Open Access Article
Open Access ArticleApplication of cold-adapted microbial agents in soil contaminate remediation: biodegradation mechanisms, case studies, and safety assessments
Jiaxin Lia,
Yujuan Wen *abc,
Zheng Fanga,
Wenqi Yanga and
Xiaoming Songa
*abc,
Zheng Fanga,
Wenqi Yanga and
Xiaoming Songa
aKey Laboratory of Regional Environment and Eco-restoration, Ministry of Education, Shenyang University, Shenyang 110044, China. E-mail: yujuanwen@syu.edu.cn; Tel: +86(24)62269636
bNortheast Geological S&T Innovation Center of China Geological Survey, Shenyang University, Shenyang 110044, China
cKey Laboratory of Black Soil Evolution and Ecological Effect, Ministry of Natural Resources, China
First published on 19th April 2024
Abstract
The microbial agent technology has made significant progress in remediating nitro-aromatic compounds (NACs), such as p-nitrophenol, 2,4-dinitrophenol, and 2,4,6-Trinitrotoluene, in farmland soil over the past decade. However, there are still gaps in our understanding of the bioavailability and degradation mechanisms of these compounds in low-temperature environments. In this review, we provide a comprehensive summary of the strategies employed by cold-adapted microorganisms and elucidate the degradation pathways of NACs pollutants. To further analyze their metabolic mechanisms, we propose using mass balance to improve our understanding of biochemical processes and refine the degradation pathways through stoichiometry analysis. Additionally, we suggest employing 13C-metabolic flux analysis to track enzyme activity and intermediate products during bio-degradation processes with the aim of accelerating the remediation of nitro-aromatic compounds, particularly in cold regions. Through a comprehensive analysis of pollutant metabolic activities and a commitment to the ‘One Health’ approach, with an emphasis on selecting non-pathogenic strains, the environmental management strategies for soil remediation could be positioned to develop and implement safe and effective measure.
Introduction
Soil environmental pollution, including heavy metals and organic contaminants, results from a combination of sources such as residual contaminants from industrial mining and textile manufacturing, plant relocation, excessive use of pesticides, and the discharge of domestic wastewater. These sources irreversibly damage the soil and inevitably affect human health. Microorganisms play a crucial role in global biogeochemical cycling by facilitating carbon fixation, nitrogen fixation, and sulfur metabolism. The microbial degradation process can be achieved either through the direct degradation of chemicals or by converting them into intermediates that are less toxic or safer. In addition, microorganisms harbor a wide range of metabolic enzymes that can be effectively utilized for the safe removal of contaminants. The soundness of biotechnology should first be demonstrated through the examination of organisms, biochemical processes, and specific chemical reactions, in order to lay the foundation for conceptualizing the technology and conducting prototype trials or scale-up experiments.1 Despite the progress, many studies fail to adequately elucidate the complex chemistry underlying microbial mechanisms, often relying solely on concentration differences between inoculated and non-inoculated samples. This leads to a significant gap in information regarding the metabolic activity supporting bioremediation claims.2 Consequently, a systematic investigation into the biodegradation process and mechanisms is of paramount importance.The utilization of microbial agents has emerged as a prominent area of investigation owing to the ubiquitous distribution of soil contamination. However, the predominant of strains selected in practical applications remain mesophilic characteristics. As one of the restricted factors in contaminant bio-remediation, if the ambient temperature is lower than the minimum growth temperature required for microorganisms, microbial dormancy ensues.3 A decreasing trend can be clearly observed in the number of dominant populations when screening microorganisms in low-temperature areas. It was found that a 10 °C decline in temperature reduced microbial activity by 2- to 3-fold, a change in the microbial degradation pattern occurred once the temperature decreased to extreme values (0 °C), and the psychrosensitive organisms resulted in microbial diversity loss.4 Strong negative changes in the state of water in low-temperature areas extensively inhibit the biochemical reactions of microorganisms, damage the cell membrane,5 alter the enzymatic reactions, and inhibit the protein synthesis, RNA transcription, and translation functions of mesophilic bacteria.6 According to the disparity between the optimal growth temperature and the upper threshold for growth, cold-active bacteria can be classified into psychrophiles and psychrotrophs.7 Psychrophiles can tolerate temperatures from 0 °C to 20 °C, and their optimum temperature is about 15 °C or even lower. Psychrotrophs can grow and reproduce at 0–5 °C, and the maximum growth temperature is higher than 20 °C.7 The distribution and survival numbers of psychrotrophs in cold environments are more than psychrophiles, but psychrophiles are more resistant to the conditions found in the polar regions. The region of Northern China has an extremely long winter with an average temperature of −2 to 4 °C, reaching as low as −20 °C in the northeast. The freezing of the surface of rivers and lakes, the withering of plants, and the freezing of the soil surface in winter significantly impact the activity of mesophilic bacteria. The adaptation of cold-adapted microorganisms in cold regions is ascribed to the evolutionary development of diverse physiological mechanisms in sub-zero environments over an extended period, which is believed to hold significant potential for extensive application prospects. At present, the application of microbial agents is widely, especially compound microbial agents, and the majority of them are thermophilic bacteria, in the aspects of regulate the retting process of flax,8 aflatoxin control and promotion of nodule nitrogen fixation,9 improved the quality of compost, accelerate composting process,10,11 in situ bioremediation of oil-contaminated soil, etc. However, there still exists a significant knowledge gap in the growth mechanisms, biodegradation mechanisms, applications, and safety assessments of cold-adapted microorganisms. Hence, the biochemical process and mechanisms shall be focused as one of the main focal points for the advancement of scientific information. Relevant assessment of mass balance and stoichiometry are critically important to support and enhance the results of the biochemical processes.
Nitro-aromatic compounds (including mono-, di- and tri-nitro-aromatic compounds) have been assigned a hazard rating (HR = 3), indicating the highly hazardous and toxicity. These compounds produced by incomplete combustion of fossil fuel or nitration reactions and are used as chemical feedstock for synthesis of explosives, pesticides, herbicides, dyes and pharmaceuticals, exhibiting considerable toxicity to human beings, fish, algae and microorganisms.12 As the intermediate of pesticides and herbicides, p-nitrophenol (PNP) plays an important role in farming. In addition, PNP was also widely used as dyes and pharmaceutical intermediates, leather preservatives, etc.13 It has already been included in the priority pollutants list and defined as a type of endocrine-disrupting chemical (EDC).14 Irrigation with wastewater containing PNP may cause crop mortality15 and poses potential health risks in animal and plant organ damage. Currently, the PNP has been detected originated from the chemical plants, printing, dyeing factories around the rivers16–18 and atmospheric fine particulate matter from coal combustion, firecracker discharge, motor vehicle exhaust, and industrial emissions.19–21 The compound 2,4-dinitrophenol (2,4-DNP) is a well-known uncoupler of respiration and oxidative phosphorylation, which are highly toxic as Priority Pollutants in the U.S. Environmental Protection Agency.22 TNT red water contaminated soil contained mainly dinitrotoluene sulfonates (DNTS) have highly water solubility and tend to migrate.23 However, the metabolic mechanism of nitro-aromatic compounds lacks a comprehensive overview. In this article, the metabolic mechanism of nitro-aromatic compounds at low temperature was elucidated using p-nitrophenol (PNP), 2,4-dinitrophenol (2,4-DNT), and 2,4,6-trinitrotoluene (TNT) as a representative compound.
This comprehensive review explores the application of microbial agent technology in the remediation of contaminated soil under cold temperature. It summarizes the degradation mechanism of low-temperature-resistant microorganisms and focuses on the biochemical process of biodegradation, utilizing the mass balance models and metabolic flux analysis cases. This provides extensive information for the present biotechnology of metabolic activity, supporting the efficient and safety development. Meanwhile, the assessment of the safety of microbial agents is purposed as the reference of bioremediation in low temperatures, providing a sustainable solution for efficiently treating soil environmental pollution.
