well the day before the transfection experiment. The transgene expression was analysed after 24 h.
3.5 Immunocytochemistry for bright field microscopy
Coverslip-adhered cells were fixed with 4% PFA in PBS for 20 min at 20 °C. The coverslips were then washed with PBS (3 × 0.5 mL, 5 min), PBS containing 50 mM glycine (0.5 mL, 20 min) and the cell plasma membranes were permeabilized with 0.05% Triton X-100 in PBS (0.5 mL, 5 min). The coverslips were soaked in PBS containing 10% (w/v) BSA for 1 h, washed with 0.2% acetylated BSA (BSA-c) in PBS (2 × 0.5 mL, 5 min) and then incubated with 6 nM (Nb-E3)2:(K3)2AuNP in 0.2% BSA-c containing 10% FCS, 0.5 mL for 1 h. Next the cells were washed with 0.2% BSA-c (2 × 0.5 mL, 5 min) and with 80 mM citrate buffer, pH 6.2 (3 × 0.5 mL, 5 min). Finally the AuNPs were made visible using a silver staining protocol modified from the Danscher method as reported previously.483.6 Pre-embedding immunolabelling and sample preparation for EM
Cells were fixed with 4% PFA in PBS for 20 min at 20 °C. The coverslips were washed with PBS (3 × 0.5 mL, 5 min), PBS containing 50 mM glycine (0.5 mL, 20 min) and the cell plasma membranes were permeabilized with 0.05% Triton X-100 in PBS (0.5 mL, 5 min). The coverslips were soaked in PBS containing 10% (w/v) BSA for 1 h, washed with 0.2% BSA-c in PBS (2 × 0.5 mL, 5 min) and then incubated with the (Nb-E3)2:(K3)2AuNP (12 nM in 0.2% BSA-c containing 10% FCS) overnight at 4 °C. The next day the cells were washed with PBS (5 × 0.5 mL, 8 min) and were post-fixed with 1% glutaraldehyde in PBS (15 min). The cells were washed with PBS (3 times) and with H2O (5 times) and the AuNPs were enlarged using R-Gent SE-EM silver enhancement reagent (Aurion, 80 min of development). The specimen was then washed with H2O (5 times) and post-fixed with 0.5% osmium tetroxide (15 min). The specimen was again washed with H2O, dehydrated with increasing concentrations of ethanol, treated with propylene oxide and flat embedded in Epon. The resin-embedded sample was sectioned into 60 nm thick slices that were deposited onto copper 200 mesh grids (Electron Microscopy Sciences). High-angle annular dark-field scanning transmission electron microscopy (HAADF-STEM) and energy dispersive X-ray spectroscopy (EDX): cellular samples were imaged using a Cs-corrected JEOL JEM-2100F scanning transmission electron microscope operating at 200 kV and equipped with a high angle annular dark field detector. EDX analysis was performed on the same instrument equipped with a JEOL silicon drift detector (DrySD60GV, sensor size 60 mm2) with a solid angle of 0.6 srad.4. Conclusion
During our investigation to precisely functionalize AuNPs with the minimal sized Nb, we discovered that the conjugation approach involving the non-covalent assembly between complementary associating peptide tags appended to the AuNP and the nanobody, respectively, yielded probes that specifically bind to GFP. These probes proved highly effective for the visualization of various GFP-protein fusions inside cells by BFM and high resolution HAADF-STEM, thus pointing out the versatility of these gold-tagged Nbs for labelling applications using two different imaging modalities. The synthesis of the probe is convenient to perform the following: the synthetic steps can be monitored in a straightforward manner and the building blocks can be separately synthesized and stored in accordance with