Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Discovery of selective, antimetastatic and anti-cancer stem cell metallohelices via post-assembly modification†
Hualong
Song
 a,
Nicola J.
Rogers
a,
Nicola J.
Rogers
 a,
Simon J.
Allison
b,
Viktor
Brabec
a,
Simon J.
Allison
b,
Viktor
Brabec
 *c,
Hannah
Bridgewater
d,
Hana
Kostrhunova
c,
Lenka
Markova
c,
Roger M.
Phillips
*b,
Emma C.
Pinder
b,
Samantha L.
Shepherd
b,
Lawrence S.
Young
d,
Juraj
Zajac
c and
Peter
Scott
*c,
Hannah
Bridgewater
d,
Hana
Kostrhunova
c,
Lenka
Markova
c,
Roger M.
Phillips
*b,
Emma C.
Pinder
b,
Samantha L.
Shepherd
b,
Lawrence S.
Young
d,
Juraj
Zajac
c and
Peter
Scott
 *a
*a
aDepartment of Chemistry, University of Warwick, Coventry CV4 7AL, UK. E-mail: peter.scott@warwick.ac.uk
bSchool of Applied Sciences, University of Huddersfield, Huddersfield, HD1 3DH, UK
cThe Czech Academy of Sciences, Institute of Biophysics, Kralovopolska 135, CZ-61265 Brno, Czech Republic
dWarwick Medical School, University of Warwick, Coventry CV4 7AL, UK
First published on 18th July 2019
Abstract
Helicates and related metallofoldamers, synthesised by dynamic self-assembly, represent an area of chemical space inaccessible by traditional organic synthesis, and yet with potential for discovery of new classes of drug. Here we report that water-soluble, optically pure Fe(II)- and even Zn(II)-based triplex metallohelices are an excellent platform for post-assembly click reactions. By these means, the in vitro anticancer activity and most importantly the selectivity of a triplex metallohelix Fe(II) system are dramatically improved. For one compound, a remarkable array of mechanistic and pharmacological behaviours is discovered: inhibition of Na+/K+ ATPase with potency comparable to the drug ouabain, antimetastatic properties (including inhibition of cell migration, re-adhesion and invasion), cancer stem cell targeting, and finally colonosphere inhibition competitive with the drug salinomycin.
Introduction
Lehn envisaged in the original report1 that helicates2–4 – self-assembling multimetallic coordination compounds – may find uses in biochemistry. Indeed, while their underlying chemistry is very different to that of the small cationic α-helical peptide units that are deployed in nature for e.g. signalling, structural and host-defence roles,5–7 some such metallofoldamers8 have similar dimensions and charge. With this in mind we have developed several classes of water-compatible, optically pure metallohelix compounds,9–11 each of which has unique properties, including a growing list of peptide-like behaviours: binding of DNA motifs,12 anticancer activity,5,11,13 and the inhibition of e.g. amyloid-β aggregation,12,14 enzyme activity15,16 and ice recrystallization.17 Thus, while we cannot expressly mimic the exquisite architectures of natural peptides, we are motivated to seek methods by which diverse metallohelices might be rapidly accessed and new biological properties discovered and optimised.18The addition of new covalent bonds to supramolecular assemblies by post-assembly modification (PAM) may be used for various purposes including locking-down dynamic structures, triggering structural transformations, or simply late-stage derivatisation to introduce new functional groups.19 The latter is attractive to us since it may facilitate phenotypic discovery, property optimisation or the elucidation of structure/activity relationships, all without the need for extensive pre-assembly ligand synthesis. Further, we may include functional groups that are incompatible with the self-assembly. However, such reactions must be clean and efficient under mild conditions, and we note that the lability or reactivity of many metallosupramolecular structures means that application of the otherwise extremely versatile copper-catalysed azide/alkyne cycloaddition (CuAAC) may be hampered by deleterious reactions20 such as displacement of the original metal by copper from the catalyst.21 In addition the new triazole units formed may bind to metals.22
We report here that our self-assembled optically pure, water-soluble triplex metallohelices8 provide excellent molecular frameworks on which to perform such late-stage modification. Through this chemistry we have discovered compounds with excellent potency against a panel of cancer cell lines, with enantioselectivity reflected in cell cycle studies, plus enhanced selectivity with respect to a panel of non-cancer cell lines in vitro. One selected compound displays a remarkable array of properties: antimetastatic (inhibition of cell migration, re-adhesion and invasion), cancer stem cell targeting, and colonosphere inhibition competitive with the drug salinomycin. Mechanistically, the compound does not induce apoptosis but appears to inhibit Na+/K+ ATPase activity with potency comparable to the drug ouabain.
Results
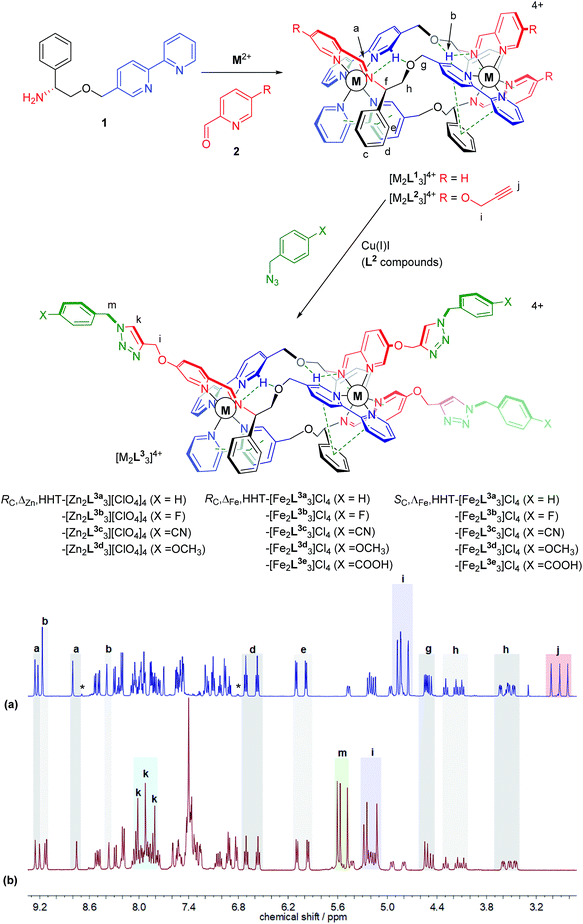
Click synthesis of new metallohelices
We recently synthesised ranges of optically pure water-soluble metallohelices in which the ligand strands run in opposing directions (the “head-to-head-to-tail” or HHT isomers).9 Of these we selected the system based on [M2L13]4+ (Fig. 1, R = H) since it possesses an appealing facially amphipathic architecture. The new metallohelix enantiomers [M2L23]4+ (M = Zn, Fe) with perchlorate and chloride counter-ions respectively, synthesised using sub-components 1 and 5-(propargyloxy)picolinaldehyde (2), are decorated with three chemically inequivalent alkyne substituents on one face of the structure.The 1H NMR spectra have several unusual features that confirm the topologically asymmetric structure [Fig. 1a]. In [Zn2L23][ClO4]4 at 293 K, three spectroscopically unique ligand environments give rise to imine singlets Ha at 9.26, 9.17 and 8.80 ppm. Two of the bpy protons Hb appear in the same region (9.22 and 9.17 ppm) as a result of intramolecular hydrogen bonds – see dotted lines in structures of Fig. 1. The third bpy proton Hb with no such interaction was found at 8.39 ppm. Similarly, the two sets of ring protons Hd and He arising from pendant phenyls taking part in bifurcated π-stacks with coordinated bpy units, appear at 6.80 and 5.90 ppm. The third phenyl group that is instead π-stacked to a coordinated pyridine has Hd and He resonances at more conventional chemical shifts of 7.11 and 6.96 ppm. At lower temperatures these signals begin to broaden, consistent with slowing of phenyl group de-coordination/rotation (ESI Fig. S3†). The rather rigid arrangement of the ligand strands leads to six distinct resonances for Hh, clustered at 4.42–4.10 ppm (apparent triplets) and 3.63–3.47 ppm (approximately doublets of doublets). Three alkyne proton singlets Hj appear at 3.0–2.8 ppm.