Microorganisms cold-adapted mechanism
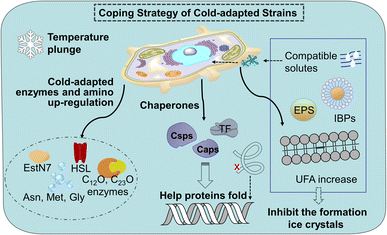
Cold stress leads to decreased fatty acid biosynthesis and metabolism and the decreased trend of membrane fluidity of mesophiles. The down-regulation of proteins involved in carbohydrate metabolism, energy production, and conversion in mesophiles will slow down the growth of microorganisms, affect the tricarboxylic acid cycle (TCA) of nitro-aromatic compounds. A sudden lowering temperature brings microorganisms cold shock reaction (CSR) and freeze can cause DNA damage, which leads to innormal transcription and translation functions. In contrast to conventional microorganisms, cold-adapted microorganisms exhibit minimal alterations in their biochemical reactions under both normothermic and low-temperature conditions. This is attributed to the enhanced adaptability of microorganisms inhabiting extreme environments towards the detrimental impacts of low temperatures on enzyme levels, protein composition and cellular membrane integrity.24 The detail of cold-adapted mechanism in strains in Fig. 1.Cold-adapted enzyme proteins showed high specificity and adjusted amino acid content. Enzymes are usually involved in the process of microbial degradation. The structure can undergo conformational changes according to the needs of catalytic activity, making up for the lack of activation energy at low temperatures.25 For example, there are phenol 2-monooxgenase, and enzymes involved in the ortho-cleavage of catechol were present in the aerobic strain AQ5-05. Cold-adapted enzymes C12O and C23O enzymes have been found in several cold-adapted phenol-degrading strains.26 It is reported that cold-adapted enzymes have ten-fold greater activity at low temperatures (below 20–30 °C) compared to their mesophilic counterparts.27 Most of the cold-adapted enzymes have high specific activity below 20–30 °C, and some enzymes such as a novel-hormone sensitive lipase (HSL) family IV cold-adapted esterase (EstN7) and the psychrophilic BpL5 (a corresponding enzyme isolated from the arctic bacterium B. pumilus) can retain the maximum activity in 5 °C.28 Meanwhile, differentially expressed genes (DEGs) in cold-adapted strains would be triggered at low temperatures. A research shows a novel cold-adapted HNAD-capable bacterium, Pseudomonas indoloxydans YY-1, for biological nitrogen removal (BNR) from wastewater at cold temperatures.29 The result showed a dramatical increase in cellular ATP levels as temperatures dropped to near-freezing, some key genes related to TCA cycle, amino acid metabolism, and cold-shock PN gene, were upregulated at 10 °C, thereby providing the essential extra energy requirement and increasing the cellular translation efficiency of strain YY-1 for cold adaptation. Currently, research on the genes responsible for the low-temperature degradation of specific pollutants remains limited, thus, further investigation is imperative. In addition, arginine, lysine, and proline residues form fewer salt bridges and fewer hydrogen bonds, while the glycine residue clusters basically have no side chains, which can provide the location of chain migration. Therefore, in terms of amino acid content, the contents of arginine, proline, and lysine decrease, while the contents of asparagine, methionine and glycine increase.30
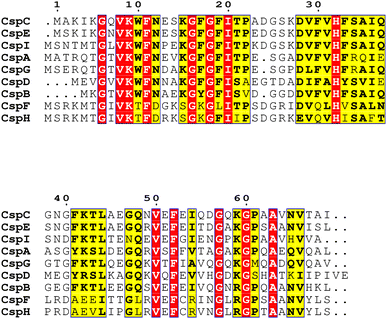
Chaperones play a role in the cell in assisting proteins to fold properly, helping them reach the correct conformation. When the temperature drops sharply, the microbial cold shock response (CSR) causes cell growth to stop for 3–6 hours, at which time most protein synthesis is blocked.31 In this period, the short-term response cold shock proteins (Csps) and long-term adapted cold acclimation proteins (Caps) at low temperatures were synthesized in large amounts.6 Caps can only be expressed by cold-loving bacteria growing at low temperatures and are not produced in milder environments. The synthesis rate of CAPs was faster than that of Csps at low temperatures. Previous studies have found that the presence of Csp is the main manifestation of the difference between cold-adapted microorganisms and mesothermal bacteria, and the rapid drop in temperature does not affect the maintenance of Csps levels. The Csp family includes 9 species from CspA to CspI.32 A conserved region in the Csp family has been found in the National Center for Biotechnology Information with obvious sequence homology, which is considered to be the target region for the degradation of cold-active microorganisms at low temperatures (Fig. 2). In the Csp family, CspD is used in nutrient deprivation and is extremely active when carbon sources are lacking in the environment. CspA, CspB, and CspG are classified as Csp Class I proteins, which are highly expressed after extreme cooling and are used to combat the cold shock response.33 CspA, CspB, CspC, and CspG are chaperones of RNA and DNA, which can help protein folding, prevent the formation of secondary structures, make cold-active microorganisms have a low protein translation error rate under low temperature conditions, and promote the effective translation and transcription function of mRNA.34 In addition to Csps, trigger factors (TF) are a specific chaperones that co-translate with almost all nascent peptides, helping proteins fold correctly during the early stages.35
At the cellular level, the membrane lipid conditions of cold-adapted microbes change, showing the ratio of branched fatty acids (BCFA) and unsaturated fatty acids (UFA) increased, the branched fatty acids grew and the main chain shortened, Unsaturated fatty acids have lower melting point, makes it easier for cell membranes to maintain fluidity and softness at low temperatures.36 Compatible solutes such as glycine, betaine, sucrose and mannitol accumulate as cryoprotectants in the cryophilic bacteria, preventing the production of cytoplasmic ice crystals and improving the survival rate of cells after thawing.37 With the decrease in temperature, cells produce a high level of extracellular polymeric substances exopolysaccharides (EPS), especially the secretion of extracellular proteins, preventing cell osmotic pressure damage and ice recrystallization.38 Meanwhile, ice binding proteins (IBPs) such as antifreeze proteins (AFPs) and ice nuclear proteins (INPs) can inhibit the growth of ice crystals. There are multiple ice binding sites on the surface of AFPs to adsorb ice crystals, inhibit their recrystallization, reduce the freezing point of the solution, delay freezing or reduce the damage caused by freezing and thawing, and help cells survive at low temperatures, where INPs prevent the fast cooling of water by inducing ice crystallization at high subzero temperatures.39 Compared to mesophiles, the coding of these cold-adapted genes ensures that cells function without slowing down the rate of chemical reactions at low temperatures. The enzymes and diversified proteins of the cold-adapted strain produce a pathway for the specific degradation of nitro-aromatic compounds. This mechanism increases the degradation ability at low temperatures and converts them into non-toxic metabolites. This cold-adapted ability of microorganism provides application value for the degradation of nitro-aromatic compounds in polar regions, high mountains, and seasonal frozen soil regions.
Nitro-aromatic compounds degradation pathway
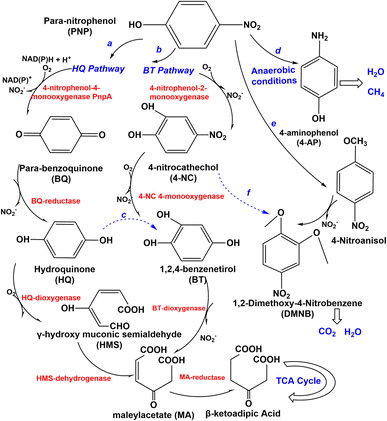
Several types of microorganisms in the environment could biodegrade nitro-aromatic compounds, including aerobic bacteria, fungi, actinomycetes, and anaerobes (Table 1), and the degradation mechanism was also different in these strains (Fig. 3 and 4). The aerobic degradation of PNP is divided into (hydroquinone) HQ pathway and 1,2,4-benzenetrio (BT) pathways. Compounds such as hydroquinone, p-nitrocatechol, or p-benzoquinone are formed before the uncomplete oxidation of p-nitrophenol to CO2.40 PNP was degraded via the hydroquinone (HQ) pathway catalyzes the hydroxylation reaction of PNP by the key enzyme is p-nitrophenol 4-monooxygenase PnpA in the initial reactions of PNP to p-benzoquinone, with the release of nitrite or nitrate.41 Then benzoquinone (BQ) converted to hydroquinone (HQ) via the BQ-reductase, which ring-opened by γ-hydroxymuconic semialdehyde (HMS) through the action of HQ-dioxygenase to form maleylacetate acid (MA), which further converted to β-ketoadipic acid by HMS-dehydrogenase, finally through the TCA cycle42 and changed in various low or non-toxic compounds. A general overview of the pathway is shown in Fig. 3. HQ pathway generally founded in Gram-negative bacteria such as Burkholderia spp. and Moraxella spp.43 The second pathway of the degradation of PNP is hydroxylated by PNP-2monooxygenase, which converts to 4-nitrocatechol (4-NC) (Fig. 3), nitro-group is releasedβ-ketoadipic acid to enter the tricarboxylic acid cycle (TCA) cycle.44 BT pathway predominates in Gram-positive bacteria and a few Gram-negative bacteria,45 such as Arthrobacter spp.46 Bacillus spp.47 and Rhodococcus spp.,48 the degradation pathway of aerobic bacteria not always follows one set pattern. It is founded that a part of Gram-negative bacteria could also degrade PNP via the 4-nitrocatechol (4-NC) pathway such as Burkholderia sp. SJ98. In addition, B. zhejiangensis CEIB S4-3 is capable of degrading both methyl parathion (MP) and PNP using both of two degradation pathways49 (Fig. 3). PNP could be degradation by the Pseudomonas aeruginosa HS-D38 strain accompanied by release of stoichiometric amount of nitrate from PNP, and hydroquinone and 1,2,4-benzenetriol were observed as the key degradation intermediates.50 Studies on the degradation of PNP by anaerobic microorganisms possible to produce carcinogenic nitroso and hydroxylamine compounds.51| Nitro-aromatic compounds | Microorganism(s) | Biodegradation pathways | |