their stability. Moreover, the linkage of the recombinant nanobody to the AuNP at the latest synthetic step (herein after the required pegylation to protect the AuNP from interacting with encountered proteins) is extremely convenient as it limits possible chemical or structural denaturation of the protein. Even if the current probes are mixtures of species (the number of dimerized E3-Nbs per (K3)2AuNP oscillates around 2), this approach offers by far more control over stoichiometry and orientation of the attached E3-Nbs than the nanoparticle functionalization via adsorption or EDC chemistry. This non-covalent conjugation approach is clearly implementable for other nanobodies and therefore unlocks the usefulness of future nanobodies targeting proteins for the individual localization of protein inside the cell by EM and possibly cryo EM at unprecedented high resolution.Author contributions
N. G., M. D., G. Z., D. S., M. B., D. I. and A. M. H. designed and performed experiments; N. G., G. Z., P. S., S. O., and O. E. conceived experiments, analysed the data and revised the manuscript; N. G. and G. Z. drafted the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the CNRS 80prime program attributed to GZ, OE and PS, the ITI Innovec (IdEx (ANR-10-IDEX-0002), SFRI (ANR-20-SFRI-0012)) and the La Ligue contre le Cancer CCIR-Est). NG was funded by a PhD fellowship from the IdEX Unistra (Université de Strasbourg and Investissement d’Avenir). Transmission electron microscopy observation was performed at Plateforme Imagerie in vitro, CNRS UPS 3156, Strasbourg. We are grateful to Robert Drillien for style-edition of the text.Notes and references
- L. Schermelleh, A. Ferrand, T. Huser, C. Eggeling, M. Sauer and O. B. P. C. Drummen, Nat. Cell Biol., 2019, 21, 72–84 CrossRef CAS PubMed.
- Z. Jiang, X. Jin, Y. Li, S. Liu, X.-M. Liu, Y.-Y. Wang, P. Zhao, X. Cai, Y. Liu, Y. Tang, X. Sun, Y. Liu, Y. Hu, M. Li, G. Cai, X. Qi, S. Chen, L.-L. Du and W. He, Nature Methods, 2020, 17, 937–946 CrossRef CAS PubMed.
- W. P. Faulk and G. M. Taylor, Immunochemistry, 1971, 8, 1081–1083 CrossRef CAS PubMed.
- E. V. Polishchuk and R. S. Polishchuk, Tissue and Cell, 2019, 57, 103–110 CrossRef CAS PubMed.
- R. C. N. Melo, E. Morgan, R. Monahan-Earley, A. M. Dvorak and P. F. Weller, Nat Protoc, 2014, 9, 2382–2394 CrossRef CAS PubMed.
- I. Orlov, A. Schertel, G. Zuber, B. Klaholz, R. Drillien, E. Weiss, P. Schultz and D. Spehner, Sci. Rep., 2015, 5, 8324 CrossRef CAS PubMed.
- M. S. Sirerol-Piquer, A. Cebrián-Silla, C. Alfaro-Cervelló, U. Gomez-Pinedo, M. Soriano-Navarro and J.-M. G. Verdugo, Micron, 2012, 43, 589–599 CrossRef CAS PubMed.
- U. Rothbauer, K. Zolghadr, S. Tillib, D. Nowak, L. Schermelleh, A. Gahl, N. Backmann, K. Conrath, S. Muyldermans, M. C. Cardoso and H. Leonhardt, Nat. Methods, 2006, 3, 887–889 CrossRef CAS PubMed.
- K. Farnaz, B. Mahdi, R. Abbas and K.-L. Fatemeh, Int. Rev. Immunol., 2018, 37, 316–322 CrossRef PubMed.
- D. Schumacher, J. Helma, A. F. L. Schneider, H. Leonhardt and C. P. R. Hackenberger, Angew. Chem., Int. Ed., 2018, 57, 2314–2333 CrossRef CAS PubMed.
- T. Hebbrecht, J. Liu, O. Zwaenepoel, G. Boddin, C. Van Leene, K. Decoene, A. Madder, K. Braeckmans and J. Gettemans, New Biotechnol., 2020, 59, 33–43 CrossRef CAS PubMed.
- S. Massa, C. Xavier, J. De Vos, V. Caveliers, T. Lahoutte, S. Muyldermans and N. Devoogdt, Bioconjug. Chem., 2014, 25, 979–988 CrossRef CAS PubMed.