In the same 1H NMR spectrum of Fig. 1 the small singlet at 8.7 ppm is assigned to the HHH isomer of this compound – the three-fold symmetric compound where all three strands run in the same direction – and on this assumption we estimate the selectivity HHT:HHH to be ca. 99%. Other small peaks consistent with the presence of this minor isomer can be seen in the baseline. At 6.8 ppm a doublet is tentatively assigned to protons of type e in the HHH isomer. It is interesting to note the absence of a triplet for type d protons in the region 6.4–6.8 ppm in this minor component; no such signal is expected since there is no phenyl-bpy π-stack in the HHH isomer. Similarly, no minor doublets for type e protons are expected around 6 ppm. As such, the appearance of these minor isomer peaks corroborates our assignments for the major isomer.
Reactions employing a range of CuAAC conditions23–25 were explored and it was found that heating [M2L23]4+ (M = Zn, Fe) with benzyl azide in the presence of catalytic copper(I) iodide for 18 h cleanly gave [M2L3a3]4+. The disappearance of the alkyne resonances in the 1H NMR spectra of [Zn2L23]4+ at ca. 3 ppm (Hj) [Fig. 1b] and 78/77 ppm (Cj/Cn) (ESI Fig. S4†) demonstrates that the reaction progresses to completion. Upon formation of the triazole moiety, three new proton singlets (Hk) are evident at ca. 8 ppm and new quaternary carbon resonances (Cl) at ca. 142 ppm are observed. In addition, three new singlets at ca. 5.6 ppm are observed in the 1H NMR spectra of both [Zn2L23]4+and [Fe2L23]4+ (and 13C at ca. 55 ppm), due to the addition of the benzyl methylene group (Hm), as well as new resonances due to the benzyl ring in the aromatic region. The NMR signals corresponding to the imine, bipyridyl and phenyl units remain unperturbed by the CuAAC reaction, demonstrating that the structure of the metallohelix architecture is preserved. The successful synthesis of all the “clicked” complexes was also confirmed by high resolution electrospray mass spectrometry (ESI Fig. S18–23†). For instance, a strong signal was observed by electrospray mass spectrometry at m/z 464.1580 Da for the tetracationic molecular ion of RC,ΔFe,HHT-[Fe2L3a3]Cl4, within 0.001 Da of the calculated value for C105H93Fe2N21O6 (m/z 464.1582 Da). Inductively-Coupled Plasma Mass Spectrometry (ICP-MS) analysis of the iron triplex metallohelices revealed that only trace amounts of copper could be detected.
Subsequently fourteen new benzyl triazole-functionalised metallohelix enantiomers were isolated in a similar manner. Remarkably, the carboxylic acid derivatives [Fe2L3e3]4+ were accessible, despite the stability of Fe carboxylates. In all cases, the triplex architecture was retained, even in the case of conventionally highly labile Zn(II). Characterizing data including NMR, MS, circular dichroism (CD), IR and microanalysis are detailed in ESI.† NMR and UV-vis experiments indicate that little decomposition of the product [Fe2L3a3]Cl4 occurred over months in aqueous solution (ESI Fig. S14, S15 and S17†).
Antiproliferative activity and cell line selectivity studies
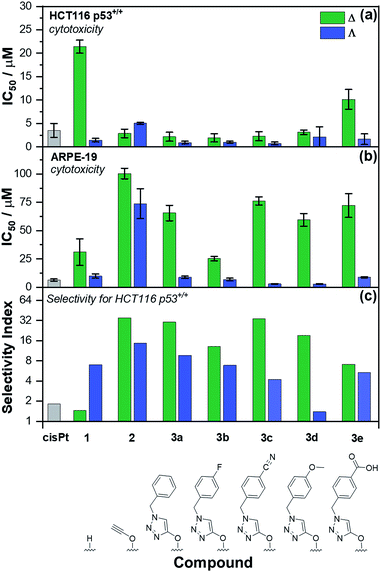
The panel of Fe(II) compounds of Fig. 1 were initially evaluated alongside cisplatin for potency against the human epithelial colorectal cancer cell line HCT116 p53+/+ (wild-type p53) and non-cancerous human epithelial retinal pigment cells (ARPE-19) (Fig. 2).26The modest potency of parent compound Δ-[Fe2L13]Cl4 against HCT116 p53+/+ leads to a poor selectivity index: the ratio of IC50 values of ARPE-19 cells to HCT116 p53+/+ cells (see Fig. 2 – note log scale). While this is higher for the Λ enantiomer, the performance of the alkyne derivatives [Fe2L23]Cl4 is much improved, with potencies similar to that of cisplatin against HCT116 p53+/+ but giving rather better selectivity indices. The “clicked” metallohelices [Fe2L3a–e3]Cl4 perform better still. They are all more potent than cisplatin against the HCT116 p53+/+ colon cancer cells, with the exception of the more moderately active tricarboxylic acid Δ-[Fe2L3e3]Cl4. These complexes also exhibit enantioselectivity; in all cases the ΛFe compounds are more active than ΔFe [Fig. 2a]. Pleasingly, the new metallohelices are all significantly less toxic to non-cancerous ARPE-19 cells with the ΔFe enantiomers being substantially less toxic than ΛFe [Fig. 2b]. The resulting selectivity indices show that the ΔFe-enantiomers are all more selective than their ΛFe analogues; both Δ-[Fe2L3a3]Cl4, and Δ-[Fe2L3c3]Cl4, have a selectivity indices for these particular cell lines of >30. On the basis that the former is the simpler compound, it was selected from the click derivatives for further study.