|---|---|---|---|
| 4-Nitrophenol (PNP) | Fungi | Phanerochaete chrysosporium | Via 4-nitroanisole (4-NA) and 1,2 dimethoxy-4-nitrobenzene (DMNB), degrade to CO2 (ref. 54) |
| Fungi | Penicillium strain Bi 7/2 | Converted to 4-nitrocatechol55 | |
| Actinomycete | Arthrobacter sp. JS443 | Via 1,2,4-benzenetriol, converted to maleylacetic acid (MA), further degraded by the β-ketoadipate pathway61 | |
| Actinomycete | Nocardioides sp. NSP41 | Degraded via a hydroquinone pathway56 | |
| Bacteria | Moraxella spp. | Converted to hydroquinone with simultaneous release of nitrite73 | |
| Bacteria | Burkholderia sp. SJ98 | Via the formation of 4-nitrocatechol (4-NC) and 1,2,4-benezenetriol (BT), and further converted into 2-hydroxy-1,4-benzoquinone (HBQ) and subsequently degraded via formation of benzoquinone (BQ) and hydroquinone (HQ)44 | |
| Bacteria | Ralstonia sp. SJ98 (ref. 62) | — | |
| Bacteria | Burkholderia zhejiangensis strain CEIB S4-3 | Hydroquinone (HQ) pathway or 4-nitrocatechol (4-NC) pathway48 | |
| Bacteria | Burkholderia cenocepacia CEIB S5-2 | Hydroquinone (HQ) pathway and benezenetriol (BT) pathway63 | |
| Anaerobe | Halophilic anaerobic Eubacteria Haloanaerobium praevalens and Sporohalobacter marismortui | Converted to 4-aminophenol (4-AP), even entirely mineralized to form methane and carbon dioxide52 | |
| 2,4-Dinitrophenol (DNP) | Anaerobe | Sulphate-reducing bacteria Dedulfovibio sp. methanogenic bacteria Methanococcus strain B | First reduced to diaminophenol, and this diaminophenol reductively deaminated to phenol64 |
| Methanococcus delatae | |||
| Methanococcus thermolithotrophicus | |||
| Bacteria | Pseudomonas sp. | Release of nitrite and nitrate ions22 | |
| Burkholderia stabilis | |||
| Rhodococcus gingshengii | |||
| Bacillus cereus | |||
| Bacteria | Rhodococcus strain XM24 (ref. 65) | — | |
| Bacteria | Rhodococcus opacus | Released nitrite in the aerobic process, and anoxic processes remove the nitrite released58 | |
| Cyanobacteria | A. variabilis NIES23 | Converted to diaminophenol66 | |
| A. cylindrica NIES19 | |||
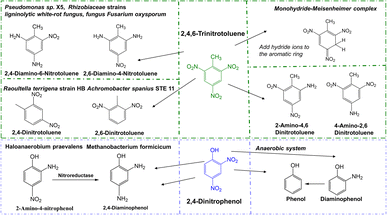
| 2,4,6-Trinitrotoluene (TNT) | Bacteria | Pseudomonas sp. X5 | Converted to 2-amino-4,6 dinitrotoluene23 |
| Bacteria | Raoultella terrigena strain HB | Aminodinitrotoluenes (2-ADNT, 4-ADNT) and diaminonitrotoluenes (2,4-DANT)60 | |
| Bacteria | Achromobacter spanius STE 11 | Formed to 2,4-dinitrotoluene, 2,6-dinitrotoluene, 2-aminodinitrotoluene (2-ADNT) and 4-aminodinitrotoluene (4-ADNT)67 | |
| Bacteria | Rhizobiaceae strains, T10, B5, and M8 | Transform TNT to hydroxylaminodinitrotoluenes (2-HADNT, 4-HADNT), aminodinitrotoluenes (2-ADNT, 4-ADNT), and diaminonitrotoluene (2,4-DANT)68 | |
| Fungi | Candida sp. AN-L15, Geotrichum sp. AN-Z4 | Formation of 2,4-dinitrotoluene and release nitrites69 | |
| Fungi | Aspergillus niger N2-2, Mucor sp. T1-1, penicillium, fusarium, trichoderma, rhizopus, botrytis, alternaria, cladosporium and trichothecium | Deeply degraded70 | |
| Fungi | Ligninolytic white-rot fungus | Main reduction products include 2-amino-4,6-dinitrotoluene (2-A-4,6-DNT), 4-amino-2,6-dinitrotoluene (4-A-2,6-DNT), 2,4-diamino-6-nitrotoluene (2,4-DA-6-NT) and 2,6-diamino-4-nitrotoluene (2,6-DA-4-NT)71,72 | |
| Fungus Fusarium oxysporum | |||
 | ||
| Fig. 4 Degradation pathway of 2,4-dinitrophenol (DNP), and 2,4,6-trinitrotoluene (TNT) in various microorganisms. | ||
PNP has been demonstrated it can be completely degraded by Haloanaerobium praevalens, Methanobacterium formicicum, and Desulfotomaculum orientis,52 and from 4-nitrocatechol as nitrite, and MA is generated by ring-opening of 1,2,4-benzenetriol (BT), then reduced to quantitatively converted to para-aminophenol (4-AP) (Fig. 3), which has lower toxicity than PNP and can sometimes be entirely mineralized to form methane and carbon dioxide.53 Furthermore, there also have some aerobic fungi and actinomycetes have been used to degrade PNP. The study of Teramoto et al., (2004) found that there are two pathways of Phanerochaete chrysosporium from white-rot basidiomycetes that were used to degradation of 4-NP.54
The conversion of 4-NP to 4-nitroanisol (4-NA) and 1,2-dimethoxy-4-nitrobenzene (DMNB) is the main pathway (Fig. 3). It is converted to DMNB and a trace amount of 4-NP is formed when 4-NA is used as an exogenous substrate. However, the possible intermediate 1-hydroxy-2-methoxy-5-nitrobenzene was not observed in the metabolism of 4-NA, so a metabolic pathway from 4-NP to DMNB with the intermediate product 4-NA could be reported. Moreover, 4-NC is probably an intermediate for DMNB formation (Fig. 3). DMNB was finally degraded to CO2 and H2O, revealing that PNP could be completely mineralized. Hofrichter et al., (1993) cultured Penicillium strain Bi 7/2 at high concentrations of phenols as a carbon source, nitrocatechols were accumulated and formed resulting from synergistically transforming 4-nitrophenols, 3-nitrophenols, and 2-nitrophenols.55 Cho et al. (2000) clearly found that the aerobic actinomycete Nocardioides NSP41 is reported to degradate phenol and PNP.56 Herman and Costerton, (1993) showed that the degradation of phenol by Actinomycete releases nitrite, which is eventually mineralized to CO2.57
The degradation microorganisms of 2,4-dinitrophenol (DNP) are prone of bacteria, which reduced to diaminophenol (2-ANP), and final reductively to phenol in anaerobe condition. It usually with the release of nitrite in the aerobic process, and further remove the nitrite released under anoxic processes.58 The degradation pathway of 2,4,6-Trinitrotoluene (TNT) is characterized by reductive reactions by microorganisms catalyse the reduction of one or two nitro-groups of TNT, forming to aminodinitrotoluenes (ANDT) and diaminonitrotoluenes (DANT). And the main metabolites and the main degradation pathway are prone to 2-amino-4,6-dinitrotoluene (2-ADNT) and 4-amino-2,6-dinitrotoluene (4-ADNT),59 and with the nitro-groups reduced finally may result in the 2,4,6-triaminotoluene (TAT) in anaerobe condition. Hydroxylamino-dinitrotoluenes (HADNTs) and tetranitroazoxytoluenes (AZTs), dinitrotoluene (DNT) also as the initial metabolites. These metabolites further converted by biotic and abiotic mechanisms to azo-, azoxy-, hydrazone- and phenol-acetyl derivatives. Another pathway is forming Meisenheimer-complexes (adducts of aromatic nitro-compounds with a nucleophile) by addition of one or two hydride ions to the aromatic ring, which often accompany the release of nitrite.60 The involvement of enzymes in catalyzing all degradation processes is widely acknowledged. However, no enzymes capable of degrading hydrocarbons or nitro compounds with cold-adapted characteristics have been identified.
Correlation between stoichiometry and microbial metabolism analysis
Stoichiometry is a method of obtaining more accurate biodegradation products through a detailed study of the degradation process. Based on the analysis of existing degradation pathways, we can master the metabolic pathways of nitro-aromatic compounds in more detail, understand the microbial degradation mode and enzyme activity changes in the entire metabolic cycle system, especially the metabolic pathway of cold-adapted microbial soil remediation, which has not been studied in such detail. In this section, mass balance (MB) and metabolic flux analysis (MFA) has been proppsal, which are used for data analysis of metabolic processes and detailed calculations at the enzyme level, respectively. Previous studies on mass balance have been widely used in wastewater treatment, and MFA mainly plays a role in the design, construction and optimization of metabolic pathways and the redirection of intracellular flow. We believe that it can be added to the soil remediation of cold-adapted microorganisms to solve the problems of low enzyme activity and high toxicity of intermediate products in cold areas, control the metabolic process of pollutants, and thus improve the efficiency of bioremediation. At the same time, when these methods were applied, microbial remediation techniques obtained more convincing data on degradation pathways.The principles of mass balance are crucial in the context of bioremediation for soil pollution. Based on the study of degradation pathways, mass metabolic balance can effectively identify the specific pollutant metabolism process. Different methods used to calculate the degradation rate in previous studies, leading to errors in the degradation rate results data and negatively affecting the readers. Failure to consider recovery factors can also lead to inaccuracies. Mass balance has been raised to improve the understandings of fate and transport of pollution, assess the potential risks posed to surface water and soil quality74 and responsible biochemical processes. The basic fundamental of mass balance are as follows:
| Xin − Xout = XAccum/rem |
| C6H5NO3 + aO2 → bCαHβOγN + cH2O + dCO2 |
where X and X0 (mg L−1, dry cell weight) represent the biomass concentration at the end and beginning of reaction (b), respectively, and S and S0 (mg L−1) represent the final and initial substrate concentration, respectively. From these equations, mass balance on each element and biomass yield from PNP as single substrate were obtained, and the result of biomass yield indicated the degradation effectiveness.