- E. Platonova, C. M. Winterflood, A. Junemann, D. Albrecht, J. Faix and H. Ewers, Methods, 2015, 88, 89–97 CrossRef CAS PubMed.
- T. Pleiner, M. Bates, S. Trakhanov, C.-T. Lee, J. E. Schliep, H. Chug, M. Böhning, H. Stark, H. Urlaub and D. Görlich, Elife, 2015, 4, e11349 CrossRef PubMed.
- G. Carrington, D. Tomlinson and M. Peckham, Mol. Biol. Cell, 2019, 30, 2737–2740 CrossRef CAS PubMed.
- B. N. G. Giepmans, Front. Cell. Neurosci., 2020, 14 Search PubMed.
- R. D. Brokx, E. Bolewska-Pedyczak and J. Gariépy, J. Biol. Chem., 2003, 278, 2327–2332 CrossRef CAS PubMed.
- M. Vigneron, F. Dietsch, L. Bianchetti, A. Dejaegere, Y. Nominé, A. Cordonnier, G. Zuber, B. Chatton and M. Donzeau, Bioconjug. Chem., 2019, 30, 1734–1744 CrossRef CAS PubMed.
- G. M. Clore, J. G. Omichinski, K. Sakaguchi, N. Zambrano, H. Sakamoto, E. Appella and A. M. Gronenborn, Science, 1994, 265, 386–391 CrossRef CAS PubMed.
- M. H. Kubala, O. Kovtun, K. Alexandrov and B. M. Collins, Protein Sci., 2010, 19, 2389–2401 CrossRef CAS PubMed.
- M. Kijanka, E. G. van Donselaar, W. H. Müller, B. Dorresteijn, D. Popov-Čeleketić, M. el Khattabi, C. T. Verrips, P. M. P. van Bergen en Henegouwen and J. A. Post, J. Struct. Biol., 2017, 199, 1–11 CrossRef CAS PubMed.
- N. Ariotti, T. E. Hall, J. Rae, C. Ferguson, K.-A. McMahon, N. Martel, R. E. Webb, R. I. Webb, R. D. Teasdale and R. G. Parton, Dev. Cell, 2015, 35, 513–525 CrossRef CAS PubMed.
- N. Ariotti, J. Rae, N. Giles, N. Martel, E. Sierecki, Y. Gambin, T. E. Hall and R. G. Parton, PLoS Biol., 2018, 16, e2005473 CrossRef PubMed.
- J. Goossens, H. Sein, S. Lu, M. Radwanska, S. Muyldermans, Y. G. J. Sterckx and S. Magez, Anal. Methods, 2017, 9, 3430–3440 RSC.
- G. Zuber, E. Weiss and M. Chiper, Nanotechnology, 2019, 30, 352001 CrossRef CAS PubMed.
- L. Boselli, E. Polo, V. Castagnola and K. A. Dawson, Angew. Chem., 2017, 129, 4279–4282 CrossRef.
- M. Zhou, C. Zeng, Y. Chen, S. Zhao, M. Y. Sfeir, M. Zhu and R. Jin, Nat. Commun., 2016, 7, 1–7 Search PubMed.
- C. J. Ackerson, P. D. Jadzinsky, G. J. Jensen and R. D. Kornberg, J. Am. Chem. Soc., 2006, 128, 2635–2640 CrossRef CAS PubMed.
- C. J. Ackerson, R. D. Powell and J. F. Hainfeld, in Methods in Enzymology, Elsevier, 2010, vol. 481, pp. 195–230 Search PubMed.
- C. Leduc, S. Si, J. Gautier, M. Soto-Ribeiro, B. Wehrle-Haller, A. Gautreau, G. Giannone, L. Cognet and B. Lounis, Nano Lett., 2013, 13, 1489–1494 CrossRef CAS PubMed.