Accordingly, the compounds Δ-[Fe2L13]Cl4, Δ-[Fe2L23]Cl4 and Δ-[Fe2L3a3]Cl4 were screened against a larger panel of cell lines of different tissue origins (colon, ovarian, cervical and breast cancers, plus a range of non-cancer). The data are given in Table 1 and the 112 selectivity indices determined (four compounds, seven cancer cell lines, four non-cancer) are plotted in Fig. 3. The metallohelices were without exception more active against each of the seven cancer cell lines than the four different non-cancer cell lines. Furthermore, they display an overwhelmingly favourable selectivity compared with the clinically used chemotherapeutic agent cisplatin. In most instances the compound Δ-[Fe2L3a3]Cl4 was the most selective.
| Cell line | Compound | ||||
|---|---|---|---|---|---|
| Δ-[Fe2L13]Cl4 | Δ-[Fe2L23]Cl4 | Δ-[Fe2L3a3]Cl4 | Cisplatin | ||
| a The experiments were performed in triplicate or quadruplicate. The cells were treated with the investigated compounds for 72 h, unless otherwise stated. The results are expressed as mean values ± SD from three or four independent experiments. b Cells were treated for 96 h. | |||||
| Cancer cell lines | HCT116 p53+/+ (colon) | 21.4 ± 1.4b | 2.9 ± 0.9b | 2.2 ± 1.0b | 3.3 ± 0.4b |
| HCT116 p53−/− (colon) | 7.7 ± 3.7b | 3.4 ± 0.2b | 3.3 ± 0.3b | 7.5 ± 0.7b | |
| A2780 (ovarian) | 6.38 ± 0.1 | 6.1 ± 0.8 | 0.9 ± 0.2 | 3.3 ± 0.2 | |
| A2780cisR (ovarian) | 4.43 ± 0.1 | 6.1 ± 0.3 | 0.24 ± 0.02 | 20 ± 3 | |
| HeLa (cervical) | 3.8 ± 0.9 | 16 ± 6 | 7.6 ± 0.5 | 14.0 ± 0.9 | |
| MCF-7 (breast) | 2.4 ± 0.4 | 16 ± 2 | 2.2 ± 0.2 | 12.9 ± 0.6 | |
| MDA-MB-231 (breast) | 7 ± 1 | 22 ± 1 | 2.1 ± 0.2 | 22 ± 2 | |
| Non-cancer cell lines | ARPE-19 (retinal) | 31 ± 12b | 100 ± 5b | 66 ± 7b | 6.4 ± 1.0b |
| MRC-5 pd30 (lung) | 31 ± 6 | 65 ± 5 | 32 ± 5 | 12 ± 1 | |
| HMF (breast) | 18 ± 5b | 14 ± 2b | 11 ± 1b | 18 ± 2b | |
| WI-38 (lung) | >100b | >100b | 16 ± 3b | 2.2 ± 0.8b | |
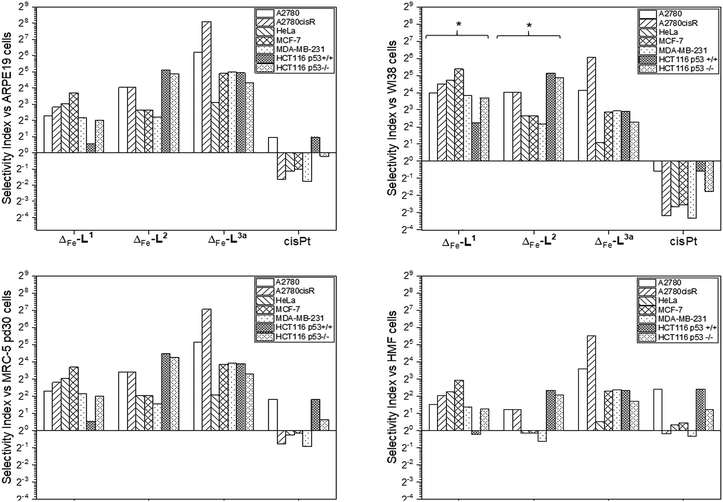 | ||
| Fig. 3 Antiproliferative selectivity indices of triplex metallohelices for cancer vs. non-cancer cells. Selectivity index = mean IC50 ratio: (non-cancer cell line/cancer cell line), IC50 values as listed in Table 1. *Minimum selectivity indices plotted as IC50 (WI-38) > 100 μM. | ||
We noted that while conversion of alkyne Δ-[Fe2L23]Cl4 to benzyltriazole Δ-[Fe2L3a3]Cl4 had little effect on potency in the HCT116 colon cancer cells, potency in the ovarian and breast cancer cell lines was increased ∼7 to 25 fold (Table 1). In contrast, activity against three of the four non-cancer cell lines was only modestly increased (by <1.3 fold in HMF, ∼1.5 fold in ARPE19 and ∼2 fold in MRC-5 pd30). The click modification resulted in a ∼4-fold increase in potency towards the cisplatin-resistant ovarian cancer cells (A2780cisR) compared to the cisplatin-sensitive parental cells indicating a lack of cross-resistance and, unsurprisingly, a distinct mechanism of action to that of the DNA “alkylator” cisplatin.
The p53 tumour suppressor gene is one of the most frequently mutated in cancer, commonly causing increased resistance to chemotherapeutic drugs. While accordingly here cisplatin was found to be >2 fold less active towards HCT116 p53−/− cells than the p53+/+ isogenic clones (Table 1), such a loss of potency was not observed for any of the Δ-metallohelices.
Mechanistic studies
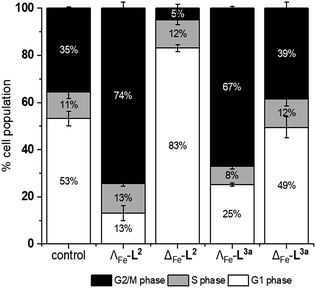
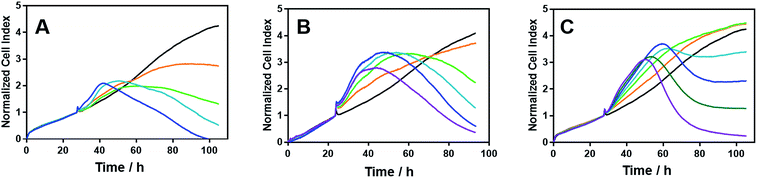
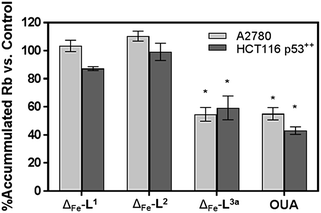
We noted that the enantiomer potencies for [Fe2L23]Cl4 (i.e. Δ > Λ) against cancer cells were reversed for [Fe2L3a3]Cl4 and the other click derivatives. We thus compared the effects of the enantiomer pairs on the cell cycle profile of asynchronously growing cells. Striking enantiomeric and structure-dependent differences were observed (Fig. 4) implying different mechanisms of action. Further, since the high selectivity index compound Δ-[Fe2L3a3]Cl4 did not induce significant alteration in the cell cycle profile, the induction of cell death via apoptosis, a target of many anticancer drug treatments,27–29 was investigated. To our surprise, induction of apoptosis was not observed via a membrane phosphatidylserine (PS) assay30 for HCT116 p53+/+ cells except after prolonged exposure at 2 × IC50 (ESI Fig. S26†).Time-dependent cellular response profiles (TCRPs) produced by impedance-based monitoring provides predictive mechanistic information for the action of small molecules.31–33 Ovarian cancer cells (A2780) that have been treated with Δ-[Fe2L13]Cl4, Δ-[Fe2L23]Cl4 and Δ-[Fe2L3a3]Cl4 show clearly distinct TCRPs (Fig. 5). For the parent metallohelix Δ-[Fe2L13]Cl4 the initial rise in Cell Index (CI) impedance signal is less apparent than for other compounds and the period of signal elevation is the shortest. For the alkyne Δ-[Fe2L23]Cl4 the CI signal increases to ca. 1.7× that of the control and the peak is relatively broad, the signal decreasing steadily over the measurement period. For the benzyl triazole derivative Δ-[Fe2L3a3]Cl4 the CI signals reach a much sharper dose-dependent maximum. A TCRP profile database search31 indicated a similarity with that for compounds that inhibit Na+/K+ stimulated ATPases; a highly conserved integral cell membrane pump expressed in virtually all cells of higher organisms that maintains ionic concentration gradients.34 An established rubidium-based assay35 subsequently showed that Δ-[Fe2L3a3]Cl4 did indeed inhibit uptake of the cation in A2780 and HCT116 p53+/+ cell lines under these conditions, by 35–47% (Fig. 6). This performance is comparable with that of the known potent Na+/K+-ATPase inhibitor ouabain36 (39–57% inhibition). In contrast Δ-[Fe2L13]Cl4 and Δ-[Fe2L23]Cl4 had little if any effect.