Metabolic flux analysis (MFA) has emerged as a potent tool for elucidating the activities of metabolic pathways, which constitute crucial determinants of cellular physiology in the field of metabolic engineering. Stable-isotope tracing techniques based on 13C-tracers are now one of the most accurate and widely used techniques (13C-metabolic flux analysis, 13C-MFA). It has been used to label the substrate to effectively improve the accuracy of metabolic flux assessment, to conduct a comprehensive and detailed analysis for study the full details of microbial metabolism.77 Based on the concentration data of substrate, metabolites, and the relative ratio of 13C, a network model of the transformation path and metabolic reaction of the substrate is constructed. Mathematical calculation is carried out to deeply understand the key metabolic reactions and their existing bottlenecks, which contribute to facilitating the optimization of microbial degradation of the substrate on the level of enzyme or gene, changing the metabolic flux distribution, and regulating the required reaction types, improve the ability of microbial degradation.78,79 In addition, 13C-MFA is also used to analyze measure fluxes and cross-feeding interactions in microbial consortia to better understand the microbial interactions,80 which means we can further enrich and verify the enzyme and intermediate products of the degradation of nitro-aromatic compounds under relevant microbial. Take together, mass balance and MFA technology could enhance the biochemical processes. The introduction of cold-adapted microorganisms combining into the contaminated sites is still in its infancy compared to the use of mesophilic microorganisms, especially under the FTCs condition still a number of limitations because of the temperature, effect of moisture content on soil structure, pollutant characteristics, poor capabilities of microbial communities in the field, lesser bioavailability of pollutants, growth conditions. Thus, understanding the metabolic mechanisms involved in the biodegradation process by cold-adapted organisms is a prerequisite for employing the inherent property of cold-adapted organisms to improve biodegradation at low temperatures. It should be considered to applicated in the cold-adapted microbial agents of nitro-aromatic compounds, clarify the metabolic mechanism and energy utilization in the degradation process to ensure the integrity of biodegradation.81
Progress in the research of microbial consortia and application restoration case
Microbial consortia
The efficiency and environmentally friendly properties of microbial agents have been widely recognized by researchers in the treatment of contaminated soil, water environments, and ecological agriculture. The change of bacterial abundance caused by the use of microbial agents and varying temperatures may be the internal mechanism of pollutant degradation, which lays the foundation for providing a healthier microbial community in polluted environments.90 Generally speaking, microbial agents can be divided into single or compound bacteria, fungi, and actinomycetes. Most studies have confirmed that microbial consortia have better efficient in degrading pollutants than single microbial agents91,92 A composed microbial agent (holding up to 700 mg L−1 PNP) as the only carbon source performed better in degrading pollution than most efficient pure bacteria available, provided a powerful basis for the development of composed biodegradation.93 Jia investigated a microbial agent composed of a mixture of cellulose-degrading and cold-adapted bacteria in equal proportions that significantly increased the composting temperature and compositing efficiency as well as the duration of high temperature during composting.94 The concept of “microbiome engineering” has been proposed to enhance microbial community performance. Metabolic functions can be divided among different strains to produce the desired biochemical products by designing and utilizing synthetic microbiomes (bottom-up microbiome engineering). Over time, community engineering can play diverse biochemical roles in the decomposition of complex substrates and optimize channel division among species to perform multiple functions simultaneously. A synthetic microbiome can degrade complex pollutants and significantly reduce the metabolic burden of each member of the synthetic consortium so that higher degradation efficiency can be obtained. Compared with strains that perform all metabolic functions, the mixed consortia composed of members with different metabolic functions can simultaneously degrade multiple pollutants and greatly reduce the metabolic burden on each species.95Immobilization of microbial agents
Microorganism immobilization technology (MIT) is a bioremediation technology that uses chemical or physical methods to confine or immobilize free microbial cells on specific carriers,96 which can improve their survival rates for the continuous use of the microbial agent. Free-state microbial inoculants are likely to lose their activity due to a lack of nutrients or during the process of degrading pollutants in contaminated areas. In contrast, immobilization technology has higher operational stability and effectively improves microbial activity and cell density.97 The immobilization provided a non-toxic matrix, protective case and create microsites for better environments for the inoculums in cold regions.98 Meanwhile, pollution removal was more efficient after MIT compared to non-immobilized at low temperature.99The traditional immobilization methods including aggregation, adsorption, embedding, cross-linking, and covalent coupling.100 In these immobilization process, the properties of carrier materials have a significant influence on the effectiveness of the immobilization reaction. We summary the various adsorption carrier101 and their characteristics in Table 2. These traditional immobilization techniques are susceptible to environmental factors or their internal reactions, resulting in low immobilization efficiency, but the limitation of a single method can be overcome by combining two immobilization methods to further enhanced the efficiency of MIT.102 In a previous work a combined adsorption-embedding method compared to the free bacteria YT-11.103 The results showed that the immobilized microspheres with cinnamon shell as the carrier exhibited enhanced diesel degradation performance (69.57% degradation) compared to free bacteria YT-11 (5 days, 48.87%), indicating that the combination of cinnamon shell and strain YT-11 is beneficial to solve the problem of marine oil pollution.
| Adsorption carrier | Category | Characteristics | ||
|---|---|---|---|---|
| Organic polymer carriers | Natural organic polymer carriers | Intracellular materials | Collagen, fibrin, hyaluronic acid | Non-toxic, good mass transfer and biocompatibility, environmentally friendly, high fixed density, low cost. Poor mechanical strength, poor durability, and poor chemical stability, and are easily decomposed by microorganisms under anaerobic conditions82 |
| Extracellular materials | Chitosan, sodium alginate, agar, carrageenan, plant debris fiber, cereal grains, and others | |||
| Synthetic polymer gels | Polyacrylamid, polylactic acid, polyamido acid, polyvinyl alcohol85 | Good antibacterial properties, strong mechanical strength, good durability, high strength, and stability. Poor mass transfer performance and are potentially toxic to microorganisms83,84 | ||
| Inorganic carriers | Activated carbon, biochar, zeolite, bentonite, diatomaceous earth, silica, fly ash, quartz sand, nylon fiber, and others | Stable, resistant to corrosion and decomposition, high mechanical strength, good water absorption, great adsorption ability, mild preparation conditions, and reusable carriers; however, cells easily fall off86,87 | ||
| Composite carriers | Polyvinyl alcohol/calcium alginate, cellulose/calcium alginate/polylactic acid | Overcome the inherent limitations of a single material and have the merits of both organic polymer carrier materials and inorganic polymer materials, but high cost88 | ||
| Polymeric membrane | Hollow fiber membrane, micrometer-sized/nano membrane materials | High bacterial density for continuous operation, long preservation time and reusability, and cells can spread inside and outside the membrane89 | ||
A new immobilization technology, layer-by-layer self-assembly (LBL) microcapsule technology is emerging. A tightly structured microcapsule covered in multilayer films is formed as a result of weak intermolecular interactions, such as the electrostatic force, hydrogen bonding, ligand bonding, and cross-linking.104 Microorganisms are released to degrade environmental contamination by several environmental factors, such as pH and temperature, that trigger a change in the structure or porosity of the shell material. This method use of natural polymers such as chitosan (CHI) and sodium alginate (ALG) in particular has become a popular research topic in recent years because of their superior compatibility and environmentally friendly.105,106 It is simple to operate and implement, achieves protection of microorganisms, and greatly improves the remediation efficiency of contaminated environments. So far, there is no field application of the LBL technology for bioremediation at low temperatures, so the method is expected to be further optimized to improve the stability of microbial agents in pollutant degradation in low temperature contaminated environments.