- D. Desplancq, N. Groysbeck, M. Chiper, E. Weiss, B. Frisch, J.-M. Strub, S. Cianferani, S. Zafeiratos, E. Moeglin, X. Holy, A. L. Favier, S. De Carlo, P. Schultz, D. Spehner and G. Zuber, ACS Appl. Nano Mater., 2018, 1, 4236–4246 CrossRef CAS.
- N. Groysbeck, A. Stoessel, M. Donzeau, E. C. da Silva, M. Lehmann, J.-M. Strub, S. Cianferani, K. Dembélé and G. Zuber, Nanotechnology, 2019, 30, 184005 CrossRef CAS PubMed.
- A. Stoessel, N. Groysbeck, L. Guyot, L. Barret, Y. Nominé, L. Nguekeu-Zebaze, A. Bender, L. Voilquin, T. Lutz, N. Pallaoro, M. Blocat, C. Deville, M. Masson, G. Zuber, B. Chatton and M. Donzeau, Bioconjug. Chem., 2020, 31, 2421–2430 CrossRef CAS PubMed.
- R. Lévy, N. T. K. Thanh, R. C. Doty, I. Hussain, R. J. Nichols, D. J. Schiffrin, M. Brust and D. G. Fernig, J. Am. Chem. Soc., 2004, 126, 10076–10084 CrossRef PubMed.
- G. Zuber, L. Zammut-Italiano, E. Dauty and J.-P. Behr, Angew. Chem., Int. Ed. Engl., 2003, 42, 2666–2669 CrossRef CAS PubMed.
- M. Chiper, N. Tounsi, R. Kole, A. Kichler and G. Zuber, J Control Release, 2017, 246, 60–70 CrossRef CAS PubMed.
- G. Vauquelin and S. J. Charlton, Br. J. Pharmacol., 2013, 168, 1771–1785 CrossRef CAS PubMed.
- H. D. Herce, M. Rajan, G. Lättig-Tünnemann, M. Fillies and M. C. Cardoso, Nucleus, 2014, 5, 590–600 CrossRef PubMed.
- B. M. Humbel, M. D. de Jong, W. H. Müller and A. J. Verkleij, Microsc. Res. Tech., 1998, 42, 43–58 CrossRef CAS PubMed.
- H. Kimura and P. R. Cook, J. Cell Biol., 2001, 153, 1341–1353 CrossRef CAS PubMed.
- T. Kanda, K. F. Sullivan and G. M. Wahl, Curr. Biol., 1998, 8, 377–385 CrossRef CAS PubMed.
- W. K. M. Lai and B. F. Pugh, Nat. Rev. Mol. Cell Biol., 2017, 18, 548–562 CrossRef CAS PubMed.
- U. Schnell, F. Dijk, K. A. Sjollema and B. N. G. Giepmans, Nat. Methods, 2012, 9, 152–158 CrossRef CAS PubMed.
- G. Freund, D. Desplancq, A. Stoessel, R. Weinsanto, A.-P. Sibler, G. Robin, P. Martineau, P. Didier, J. Wagner and E. Weiss, J. Mol. Recognit., 2014, 27, 549–558 CrossRef CAS PubMed.
- X. Liu, M. Atwater, J. Wang and Q. Huo, Colloids Surf., B, 2007, 58, 3–7 CrossRef CAS PubMed.
- C. Chandrashekhar, B. Pons, C. D. Muller, N. Tounsi, R. Mulherkar and G. Zuber, Acta Biomater., 2013, 9, 4985–4993 CrossRef CAS PubMed.
- J. B. Gossart, E. Pascal, F. Meyer, E. Heuillard, M. Gonçalves, F. Gossé, E. Robinet, B. Frisch, C. Seguin and G. Zuber, Global Challenges, 2017, 1, 1700013 CrossRef PubMed.
- R. W. Burry, D. D. Vandre and D. M. Hayes, J. Histochem. Cytochem., 1992, 40, 1849–1856 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00256b |
| This journal is © The Royal Society of Chemistry 2021 |