Antimetastatic properties of metallohelices
Colorectal cancer is one of the four most common causes of cancer deaths,37 and in 90% of instances mortality is ascribed to metastasis, for which there are currently no effective treatments.38,39 Ouabain has been reported to inhibit cancer cell migration40–43 and to possess antimetastatic activity through its inhibition of Na+/K+ ATPase.43 On the basis of the mechanistic discoveries above we investigated the effects of Δ-[Fe2L3a3]Cl4 on important steps in the process of metastasis44–46 in the colon cancer cell line HCT116 p53+/+.We modelled the detachment of cancer cells from a primary tumour by an in vitro assay of cell resistance to trypsinization.47–49 HCT116 p53+/+ cells grown in monolayer were treated with the investigated compounds for 3 h and then subjected to a diluted trypsin solution. The number of cells that resisted the treatment (i.e. remained attached to the surface) was evaluated by the sulforhodamine B (SRB) assay. Treatment with Δ-[Fe2L13]Cl4 reduced detachment only at higher concentrations [top panel in Fig. 7] and Δ-[Fe2L23]Cl4 had no significant effect. In contrast, treatment of cells with Δ-[Fe2L3a3]Cl4 significantly impeded their detachment.
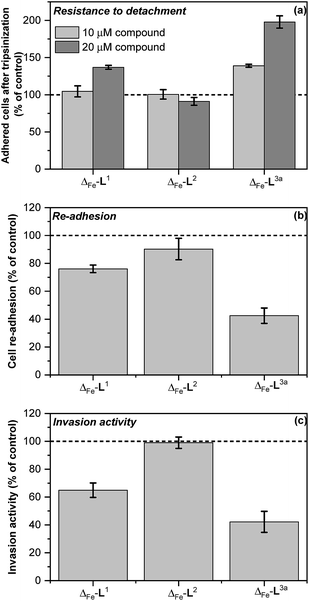 | ||
| Fig. 7 In vitro antimetastatic activity of metallohelices (a) effects of the metallohelices on cancer cell adhesion and resistance to detachment through use of diluted trypsin enzyme; cells were treated with the indicated metallohelices at 10 μM and 20 μM respectively for 3 h prior to addition of trypsin (b) cell re-adhesion assay; cells detached following trypsinization (and 3 h pre-treatment with the metallohelices at 10 μM) were assayed for their ability to re-adhere within 30 min, (c) cell invasion assay; cells were treated with the metallohelices at equitoxic (2 × IC50) concentration for 2 h, followed by assessment of invasion through Matrigel (see Methods in ESI†).The results are expressed as the mean ± SD from three independent experiments. Asterisks indicate a significant difference from untreated control (100%) with *p < 0.05 or **p < 0.001 calculated by using 2 way ANOVA. | ||
Re-attachment of cancer cells to tissue at a ‘new’ site in the body (the site of secondary tumour formation)50 was modelled in a re-adhesion assay.51,52 Cells were treated with 10 μM compound for 3 h (a non-toxic dose), detached with trypsin and re-seeded at a density of 2 × 104 cells per well. The number of cells attached after 30 min incubation was determined by the SRB assay [middle panel in Fig. 7]. Compounds Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4 reduced cell re-adhesion by 24% and 58% respectively.
Cancer cell invasion defines the ability of cells to infiltrate tissue. Using a Matrigel™ transwell assay53,54,70 (see ESI†) it was shown that while Δ-[Fe2L23]Cl4 had no effect, Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4 reduced HCT116 p53+/+ invasive ability by 35% and 58% [bottom panel in Fig. 7].
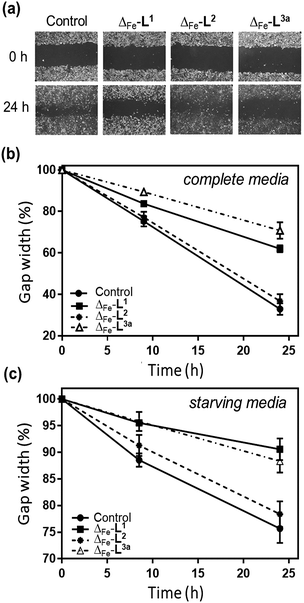
The overall ability of the compounds to influence cancer cell migration and invasion was assessed by wound healing assay (scratch gap closure)55,72 [Fig. 8a]. While in a control sample a scratch in a monolayer of HCT116 p53+/+ cells had healed in 24 h to 33%, the presence of non-toxic doses of Δ-[Fe2L13]Cl4 or Δ-[Fe2L3a3]Cl4 suppressed healing, leaving 62% and 71% respectively of the wound open. Cells treated with Δ-[Fe2L23]Cl4 resembled the control.