Case study on restoration using cold-adapted microbial agents
Over the years, researchers have been trying to in nitro-aromatic compounds of biodegradation find a breakthrough in cold environment. Li et al. (2020) used Pseudomonas sp. ZL to perform bioaugmentation on PNP contaminated soil at 10 °C, and optimized the conditions pH was 8.0, 0.5% NaCl, and 1 g L−1 NH4NO3 of the strain. It was concluded that the degradation rate of PNP could reach 99.94% in 83 h when strain concentration was 225.92 mg L−1 and concentration was 100.34 mg L−1. In addition, the maximum tolerance of Pseudomonas sp. ZL to degraded PNP was 303.71 mg L−1.107 Two indigenous bacteria from the aniline and nitrobenzene contaminated site named Delftia sp. lp09 (KR673339) and Pseudomona sp. wyj10 (KR 673340), immobilized by the vinylon fibers, 85% degradation of 150 mg L−1 PNP in 40 d under 10 °C without extra cost. It demonstrated the cold-adapted capacity, suitable for biodegradation in underground environments all year round without any increase in production.108 Wang et al. (2021) showed that the degradation of 5 mM PNP by the Antarctic psychrophile Psychrobacter sp. ANT206 cultivated at 16 °C had a biodegradation rate of 92.47%, which was higher than the biodegradation rate at 20 °C (80.59%), and a shorter fermentation time (54 h), indicating that the strain had a higher PNP biodegradation rate at lower temperatures.109 In the same study, an analysis of the degradation efficiency of the in-frame deletion mutant of psntr (Δpsntr-ANT206) and Psychrobacter sp. ANT206 under 5 mM and 10 mM PNP treatment showed that the survival rate of Δpsntr-ANT206 was reduced by 68.26% and 48.37%, respectively, showing that the cold-adapted reductase PsNTR contributed to the bioreduction of nitrobenzene and PNP and was capable of degrading PNP in a low-temperature environment, which indirectly revealed that the metabolic capacity at low temperatures was enhanced due to the cold-adapted reductase PsNTR. By extension, the cold-adapt gene PsNTR was cloned in Escherichia coli for heteroexpression, and degradation of 5.0 mM nitrobenzene. It was found that the maximum bioreduction rate of the recombinant strain was 3.03 mM h−1 at 16 °C.110 The application of MIT has shows great result at low temperature. Sreenivasulu et al. (2012) PNP was degraded by 3% calcium alginate embedding, and PNP could be completely degraded by B. pantothenticus under static culture at 4 °C for 30 h.47 However, the degradation of nitro-aromatic compounds by cold-tolerant strains are biased. Conducted toxicity tests on monoaromatic phenolic related compounds using 32 cold-tolerant yeast strains isolated from alpine habitats at the Brenner pass in Austria at 10 °C. The result showed that cold-tolerant yeast such as basidiomycota spp. and psychrophilic Microbotryomycet-idea up to 80% degradability of low toxic compounds, while compounds with nitro or methyl groups in the aromatic ring (such as p-nitrotoluene and nitrobenzene) have no degradation function.111 In addition, the degradation temperature of the above nitro-aromatic compounds at low temperatures is still high. We found the strengths of cold-adapted microorganisms in the degradation of organic compounds, it is worthy of our reference on nitro-phenolic compounds. Sepehr et al. (2019) isolated a strain named Pseudomonas sp. ATR208, which is capable of using phenol as a sole source of carbon and for phenol degradation from soils of Altai Mountains in the south of Siberia.112 An extensive degradation ability at both the psychrophilic and mesophilic temperature conditions of a psychrotolerant Rahnella sp. PFF2 was capable of growth at a temperature ranging from 4 °C to 37 °C. A degrade 100% (50 μg mL−1) of profenofos (PFF), an organophosphate insecticide, within 14 days at 20 °C and After 16 days and after 20 days no detectable peak was observed in the samples incubated at 28 °C and 15 °C respectively on HPLC chromatogram, which may capable of as the dominant strain of FTCs conditions.113 N-methylated carbamates are synthetic pesticides that are widely used for the management of a variety of pests of broad spectral nature, which induced environmental concern and possible exposure to humans and other organisms. However, it is difficult to degrade at low temperatures. In Fareed et al. (2019) study, a bacterial strain from the N-methylated carbamates used for crop protection in a cold environment, identified as Pseudomonas plecoglossicida strain (TA3), which immobilized microbial agent can degrade 80% carbamates at 4 °C, up tp 20% more than the free cells, demonstrating the efficient of the immobilization technology at low temperature.114 In practice, the cold-adapted strains at lower temperatures are mainly used to remediate petroleum hydrocarbon soil pollution. Totubaeva et al. (2023) repaired the oil spill soil contamination site in extremely high-mountainous conditions of Kyrgyz Republic through biostimulation (BS) and bioaugmentation (BA).115 The results showed that the total number of bacteria increased by both two methods, and the BS treatment showed the highest total petroleum hydrocarbons biodegradation percentage 62.78% while the degradation ability of BS + BA decreased, and the total number of bacteria decreased, indicating the appropriate method depends on the specific environmental condition. Petroleum hydrocarbon as sole carbon source was degraded by a novel cold-adapted bacteria (Arthrobacter sp. S2TR-06), which could produce cold-active degradative enzymes (xylene monooxygenase was able to transform p-xylene into a p-cresol intermediate, and catechol 2, 3-dioxygenase cleaved the ring) at 4 °C.116 Furthermore, a cold-adapted consortia have been found for degradation of petroleum hydrocarbons in polar regions at 3 °C with immobilized technology. A removal efficiencies of 57∼58%, increased by a factor of 2 to 7 compared to the untreated biopile.117 Two psychrotolerant Arthrobacter spp. strains AQ5-05 and AQ5-06 degrading 41.0% and 47.5% of diesel at degrading diesel up to 3% (v/v) over 7 days at 10 °C, respectively.118 However, due to the consist of the complex mixture of diesel oil, along with heavy metals including mercury (Hg) and silver (Ag), greatly impeded the growth and reduced to 4% of the initial diesel amount in the presence of either heavy metal ion.The above removal of soil pollution of cold-adapted could significantly improve their degradation efficiency after selecting appropriate methods, but their degradation function still could not meet the requirements of complete degradation and lower temperatures. At the same time, the complex pollution in the environment increases the difficulty of low temperature biological treatment. The changes in soil environment in low temperature environment are more complex, facing the dynamic changes of soil pH, moisture, microbial community interaction, etc. Therefore, the isolated cold-tolerant bacteria still need to be further optimized to improve their efficiency and universality.
Safety assessment of cold-adapted microbial agents
As mentioned above, anaerobic degradation of PNP will produce carcinogenic nitroso and hydroxylamine compounds, and incomplete mineralization of 4-AP will still have mutagenicity and teratogenic effects. These intermediates may also be absorbed and accumulate in the plants, decrease the absorption and transport of nutrients by plants, and hinder the occurrence of photosynthesis and transpiration. The soil microbial community structure will be changed by microbial agents, influencing the organisms that depend on soil for survival, reducing the diversity of the microbial community, and inhibiting the beneficial effects of microorganisms and plants. Especially in cold sites, eliminating organic contaminants has become an enormous challenge, and raising soil temperature is highly impracticable.119 In this case, biological methods can be considered environmentally friendly alternatives for the remediation of soil contamination. However, with the development of microbial agent technology, the safety of the microbial agent in remediating soil pollution needs to be tested. In general, the assessment of the safety of microbial agents needs to consider the following three points.(1) The completeness of the effect of microbial remediation, meaning the form and toxic accumulation of metabolic intermediates when compared to the remediation of pollution with physical and chemical methods. Host human-made compounds are designed to be extremely resistant to biodegradation. For example, remediation of EDHC synthetic chemicals has limitations including high cost, low efficiency, and toxic by-products.120 Therefore, we should treat from the source first. It is important to pay attention to enrichment culture, that means, focusing on the mineralization of the chemical as a goal, instead of the intermediate products which may be more toxic.121 Meanwhile, enrichment culture needs to satisfy the threshold concentration value (between the maintenance energy and toxicity) to achieve metabolism.122 In the design of the experiment, the control group (without inoculation or sterilized inoculum) should be included to show the relevance between microorganisms and substrate concentration change. An abiotic control must be set up in parallel to the biologically active treatments so that chemical stability can be evaluated without biases from chemical transformation. As an effective remediation tool, compound microbial agents have a wide range of enzymes and multiple degradation pathways to combat different contaminant intermediates, but the co-metabolism functions should be clarified.123 Secondly, it is vital to pay more attention on the metabolic process. Mass balance and 13C-MFA are useful to better understand the ability of biodegradation and trace the fate of compounds, and control the whole process. Most reports concentrate on the total concentration, which makes no sense, we should understand the bioavailable fraction of the total concentration (effective concentration) to evaluate the ecological toxicity and risk. Similarly, there is still a gap between biodegradation and bioremediation, and a laboratory success on degradation cannot be quickly and simply treated as a claim of EB for bioremediation in application.
(2) Toxicity and pathogenicity assessment. Whether the strains contained in the microbial agents are identified as pathogenic bacteria or destroy the diversity of the soil microbial community soil should also be considered. Shen et al. (2021) studied the effects of three different microbial agents on the bacterial community of inter-root maize soil.124 The results showed that all three microbial agents were able to improve the bacterial community of maize inter-root soil by increasing potentially beneficial bacteria and reducing harmful bacteria compared with the control and could be used as soil conditioners. The removal rate of 800 mg L−1 phenol by Acinetobacter radioresistens APH1 reached 82.6% within 24 h. APH1 rapidly changed the microbial community structure, but after 20 days, APH1 lacked nutrients and the microbial structure showed a trend similar to that of the original soil. It shows that there is no ecological occupation effect, and the microorganisms return to original state after the degradation of the degrading bacteria.125 The strains of Pseudomonas sp. WBC-3, Cuviavidus necator JMP134, and Alcaligenes sp. NyZ215 degrade PNP isomers, respectively. The study showed that the enhanced bacterial community affects the local microbial community in soil. However, the effect on species uniformity was small, and the inoculated strains remained stable during incubation and utilized local resources in the soil after removal of pollutants, thus establishing a new community structure.126 Table 3 lists the cases of degradation of nitro-aromatic compounds by nonpathogenic microorganisms. We find that current studies mainly focus on mesophile, while there are few studies on the safety and degradation ability of cold-adapted strains.
| Nitrophenol compounds | Nonpathogenic microorganisms | Environmental conditions | Effecacy of degradation (%) | Reference |
|---|---|---|---|---|
| PNP, 400 mg L−1 | Actinomycetes A5 | 30 °C, pH = 7, 48 h | 62.00% | 132 |
| PNP, 100 μM | Pseudomonas sp. YTK17 | 25 °C, 6 h | 99.00% | 133 |
| PNP, 720 μM | Rhodococcus opacus YTK32 | 25 °C, 6 h | 50.00% | 133 |
| PNP, 100.34 mg L−1 | Pseudomonas sp. ZL | pH = 8.0, 0.5% NaCl, 1 g L−1 NH4NO3, 83 h | 99.94% | 107 |
| PNP, 100 mg L−1 | Ochrobactrum sp. B2 | 30 °C, pH = 10, 16 h | Completely degradation | 134 |
| PNP, 700 mg L−1 | Brevibacterium sp. PNP1 (MH169212), Pseudomonas sp. PNP2 (MH169213), Agromyces mediolanus PNP3 (MH169214), Microbacterium oxydans PNP4 (MH169215) | Yeast extract 2 g L−1, 32 °C, pH = 9.5, 78 h | 99.12% | 93 |
| 2,6-Dichloro-4-nitrophenol, 0.4 mM | Cupriavidus sp. CNP-8 | 30 °C, pH = 7.0, 35 h | Completely degradation | 135 |
| 2,4-Dinitroanisole, 1350 mg kg−1 | ocardioides sp. JS1661 | 30 °C, pH = 6.5 | Completely degradation | 136 |
Psychrophilic, psychrotolerant and psychrosensitive taxa were characterised by an increasing, a stable and a decreasing abundance, respectively, as the sustained drop in temperature. The variation of the microbial communities was particularly significant by a biological adaptation and inhibition at 0 °C, respectively. Psychrophilic played a major role at 0 °C, however, psychrosensitive was completely inhibited at 0 °C and became more abundant with increasing temperature. At low temperatures, the microbial community succession was delayed due to the different abilities of microorganisms to grow and utilize substrate, leading to changes in microbial community composition, and when temperatures decline to extreme values (0 °C), the incapability of psychrosensitive organisms results in a loss of microbial diversity and major shifts in biodegradation patterns.127 Typically, when bioremediation faces cold environmental changes, species richness will be reduced in an area. However, a decrease in species richness may not lead to a decrease in biodegradability, because of the specialization of the bacterial community through selection of the most-adapted microorganisms to degradation pollution. Zhang et al. (2018) researched the degradation of bioaugmentation of bis (2-ethylhexyl) phthalate (DEHP) in a sequencing batch reactor at low temperature (10 °C).128 With lower temperature, the changing trend of Rhodococcus was the main difference in microbial community dynamics between the bioaugmented and noninoculated control reactors. Despite the decreased biodiversity, the DEHP degradation efficiency was significantly improved compared with non inoculated control reactors (79.8 and 36.4%, respectively). The survival and activity of the inoculated cultured microbial consortium are often limited by extreme environmental changes. In a previous study, two compound agents were isolated: C2PL05 and BOS08. The degradation effect of C2PL05 was found to be much less than that of BOS08 under low temperatures.129 Therefore, in this situation, the degradation strains were screened from non-contaminated sites to improve the degradation effect in a low-temperature environment, as the positive effects of the selected microbial agents outweighed the negative effects, although the microbial community composition was altered. Bioaugmentation with natural microbial consortia, such as BOS08It, maybe the best choice when trade-off occurs between the degradation effect and microbial community diversity and when indigenous microorganisms are not degrading PAHs, especially at low temperatures. In Jesus et al. (2021) study, freeze-thaw cycles (−20–4 °C) no decrease the potential activity of community in contaminated soils, this finding reveal that the microbial population is highly resistant to intracellular ice crystallization and cell damage.130 However, Liu et al.(2020) study shows a better removal rater under unfrozen condition, the share OTUs are different between unfrozen and FTCs condition, indicated some specific bacterial communities have changed.131
As the foundation of the “one healthy” environment for humans, animals, and plants, soil microbial communities connect all parts of the ecosystem between different organisms, and this interconnection is essential. Soil microbial diversity, such as the diversity of microbial communities; human immune systems; plant growth; soil habitats; and even global climate change are positively correlated with the “one healthy” environment.137 Environmental and human health effects of adding microbial agents. When the strain has direct or indirect contact with the human body, the strain multiplies or benefits in the human body, competition with other bacteria and nutrients, adverse reactions to the human body and other related issues should be given priority. Risks during the release of microbiotics should be precisely conceived and monitored.138 Zeng et al. (2019) used a human health risk assessment evaluation model recommended by the US Environmental Protection Agent, providing three exposure pathways to calculate the average daily exposure doses to describe the human health hazard of soil heavy mental and organic matter pollution, including ingestion, inhalation, and dermal adsorption, and systematic characterization of the non-carcinogenic and carcinogenic risk of farmland pollution to adults and children.139 We also enable using natural organisms in soil ecosystems to develop ecotoxicological tests such as phytotoxicity test, animal toxicity test, microbial toxicity test, and risk to human health to assessment the ecological risks.140
(3) Risk of horizontal gene transfer. Horizontal gene transfer is often mediated by plasmids, and risks are common in antibiotic use.141 Pollutants such as heavy metals, microplastics, and antibiotics promote the transfer of antibiotic resistance genes among microorganisms, resulting in the enhancement of antibiotic resistance.142 Such pollutants are easy to migrate from the water environment and accumulate to infect the human body. According to the cognition of Antibiotic resistance genes (ARGs)143 the spatial structure of soil biofilm, the composition of extracellular polymers and the interactions between populations play an important role in the metastasis of ARGs. It is reasonably proposed that microbial agents will evolve into new resistant strains through gene mutation or horizontal transfer of related degrading genes, thereby losing their biodegradability or adversely effect on organisms. The transfer of degraded genes from soil to plant systems and in bacteria from soil to the human body may pose a threat to human health.
Although the microbial agent method is safer than other methods, it still has shortcomings such as the long time period required for remediation, unsuitability for heavily contaminated and low-permeability soils, and susceptibility to environmental conditions. We speculate that the degradation intermediates by microbial agent treatment from incomplete degradation of pollution will have adverse effects on human health. t is easy to obtain obvious results, but quite different from the natural condition which has the uncertainty of environmental changes, especially in the freeze-thaw cycles freeze-thaw cycles (FTCs) conditions. Thus, it is necessary to systematically study the changes in the addition of microbial agents in the natural condition, track the microbial community richness and living conditions of microbial agents in the soil environment, and provide scientific data and a theoretical basis for the evaluation of the potential risk of indigenous microorganisms (conditional pathogenic bacteria) and degradability, and gradually solve the problem of biosafety. Several methods are described in the literature, including combining microbial agents and electro-remediation, and the combination of nano-remediation technology and biotechnology have both been found to significantly improve the removal rate of environmental pollution.109 Nanoremediation technology is still in the laboratory research stage, with quite promising application prospects. Furthermore, research with no obvious negative impact on the ecological environment or organisms should be considered as a future research direction in the process of microbial agent development.
Conclusion and future perspectives
Soil remediation under cold conditions is mostly applied in the polar region, and the extraction and application of strains are concentrated at the laboratory stage. On the one hand, we have to face the instability of soil physicochemical properties such as pH, soil texture, soil moisture content, soil microbial biomass carbon, and freeze-thaw cycles, which may prolong the degradation rate. On the other, forming more toxic intermediates may cause secondary damage to the ecosystem. To this end, we expect to improve strain activity and degradation efficiency through immobilization technology, aiming to optimize biochemical processes by detailed catabolic pathways using stoichiometric methods such as mass balance and metabolic flux analysis, further understand the conversion and utilization of metabolites and energy during biodegradation under cold environment, ensuring the safety of microbial agent technology.Author contributions
Jiaxin Li: conceptualization, writing – original draft. Yujuan Wen: writing – review & editing, supervision. Zheng Fang: writing. Wenqi Yang: writing. Xiaoming Song: writing – review & editing, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Northeast Geological S&T Innovation Center of China Geological Survey (No. QCJJ2022-22), National Key R&D Program of China (No. 2019YFC1804805), Key R&D Program of Liaoning Province (No. 2022020338-JH2/1013), the National Natural Science Foundation of China (No. 42172284, 41703125), Liao Ning Revitalization Talents Program (XLYC1807259), Young and Middle-aged Scientific and Technological Innovation Talents Project of Shenyang (RC230297). We thank LetPub (http://www.letpub.com) for its linguistic assistance during the preparation of this manuscript.Notes and references
- X. Liu, L. Shi and J. D. Gu, Biotechnol. Adv., 2018, 36, 1815–1827 CrossRef CAS PubMed.