Cancer stem cell (CSC) targeting
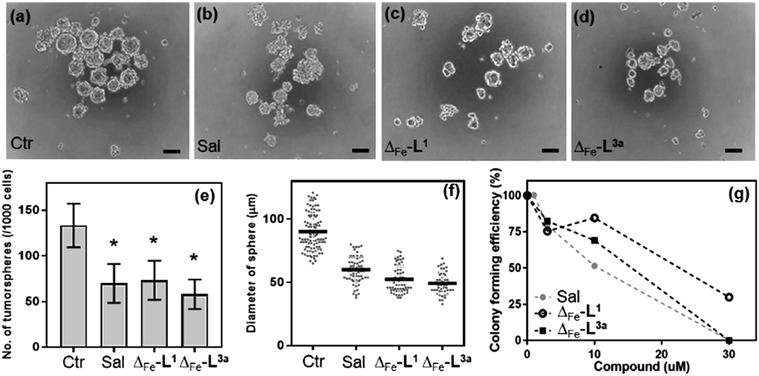
CSCs56 represent a reservoir of resistant and highly aggressive cancer cells that can remain when the bulk of a tumour has been eradicated, leading to formation of secondary or tertiary tumours.57–60 The failure to eliminate or inhibit CSCs is thereby a major cause of failure of existing anti-cancer treatments61–63 and is a key challenge to improving patient outcomes.64–67The HCT116 p53+/+ cells68,69 possess a cancer stem cell fraction capable of forming colonospheres from single cells, and have been utilized in several studies as a CSC model.70–72 The HCT116 p53+/+ cells were treated with Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4 at their respective IC30 concentrations for 72 h, and subsequently cultured as single cell suspensions in serum-free media. While both Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4 were found to inhibit colonosphere formation in HCT116 p53+/+ under these conditions, Δ-[Fe2L3a3]Cl4 in particular was significantly more effective than the CSC-selective73–76 drug salinomycin (ESI Table S3 and Fig. S27, S28†). Subsequently, significant colonosphere inhibition by both Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4 was also observed in CSC-enriched cells (HCT116.CD133+). The compounds were equally or slightly more effective than salinomycin in reduction in the number and average size of colonospheres [Fig. 9a–f].
Next, we compared the effects of Δ-[Fe2L13]Cl4, Δ-[Fe2L3a3]Cl4 and salinomycin on HCT116.CD133+ and HCT116.CD133− cells (Fig. 9 and ESI Fig. S30†) these data indicate that Δ-[Fe2L13]Cl4 and salinomycin effectively inhibit colonosphere formation from HCT116.CD133+ cells, but do not significantly inhibit colonosphere formation from HCT116.CD133− cells. This result can be interpreted to mean that Δ-[Fe2L13]Cl4 and salinomycin are CSC-selective agents. On the other hand, Δ-[Fe2L3a3]Cl4 also effectively inhibits colonosphere formation from both HCT116.CD133+ and HCT116.CD133− cells, although being less effective in CD133− negative cells. The reasonable explanation of the latter result is that Δ-[Fe2L3a3]Cl4 exhibits a lower selectivity for CSCs than Δ-[Fe2L13]Cl4 and salinomycin being able to effectively kill both differentiated cancer cells and CSCs. As effective cancer treatments must attack both rapidly-dividing differentiated (non-stem) cancer cells and CSCs, Δ-[Fe2L3a3]Cl4 appears to be a promising candidate compound able to overcome limitations connected with the use of a number of conventional chemotherapeutics.
The data in Fig. 9g arise from a clonogenic assay that examines the capability of a single cell to grow into a large colony through clonal expansion.77 CSC-enriched HCT116.CD133+ cells incubated for 48 h with 30 μM Δ-[Fe2L3a3]Cl4, exhibited no surviving cells after being allowed to grow for 8 d; an effect comparable to that of conventional salinomycin. Δ-[Fe2L13]Cl4 was less effective.
The selective anti-cancer stem cell effects of Δ-[Fe2L3a3]Cl4 were further demonstrated (ESI Table S4†). The IC50 of 1.21 ± 0.25 μM in CSC-enriched HCT116.CD133+ is around half in HCT116 p53+/+ under the same conditions; a better differential than that observed with salinomycin.
The studies aimed at the mechanism of action of the investigated metallohelices on CSCs is in progress in our laboratories and will be published in a separate article. Nevertheless, we demonstrate in our manuscript that Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4 were equally or slightly more effective in killing CSCs than CSC-selective salinomycin (ESI Fig. S30†). It was shown recently76 that nucleolin is likely a salinomycin-binding target and a critical regulator involved in human neuroblastoma CSC activity. It is, therefore, possible, due to the similar effectivity of the investigated metallohelices (Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4) and salinomycin to kill HCT116.CD133+ CSCs, that their binding to nucleolin may also be responsible for the anticancer and anti-CSC like cell activities of Δ-[Fe2L13]Cl4 and Δ-[Fe2L3a3]Cl4.
Conclusions
One of the key advantages of our metallohelix assemblies is their great stability with respect to dissociation or hydrolysis. This sort of stability will be necessary in order for any such compound to find its way into clinical use, and has here allowed the use of an extremely efficient CuAAC post-assembly modification of the triplex alkynyl enantiomers [M2L23]. This reaction gave rapid access to a new range of functionalised compounds and led to the discovery of Δ-[Fe2L3a3]Cl4; a compound with an unusual combination of pharmacological properties.In a study involving an unusually wide range of cancer and non-cancer cell types, the new “clicked” compounds demonstrated enhanced potency. More importantly however the selectivity for cancerous over non-cancerous cells was greatly improved; this bodes well for the development of compound with a wider therapeutic window than conventional chemotherapeutic agents such as cisplatin. Further, in contrast to observations for conventional drugs,78 a p53 mutated cell line did not show resistance to the compound.
While some part of the observed selectivity of Δ-[Fe2L3a3]4+ probably arises from electrostatic targeting of the anionic outer leaflet of cancer cells (as is proposed for cationic anticancer peptides79–82) the enantiomer effects observed here point to greater subtlety. The Δ compound is much more selective than Λ for cancer cells, and this is reflected in a remarkable difference in cell cycle arrest observations. It was also discovered that Δ-[Fe2L3a3]Cl4 inhibits Na+/K+ ATPase activity with potency comparable to that of the conventional inhibitor ouabain.
These mechanistic observations led to the discovery of a remarkable array of properties of Δ-[Fe2L3a3]Cl4 alongside its high potency and selectivity. The compound suppresses key metastatic capacities of cancer cells, reducing their ability to detach from other cells, migrate, invade and re-adhere elsewhere. Compound Δ-[Fe2L3a3]Cl4 exhibits selective toxicity for colon cancer stem cell-enriched cell populations, challenging some of the most selective compounds of any kind identified to date. Additionally, Δ-[Fe2L3a3]Cl4 inhibits the formation of colonospheres by specifically targeting CD133-positive, CSC-like cells. Thus Δ-[Fe2L3a3]Cl4 is identified as a potential lead compound for investigation as a selective anti-cancer, anti-metastatic and CSC-targeting drug.
More generally, while the precise molecular basis for the difference between the benzyltriazolyl-clicked compounds and the parent enantiomers is not known, it is clear that these discoveries would be much harder to unearth without access to post-assembly modification. Further, we now know that metallohelices, like conventional medicinal compounds, respond to chemical modification in such a manner as to facilitate optimization of biological and physicochemical properties.