- J. D. Gu, Appl. Environ. Biotechnol., 2020, 5, 28–33 Search PubMed.
- X. Y. Xie, Y. Zhao, Q. H. Sun, X. Q. Wang, H. Y. Cui, X. Zhang, Y. J. Li and Z. M. Wei, Bioresour. Technol., 2017, 238, 39–47 CrossRef CAS PubMed.
- L. Vergeynst, K. U. Kjeldsen, P. Lassen and S. Rysgaard, J. Hazard. Mater., 2018, 353, 127–134 CrossRef CAS PubMed.
- H. L. Ayala-del-Río, P. S. Chain, J. J. Grzymski, M. A. Ponder, N. Ivanova, P. W. Bergholz, G. Di Bartolo, L. Hauser, M. Land, C. Bakermans, D. Rodrigues, J. Klappenbach, D. Zarka, F. Larimer, P. Richardson, A. Murray, M. Thomashow and J. M. Tiedje, Appl. Environ. Microb., 2010, 76, 2304–2312 CrossRef PubMed.
- Y. Zhang and C. A. Gross, Annu. Rev. Genet., 2021, 55, 377–400 CrossRef CAS PubMed.
- A. Rizvi, B. Ahmed, M. S. Khan, S. Umar and J. Lee, Microorganisms, 2021, 9, 2451 CrossRef CAS.
- Z. Wu, J. Kang, C. Zhang, W. Zhang and J. Ge, Bioresour. Technol., 2023, 385, 129451 CrossRef CAS.
- P. Ji, H. Gu, M. Wen, H. Cai, J. Zhu, X. Yue, Q. Zhang and P. Li, Oil Crop Sci., 2023, 8, 143–148 CrossRef.
- X. Liu, X. Sun, J. Li, X. Yan, Y. Wei, S. Zheng, M. Chen, H. Kan, W. Wang, S. Li and D. Xu, Process Biochem., 2023, 134, 101–111 CrossRef CAS.
- F. Li, H. Ghanizadeh, G. Cui, J. Liu, S. Miao, C. Liu, W. Song, X. Chen, M. Cheng, P. Wang, Y. Zhang and A. Wang, Bioresour. Technol., 2023, 374, 128765 CrossRef CAS PubMed.
- M. Kulkarni and A. Chaudhari, J. Environ. Manage., 2007, 85, 496–512 CrossRef CAS PubMed.
- A. Schackmann and R. Muêller, Appl. Microbiol. Biotechnol., 1991, 34, 809–813 CrossRef CAS.
- Y. Tan, Z. Sun, H. Meng, Y. Han, J. Wu, J. Xu, Y. Xu and X. Zhang, Sep. Purif. Technol., 2019, 215, 217–226 CrossRef CAS.
- X. Zhang, Y. S. Yang, Y. Lu, Y. J. Wen, P. P. Li and G. Zhang, Water Res., 2018, 144, 616–627 CrossRef CAS PubMed.
- J. Chen, E. Wei and Q. Xian, Chin. J. Chromatogr., 2014, 32, 843 CrossRef CAS PubMed.
- S. Kuang, Q. Le, J. Hu, Y. Wang, N. Yu, X. Cao, M. Zhang, Y. Sun, W. Gu, Y. Yang, Y. Zhang, Y. Li, H. Liu and X. Yan, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2020, 228, 108638 CAS.
- Z. Yan, W. Wang, J. Zhou, X. Yi, J. Zhang, X. Wang and Z. Liu, Chemosphere, 2015, 128, 28–35 CrossRef CAS PubMed.
- C. Gu, S. Cui, X. Ge, Z. Wang, M. Chen, Z. Qian, Z. Liu, X. Wang and Y. Zhang, Environ. Res., 2022, 212, 113255 CrossRef CAS PubMed.
- M. Li, X. Wang, C. Lu, R. Li, J. Zhang, S. Dong, L. Yang, L. Xue, J. Chen and W. Wang, Sci. Total Environ., 2020, 714, 136760 CrossRef CAS PubMed.
- Y. Liang, X. Wang, S. Dong, Z. Liu, J. Mu, C. Lu, J. Zhang, M. Li, L. Xue and W. Wang, Chemosphere, 2020, 250, 126256 CrossRef CAS PubMed.
- P. Karlova, M. Halecky, J. Paca, M. Stiborova and E. I. Kozliak, Clean Technol. Environ. Policy, 2015, 17, 287–291 CrossRef CAS.
- W. Xu, Q. Zhao, Z. Li, X. Lu, S. Han and Z. Ye, J. Cleaner Prod., 2019, 214, 782–790 CrossRef CAS.
- S. K. Mishra and K. D. Yadav, Bioresour. Technol. Rep., 2021, 16, 100859 CrossRef CAS.
- S. Jin, Y. Wang and X. Zhao, Microb. Pathog., 2022, 169, 105652 CrossRef CAS PubMed.
- G. L. Y. Lee, N. N. Zakaria, H. Futamata, K. Suzuki, A. Zulkharnain, N. A. Shaharuddin, P. Convey, K. N. M. Zahri and S. A. Ahmad, Catalysts, 2022, 12, 1422 CrossRef CAS.
- S. Miri, M. Naghdi, T. Rouissi, S. Kaur Brar and R. Martel, Crit. Rev. Environ. Sci. Technol., 2019, 49, 553–586 CrossRef CAS.
- Y. Liu, K. Jia, H. Chen, Z. Wang, W. Zhao and L. Zhu, Bioprocess Biosyst. Eng., 2023, 46, 1399–1410 CrossRef CAS.
- Y. Guo, Y. Wang, Z. Zhang, F. Huang and S. Chen, Int. Biodeterior. Biodegrad., 2018, 134, 16–24 CrossRef CAS.
- V. Gupta, P. Bhaskar, J. Thoudam, S. Bisht, A. Sharma and R. Tripathi, TABCJ, 2023, 54–68 Search PubMed.
- T. Yu, R. Keto-Timonen, X. Jiang, J.-P. Virtanen and H. Korkeala, Int. J. Mol. Sci., 2019, 20, 14 Search PubMed.
- S. Liu, Y. Ma, Y. Zheng, W. Zhao, X. Zhao, T. Luo, J. Zhang and Z. Yang, J. Microbiol. Biotechnol., 2020, 30, 187–195 CrossRef CAS PubMed.
- S. Phadtare, J. Alsina and M. Inouye, Curr. Opin. Microbiol., 1999, 2, 175–180 CrossRef CAS.
- Y. H. Jung, J. Uh, K. Lee and H. Im, Biochem. Biophys. Res. Commun., 2018, 496, 374–380 CrossRef CAS PubMed.
- S. K. Gupta, S. Kataki, S. Chatterjee, R. K. Prasad, S. Datta, M. G. Vairale, S. Sharma, S. K. Dwivedi and D. K. Gupta, J. Cleaner Prod., 2020, 258, 120351 CrossRef CAS.
- N. Hassan, A. M. Anesio, M. Rafiq, J. Holtvoeth, I. Bull, A. Haleem, A. A. Shah and F. Hasan, Front. Microbiol., 2020, 11, 824 CrossRef PubMed.
- A. Casanueva, M. Tuffin, C. Cary and D. A. Cowan, Trends Microbiol., 2010, 18, 374–381 CrossRef CAS PubMed.
- J. Liu, S. Wu, L. Feng, Y. Wu and J. Zhu, Food Microbiol., 2023, 112, 104214 CrossRef CAS PubMed.
- H. Liu, G. Zheng, Z. Chen, X. Ding, J. Wu, H. Zhang and S. Jia, Genes, 2023, 14, 158 CrossRef CAS PubMed.
- C. S. Rodrigues and L. M. Madeira, Environ. Technol. Innovation, 2021, 21, 101265 CrossRef CAS.
- X. Q. Chi, J. J. Zhang, S. Zhao and N. Y. Zhou, Environ. Pollut., 2013, 172, 33–41 CrossRef CAS PubMed.
- J. Xu, B. Wang, W. Zhang, F. J. Zhang, Y. Deng, Y. Wang, J. J. Gao, Y.-S. Tian, R. H. Peng and Q. H. Yao, AMB Express, 2021, 11, 124 CrossRef CAS PubMed.
- A. Ghosh, M. Khurana, A. Chauhan, M. Takeo, A. K. Chakraborti and R. K. Jain, Environ. Sci. Technol., 2010, 44, 1069–1077 CrossRef CAS PubMed.
- A. Chauhan, G. Pandey, N. K. Sharma, D. Paul, J. Pandey and R. K. Jain, Environ. Sci. Technol., 2010, 44, 3435–3441 CrossRef CAS PubMed.
- S. P. Sam, H. T. Tan, K. Sudesh, R. Adnan, A. S. Y. Ting and S. L. Ng, J. Environ. Chem. Eng., 2021, 9, 105420 CrossRef CAS.
- J. Wang, L. Ren, Y. Jia, N. Ruth, Y. Shi, C. Qiao and Y. Yan, Toxicol. Environ. Chem., 2016, 98, 226–240 CrossRef CAS.