Experimental
Full details of synthesis, characterization and anti-cancer experiments are provided in the electronic ESI.† Outlines of key procedures are detailed in the following.Synthesis of water-soluble alkyne-functionalised triplex metallohelices
Anhydrous iron(II) chloride (2 equiv.) was added to a stirred solution of the desired chiral amine, 2-([2,2′-bipyridin]-5-ylmethoxy)-1-phenylethan-1-amine (3 equiv.) and 5-(prop-2-yn-1-yloxy)picolinaldehyde (3 equiv.) in methanol (20 ml) at ambient temperature to give a purple solution that was then heated to reflux (85 °C) for 48 h. The mixture was allowed to cool to ambient temperature, filtered through a Celite plug, and the solvents were removed in vacuo to give HHT-[Fe2L23]Cl4 as a dark purple solid (>95% yield).Post-assembly modification of triplex metallohelices
HHT-[Fe2L23]Cl4 (1 equiv.) and the chosen benzyl azide (4.5 equiv.) were dissolved in methanol (10 ml) in the presence of copper(I) iodide (0.1 equiv.). The reaction mixture was heated at 65 °C for 18 h under inert argon atmosphere. After cooling to ambient temperature, the suspension was filtered to remove copper salts and the purple product HHT-[Fe2L33]Cl4 was isolated by the addition of ethyl acetate. NMR, infrared and high-resolution mass spectrometric data were consistent with the proposed formulations. CD spectra of enantiomers in methanol were equal and opposite.Antiproliferative activity (MTT assay)
The human ovarian carcinoma cisplatin-sensitive A2780 cells, cisplatin-resistant A2780cisR (a cisplatin-resistant variant of A2780 cells), human cervical carcinoma HeLa cells, human breast cancer MCF-7 cells and human colorectal carcinoma cells HCT-116 (for experiments performed at the Czech Academy of Sciences) were kindly supplied by Professor B. Keppler, University of Vienna (Austria). Human colon carcinoma cells expressing p53 (HCT116 p53+/+) were a kind gift of Dr M. Brazdova, Institute of Biophysics, Brno (Czech Republic). Highly invasive breast carcinoma MDA-MB-231 cells and human MRC-5 pd30 cells derived from normal lung tissue were purchased from the European collection of authenticated cell cultures (ECACC) (Salisbury, UK). Isogenic clones of p53+/+ and p53−/− HCT116 colon carcinoma cells for experiments performed at the University of Huddersfield were a kind gift from Bert Vogelstein (Johns Hopkins University, Maryland, USA). ARPE19 and WI38 cells were both purchased from ATCC (American Type Culture Collection) and HMF cells were purchased from ScienCell Research Laboratories, Inc.Cells were incubated in 96-well plates in complete cell medium, containing DMEM supplemented with 10% fetal calf serum and L-glutamine (2 mM). Plates were incubated for 24 h at 37 °C in an atmosphere of 5% CO2, before drug exposure, then incubated for 96 h with drug. A volume of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg ml−1) was added to each well and incubated for a further 4 h. These solutions were removed, dimethyl sulfoxide was added to each well, and the absorbance at 540 nm was read using a Thermo Scientific Multiskan EX microplate photometer. The IC50 values were determined from a plot of percentage cell survival against drug concentration (in μM). All assays were conducted in triplicate and the mean IC50 ± standard deviation was determined.
Cell cycle assay
PBS (300 μL) containing propidium iodide (50 μg ml−1) and RNAse A (80 μg ml−1) was added to drug-treated cells before incubation for 30 min and FACS analysis. The assay was repeated four times with each compound and the mean percentage of cells in each phase ± standard deviation was determined. Red fluorescence was observed at 488 nm excitation by flow cytometry and data were analysed using Flowjo V10.Time-dependent cellular response profiling
The impedance monitoring of cell growth was performed using an xCELLigence RTCA SP Real-time cell analyser. Cells were added and grown for 22–28 h, before tested compounds were added to the medium at varying concentrations. The impedance was measured for an additional 80 h.Rubidium-based assay
Cells were seeded in 6-well plates and incubated for 24 h at 37 °C in an atmosphere of 5% CO2. Cells were then incubated with drug for 6 h, and subsequently the medium was removed and cells were washed with PBS. Cells were then incubated with RbCl (5.4 mM) for 3 h, washed and counted. Rubidium content was determined by ICP-MS.Wound healing assay
Cells were seeded in 12-well plates and incubated in complete medium (10% FBS/DMEM/gentamycin) at 37 °C in an atmosphere of 5% CO2. The medium was removed and the bottom of the well was scratched with a 10 μL pipette tip (550–650 μm gaps). Wells were washed to remove detached cells before drug was added, dissolved in complete or starving medium, and plates were incubated for 24 h. Images were taken at several time intervals post scratching, and automated analysis was performed using TScratch software (MATLAB).Inhibition of colonosphere formation
Cells were treated with drug (IC30 concentration) for 72 h, washed, harvested with StemPro Accutase, and plated in ultra-low attachment 96-well culture plates (300 cells per well). Cells were cultured in for 6 days without disturbing the plates or replenishing the medium and the number/size of spheres were determined using an inverted microscope.Clonogenic assays
Cells were seeded in ultra-low attachment 6-well plates and cultured for 4 days, to allow pre-spheroid formation. Cells were treated with drug for 48 h, and dissociated into single cell suspensions using StemPro Accutase. Single cells were seeded in normal 6-well plates (3000 cells per well) and cultured for 8 days. Methylene blue solution (1% in water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol 1
methanol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added for 30 min to stain the formed colonies, excess was washed out, and colonies containing >50 cells were counted.
1) was added for 30 min to stain the formed colonies, excess was washed out, and colonies containing >50 cells were counted.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
PS thanks the China Scholarships Council and the University of Warwick for a PhD Scholarship, and Warwick's Institute of Advanced Study for an Early Career Fellowship (HS). VB, HK, LM and JZ thank the Czech Science Foundation and Ministry of Education of the Czech Republic for grants 17-05302S and LTC17003, respectively and to Dr K. Soucek for enabling them to use RTCA equipment (xCELLigence RTCA SP, Roche).Notes and references
- J. M. Lehn, A. Rigault, J. Siegel, J. Harrowfield, B. Chevrier and D. Moras, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 2565–2569 CrossRef CAS PubMed.
- C. Piguet, G. Bernardinelli and G. Hopfgartner, Chem. Rev., 1997, 97, 2005–2062 CrossRef CAS PubMed.
- C. Piguet, M. Borkovec, J. Hamacek and K. Zeckert, Coord. Chem. Rev., 2005, 249, 705–726 CrossRef CAS.
- M. Albrecht, Chem. Rev., 2001, 101, 3457–3498 CrossRef CAS PubMed.
- R. E. Hancock and R. Lehrer, Trends Biotechnol., 1998, 16, 82–88 CrossRef CAS PubMed.
- K. L. Brown and R. E. Hancock, Curr. Opin. Immunol., 2006, 18, 24–30 CrossRef CAS PubMed.
- F. Schweizer, Eur. J. Pharmacol., 2009, 625, 190–194 CrossRef CAS PubMed.
- G. Maayan and M. Albrecht, Metallofoldamers: Supramolecular Architectures from Helicates to Biomimetics, John Wiley & Sons, Ltd, 2013 Search PubMed.
- A. D. Faulkner, R. A. Kaner, Q. M. A. Abdallah, G. Clarkson, D. J. Fox, P. Gurnani, S. E. Howson, R. M. Phillips, D. I. Roper, D. H. Simpson and P. Scott, Nat. Chem., 2014, 6, 797–803 CrossRef CAS PubMed.
- S. E. Howson, A. Bolhuis, V. Brabec, G. J. Clarkson, J. Malina, A. Rodger and P. Scott, Nat. Chem., 2012, 4, 31–36 CrossRef CAS PubMed.
- R. A. Kaner, S. J. Allison, A. D. Faulkner, R. M. Phillips, D. I. Roper, S. L. Shepherd, D. H. Simpson, N. R. Waterfield and P. Scott, Chem. Sci., 2016, 7, 951–958 RSC.
- A. Zhao, S. E. Howson, C. Zhao, J. Ren, P. Scott, C. Wang and X. Qu, Nucleic Acids Res., 2017, 45, 5026–5035 CrossRef CAS PubMed.
- V. Brabec, S. E. Howson, R. A. Kaner, R. M. Lord, J. Malina, R. M. Phillips, Q. M. A. Abdallah, P. C. McGowan, A. Rodger and P. Scott, Chem. Sci., 2013, 4, 4407–4416 RSC.
- M. Li, S. E. Howson, K. Dong, N. Gao, J. Ren, P. Scott and X. Qu, J. Am. Chem. Soc., 2014, 136, 11655–11663 CrossRef CAS PubMed.
- J. Malina, P. Scott and V. Brabec, Nucleic Acids Res., 2015, 43, 5297–5306 CrossRef CAS PubMed.
- J. Malina, P. Scott and V. Brabec, Dalton Trans., 2015, 44, 14656–14665 RSC.
- D. E. Mitchell, G. Clarkson, D. J. Fox, R. A. Vipond, P. Scott and M. I. Gibson, J. Am. Chem. Soc., 2017, 139, 9835–9838 CrossRef CAS PubMed.
- R. A. Kaner and P. Scott, Future Med. Chem., 2015, 7, 1–4 CrossRef CAS PubMed.
- M. Meldal and C. W. Tornoe, Chem. Rev., 2008, 108, 2952–3015 CrossRef CAS PubMed.
- D. Gonzalez Cabrera, B. D. Koivisto and D. A. Leigh, Chem. Commun., 2007, 4218–4220 RSC.
- Z. Ma, S. Han, R. Hopson, Y. Wei and B. Moulton, Inorg. Chim. Acta, 2012, 388, 135–139 CrossRef CAS.
- S. E. Howson, G. J. Clarkson, A. D. Faulkner, R. A. Kaner, M. J. Whitmore and P. Scott, Dalton Trans., 2013, 42, 14967–14981 RSC.
- F. Xie, K. Sivakumar, Q. Zeng, M. A. Bruckman, B. Hodges and Q. Wang, Tetrahedron, 2008, 64, 2906–2914 CrossRef CAS.
- B. H. Lipshutz and B. R. Taft, Angew. Chem., Int. Ed., 2006, 45, 8235–8238 CrossRef CAS PubMed.
- L. Ackermann, H. K. Potukuchi, D. Landsberg and R. Vicente, Org. Lett., 2008, 10, 3081–3084 CrossRef CAS PubMed.
- K. Dunn, A. Aotaki-Keen, F. Putkey and L. Hjelmeland, Exp. Eye Res., 1996, 62, 155–170 CrossRef CAS PubMed.
- J. A. Hickman, Cancer Metastasis Rev., 1992, 11, 121–139 CrossRef CAS PubMed.
- C. M. Pfeffer and A. T. Singh, Int. J. Mol. Sci., 2018, 19, 448 CrossRef PubMed.
- S. Elmore, Toxicol. Pathol., 2007, 35, 495–516 CrossRef CAS PubMed.
- G. Koopman, C. Reutelingsperger, G. Kuijten, R. Keehnen, S. Pals and M. Van Oers, Blood, 1994, 84, 1415–1420 CAS.
- Y. A. Abassi, B. Xi, W. Zhang, P. Ye, S. L. Kirstein, M. R. Gaylord, S. C. Feinstein, X. Wang and X. Xu, Chem. Biol., 2009, 16, 712–723 CrossRef CAS PubMed.
- V. Novohradsky, J. Yellol, O. Stuchlikova, M. D. Santana, H. Kostrhunova, G. Yellol, J. Kasparkova, D. Bautista, J. Ruiz and V. Brabec, Chem.–Eur. J., 2017, 23, 15294–15299 CrossRef CAS PubMed.
- B. Liskova, L. Zerzankova, O. Novakova, H. Kostrhunova, Z. Travnicek and V. Brabec, Chem. Res. Toxicol., 2012, 25, 500–509 Search PubMed.
- M. V. Clausen, F. Hilbers and H. Poulsen, Front. Physiol., 2017, 8, 371 Search PubMed.
- D. S. Gill, R. Gill, D. Wicks, S. Despotovski and D. Liang, Assay Drug Dev. Technol., 2004, 2, 535–542 CrossRef PubMed.
- G. Lees, A. Lehmann, M. Sandberg and A. Hamberger, Neurosci. Lett., 1990, 120, 159–162 CrossRef CAS PubMed.
- R. Siegel, C. DeSantis, K. Virgo, K. Stein, A. Mariotto, T. Smith, D. Cooper, T. Gansler, C. Lerro and S. Fedewa, Ca-Cancer J. Clin., 2012, 62, 220–241 CrossRef PubMed.
- A. M. Maffione, E. Lopci, C. Bluemel, F. Giammarile, K. Herrmann and D. Rubello, Eur. J. Nucl. Med. Mol. Imaging, 2015, 42, 152–163 CrossRef CAS PubMed.
- S. Salah, F. Ardissone, M. Gonzalez, P. Gervaz, M. Riquet, K. Watanabe, J. Zabaleta, D. Al-Rimawi, S. Toubasi, E. Massad, E. Lisi and O. H. Hamed, Ann. Surg. Oncol., 2015, 22, 1844–1850 CrossRef PubMed.
- N. Liu, Y. Li, S. Su, N. Wang, H. Wang and J. Li, Oncol. Lett., 2013, 6, 475–479 CrossRef PubMed.
- Y. Xiao, C. Meng, J. Lin, C. Huang, X. Zhang, Y. Long, Y. Huang and Y. Lin, Oncol. Lett., 2017, 14, 6678–6684 Search PubMed.
- V. Pongrakhananon, P. Chunhacha and P. Chanvorachote, PLoS One, 2013, 8, e68623 CrossRef CAS PubMed.
- M. A. Khajah, P. M. Mathew and Y. A. Luqmani, PLoS One, 2018, 13, e0193779 CrossRef PubMed.
- N. Riggi, M. Aguet and I. Stamenkovic, Annu. Rev. Pathol.: Mech. Dis., 2018, 13, 117–140 CrossRef CAS PubMed.
- S. Valastyan and R. A. Weinberg, Cell, 2011, 147, 275–292 CrossRef CAS PubMed.
- V. Kaushik, N. Azad, J. S. Yakisich and A. K. V. Iyer, Cell Death Discovery, 2017, 3, 17009 CrossRef PubMed.
- A. Wells, J. Grahovac, S. Wheeler, B. Ma and D. Lauffenburger, Trends Pharmacol. Sci., 2013, 34, 283–289 CrossRef CAS PubMed.
- C. L. Buchheit, K. J. Weigel and Z. T. Schafer, Nat. Rev. Cancer, 2014, 14, 632 CrossRef CAS PubMed.
- G. Kirfel, A. Rigort, B. Borm and V. Herzog, Eur. J. Cell Biol., 2004, 83, 717–724 CrossRef PubMed.
- N. Makrilia, A. Kollias, L. Manolopoulos and K. Syrigos, Cancer Invest., 2009, 27, 1023–1037 CrossRef CAS PubMed.
- V. Novohradský, A. Bergamo, M. Cocchietto, J. Zajac, V. Brabec, G. Mestroni and G. Sava, Dalton Trans., 2015, 44, 1905–1913 RSC.
- J. Pracharova, V. Novohradsky, H. Kostrhunova, P. Štarha, Z. Trávníček, J. Kasparkova and V. Brabec, Dalton Trans., 2018, 47, 12197–12208 RSC.
- A. Bergamo, A. Masi, M. A. Jakupec, B. K. Keppler and G. Sava, Met.-Based Drugs, 2009, 2009, 1–9 CrossRef PubMed.
- F. P. Intini, J. Zajac, V. Novohradsky, T. Saltarella, C. Pacifico, V. Brabec, G. Natile and J. Kasparkova, Inorg. Chem., 2017, 56, 1483–1497 CrossRef CAS PubMed.
- J. E. Jonkman, J. A. Cathcart, F. Xu, M. E. Bartolini, J. E. Amon, K. M. Stevens and P. Colarusso, Cell Adhes. Migr., 2014, 8, 440–451 CrossRef PubMed.
- J. Marx, Science, 2007, 317, 1029–1031 CrossRef CAS PubMed.
- P. B. Gupta, C. L. Chaffer and R. A. Weinberg, Nat. Med., 2009, 15, 1010 CrossRef CAS PubMed.
- S. P. McDermott and M. S. Wicha, Mol. Oncol., 2010, 4, 404–419 CrossRef CAS PubMed.
- Z. Haixia, W. Haotong, Z. Junheng, Y. Pei, X. Lixiao, J. Pan, G. Jin, W. Hua and Z. Yan, Stem Cells, 2013, 31, 433–446 CrossRef PubMed.
- J. Kaiser, Science, 2015, 347, 226–229 CrossRef CAS PubMed.
- J. Chen, X. Cao, Q. An, Y. Zhang, K. Li, W. Yao, F. Shi, Y. Pan, Q. Jia, W. Zhou, F. Yang, F. Wei, N. Wang and B. Yu, Nat. Commun., 2018, 9, 1406 CrossRef PubMed.
- J.-H. Park, S. Chung, Y. Matsuo and Y. Nakamura, MedChemComm, 2017, 8, 73–80 RSC.
- M. Dean, T. Fojo and S. Bates, Nat. Rev. Cancer, 2005, 5, 275–284 CrossRef CAS PubMed.
- S.-s. Zhang, Z.-p. Han, Y.-y. Jing, S.-f. Tao, T.-j. Li, H. Wang, Y. Wang, R. Li, Y. Yang and X. Zhao, BMC Med., 2012, 10, 85 CrossRef PubMed.
- D. Thomas, P. S. Thiagarajan, V. Rai, O. Reizes, J. Lathia and T. Egelhoff, Oncotarget, 2016, 7, 47586 CrossRef PubMed.
- E. Batlle and H. Clevers, Nat. Med., 2017, 23, 1124 CrossRef CAS PubMed.
- H. Qin, C. Zhao, Y. Sun, J. Ren and X. Qu, J. Am. Chem. Soc., 2017, 139, 16201–16209 CrossRef CAS PubMed.
- X. Chen, H. Guan, X. Liu, D. Xie, Y. Wang, T. Ma, B. Huang and P. Zhou, Oncol. Lett., 2018, 16, 431–438 Search PubMed.
- T. M. Yeung, S. C. Gandhi, J. L. Wilding, R. Muschel and W. F. Bodmer, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 3722–3727 CrossRef CAS PubMed.
- S. S. Kanwar, Y. Yu, J. Nautiyal, B. B. Patel and A. P. N. Majumdar, Mol. Cancer, 2010, 9, 212 CrossRef PubMed.
- E. Chung, I. Oh and K. Y. Lee, Ann. Surg. Treat. Res., 2016, 90, 183–193 CrossRef CAS PubMed.
- S. Shaheen, M. Ahmed, F. Lorenzi and A. S. Nateri, Stem Cell Rev. Rep., 2016, 12, 492–499 CrossRef CAS PubMed.
- C. Naujokat and R. Steinhart, J. Biomed. Biotechnol., 2012, 2012, 1–17 CrossRef PubMed.
- S. Zhou, F. F. Wang, E. T. Wong, E. Fonkem, T. C. Hsieh, J. M. Wu and E. X. Wu, Curr. Med. Chem., 2013, 20, 4095–4101 CrossRef CAS PubMed.
- P. B. Gupta, T. T. Onder, G. Jiang, K. Tao, C. Kuperwasser, R. A. Weinberg and E. S. Lander, Cell, 2009, 138, 645–659 CrossRef CAS PubMed.
- F. Wang, S. Zhou, D. Qi, S.-H. Xiang, E. T. Wong, X. Wang, E. Fonkem, T.-c. Hsieh, J. Yang, B. Kirmani, J. B. Shabb, J. M. Wu, M. Wu, J. H. Huang, W.-H. Yu and E. Wu, J. Am. Chem. Soc., 2019, 141, 3613–3622 CrossRef CAS PubMed.
- V. Rajendran and M. V. Jain, in Cancer Stem Cells. Methods in Molecular Biology, ed. G. Papaccio and V. Desiderio, Humana Press, New York, NY, 2018, vol. 1692 Search PubMed.
- K. Hientz, A. Mohr, D. Bhakta-Guha and T. Efferth, Oncotarget, 2016, 8, 8921–8946 Search PubMed.
- C. Sinthuvanich, A. S. Veiga, K. Gupta, D. Gaspar, R. Blumenthal and J. P. Schneider, J. Am. Chem. Soc., 2012, 134, 6210–6217 CrossRef CAS PubMed.
- D. Gaspar, A. S. Veiga, C. Sinthuvanich, J. P. Schneider and M. A. R. B. Castanho, Biochemistry, 2012, 51, 6263–6265 CrossRef CAS PubMed.
- T. Iwasaki, J. Ishibashi, H. Tanaka, M. Sato, A. Asaoka, D. Taylor and M. Yamakawa, Peptides, 2009, 30, 660–668 CrossRef CAS PubMed.
- H. S. Lee, C. B. Park, J. M. Kim, S. A. Jang, I. Y. Park, M. S. Kim, J. H. Cho and S. C. Kim, Cancer Lett., 2008, 271, 47–55 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Data created during this study are openly available from the University of Warwick Research Archive Portal (WRAP) (http://wrap.warwick.ac.uk). See DOI: 10.1039/c9sc02651g |
| This journal is © The Royal Society of Chemistry 2019 |