- C. Sreenivasulu, M. Megharaj, K. Venkateswarlu and R. Naidu, Int. Biodeterior. Biodegrad., 2012, 68, 24–27 CrossRef CAS.
- S. R. Subashchandrabose, K. Venkateswarlu, K. Krishnan, R. Naidu, R. Lockington and M. Megharaj, J. Hazard. Mater., 2018, 347, 176–183 CrossRef CAS.
- E. C. Popoca-Ursino, F. Martínez-Ocampo, E. Dantán-González, E. Sánchez-Salinas and Ma. L. Ortiz-Hernández, Biodegradation, 2017, 28, 351–367 CrossRef CAS.
- Y. Zheng, D. Liu, H. Xu, Y. Zhong, Y. Yuan, L. Xiong and W. Li, Int. Biodeterior. Biodegrad., 2009, 63, 1125–1129 CrossRef CAS.
- M. R. Haghighi-Podeh and S. K. Bhattacharya, Water Sci. Technol., 1996, 34, 345–350 CrossRef CAS.
- P. K. Arora, A. Srivastava and V. P. Singh, J. Hazard. Mater., 2014, 266, 42–59 CrossRef CAS PubMed.
- A. Oren, P. Gurevich and Y. Henis, Appl. Environ. Microbiol., 1991, 57, 3367–3370 CrossRef CAS PubMed.
- H. Teramoto, H. Tanaka and H. Wariishi, Appl. Microbiol. Biotechnol., 2004, 66, 312–317 CrossRef CAS PubMed.
- M. Hofrichter, T. Gtinther and W. Fritsche, Biodegradation, 1993, 3, 415–421 CrossRef CAS.
- Y.-G. Cho, S.-K. Rhee and S.-T. Lee, Biodegradation, 2000, 11, 21–28 CrossRef CAS PubMed.
- D. C. Herman and J. W. Costerton, Appl. Environ. Microbiol., 1993, 59, 340–343 CrossRef CAS PubMed.
- V. L. Gemini, A. Gallego, V. Tripodi, D. Corach, E. I. Planes and S. E. Korol, Int. Biodeterior. Biodegrad., 2007, 60, 226–230 CrossRef CAS.
- D. Appel, J. S. Strehse, H.-J. Marti and E. Maser, Mar. Pollut. Bull., 2018, 135, 1072–1078 CrossRef CAS PubMed.
- H. Claus, T. Bausinger, I. Lehmler, N. Perret, G. Fels, U. Dehner, J. Preuß and H. König, Biodegradation, 2006, 18, 27–35 CrossRef PubMed.
- R. K. Jain, J. H. Dreisbach and J. C. Spain, Appl. Environ. Microbiol., 1994, 60, 3030–3032 CrossRef CAS PubMed.
- D. Paul, R. Singh and R. K. Jain, Environ. Microbiol., 2006, 8, 1797–1804 CrossRef CAS PubMed.
- M. Ortiz-Hernández, Y. Gama-Martínez, M. Fernández-López, M. L. Castrejón-Godínez, S. Encarnación, E. Tovar-Sánchez, E. Salazar, A. Rodríguez and P. Mussali-Galante, Environ. Sci. Pollut. Res., 2021, 28, 42414–42431 CrossRef PubMed.
- R. Boopathy, C. F. Kulpa and J. Manning, Bioresour. Technol., 1998, 63, 81–89 CrossRef CAS.
- F. Hu, L. Yang, Z. Wang and J. Wang, Genes Genomics, 2021, 43, 829–835 CrossRef CAS PubMed.
- T. Hirooka, H. Nagase, K. Hirata and K. Miyamoto, Biochem. Eng. J., 2006, 29, 157–162 CrossRef CAS.
- B. Gumuscu and T. Tekinay, Int. Biodeterior. Biodegrad., 2013, 85, 35–41 CrossRef CAS.
- M. Labidi, D. Ahmad, A. Halasz and J. Hawari, Can. J. Microbiol., 2001, 47, 559–566 CrossRef CAS PubMed.
- A. M. Ziganshin, A. V. Naumov, E. S. Suvorova, E. A. Naumenko and R. P. Naumova, Microbiology., 2007, 76, 676–682 CAS.
- L. Kutateladze, N. Zakariashvili, I. Khokhashvili, M. Jobava, T. Alexidze, T. Urushadze and E. Kvesitadze, Eurobiotech J., 2018, 2, 39–46 CrossRef.
- B. V. Aken, M. Hofrichter, K. Scheibner, A. I. Hatakka, H. Naveau and S. N. Agathos, Biodegradation, 1999, 10, 83–91 CrossRef.
- C. F. Hoehamer, N. L. Wolfe and K. E. L. Eriksson, Int. J. Phytorem., 2006, 8, 95–105 CrossRef CAS PubMed.
- J. C. Spain and D. T. Gibson, Appl. Environ. Microbiol., 1991, 57, 812–819 CrossRef CAS PubMed.
- I. Martínez-Alcalá, J. M. Guillén-Navarro and C. Fernández-López, Chem. Eng. J., 2017, 316, 332–340 CrossRef.
- D. K. Kanaujiya and K. Pakshirajan, Bioresour. Technol., 2022, 344, 126172 CrossRef CAS PubMed.
- Y. Xie, N. Chen, Z. Liang, Y. Huang and H. Shim, J. Water Process Eng., 2023, 53, 103862 CrossRef.
- M. Kohlstedt and C. Wittmann, Metab. Eng., 2019, 54, 35–53 CrossRef CAS PubMed.
- M. R. Antoniewicz, Metab. Eng., 2021, 63, 2–12 CrossRef CAS PubMed.
- P. I. Nikel, T. Fuhrer, M. Chavarría, A. Sánchez-Pascuala, U. Sauer and V. De Lorenzo, ISME J., 2021, 15, 1751–1766 CrossRef CAS PubMed.
- M. R. Antoniewicz, Curr. Opin. Biotechnol., 2020, 64, 230–237 CrossRef CAS PubMed.
- M. R. Antoniewicz, Metab. Eng., 2021, 63, 2–12 CrossRef CAS PubMed.
- J. He, W. Zhang, X. Ren, L. Xing, S. Chen and C. Wang, Environ. Eng. Sci., 2019, 36, 604–613 CrossRef CAS.
- Z. B. Bouabidi, M. H. El-Naas and Z. Zhang, Environ. Chem. Lett., 2019, 17, 241–257 CrossRef CAS.
- A. Partovinia and B. Rasekh, Crit. Rev. Environ. Sci. Technol., 2018, 48, 1–38 CrossRef CAS.
- S. I. Mulla, M. P. Talwar, Z. K. Bagewadi, R. S. Hoskeri and H. Z. Ninnekar, Chemosphere, 2013, 90, 1920–1924 CrossRef CAS PubMed.
- E. D. Yushkova, E. A. Nazarova, A. V. Matyuhina, A. O. Noskova, D. O. Shavronskaya, V. V. Vinogradov, N. N. Skvortsova and E. F. Krivoshapkina, J. Agric. Food Chem., 2019, 67, 11553–11567 CrossRef CAS PubMed.
- Q. Nie and F. Gu, Adv. Mater. Res., 2010, 113, 301–304 Search PubMed.
- Y. Jiang, F. Yang, M. Dai, I. Ali, X. Shen, X. Hou, S. S. Alhewairini, C. Peng and I. Naz, Chemosphere, 2022, 286, 131721 CrossRef CAS PubMed.
- H. Gao, J. Lu, Y. Jiang, Y. Fang, Y. Tang, Z. Yu, W. Zhang, F. Xin and M. Jiang, Biofuels, Bioprod. Biorefin., 2021, 15, 1160–1173 CrossRef CAS.
- S. Chen, W. Zhong, Z. Ning, J. Niu, J. Feng, X. Qin and Z. Li, Bioresour. Technol., 2023, 368, 128302 CrossRef CAS PubMed.
- Z. Zhang, L. Gai, Z. Hou, C. Yang, C. Ma, Z. Wang, B. Sun, X. He, H. Tang and P. Xu, Bioresour. Technol., 2010, 101, 8452–8456 CrossRef CAS PubMed.
- J. Liang, Y. Jin, X. Wen, J. Mi and Y. Wu, Environ. Pollut., 2020, 265, 114202 CrossRef CAS PubMed.
- P. Sarkar and A. Dey, J. Environ. Chem. Eng., 2020, 8, 104347 CrossRef CAS.
- X. Jia, X. Qin, X. Tian, Y. Zhao, T. Yang and J. Huang, J. Environ. Chem. Eng., 2021, 9, 105294 CrossRef CAS.
- H. Hu, M. Wang, Y. Huang, Z. Xu, P. Xu, Y. Nie and H. Tang, mLife, 2022, 1, 382–398 CrossRef.
- X. G. Fu, K. Shi, J. L. Xue, C. Chen, Y. Bai, Y. L. Qiao, Y. X. Liu, X. M. Hu, Y. Gao and H. Yu, Pet. Sci., 2021, 18, 1543–1550 CrossRef CAS.
- S. H. Liu, G. M. Zeng, Q. Y. Niu, Y. Liu, L. Zhou, L. H. Jiang, X. Tan, P. Xu, C. Zhang and M. Cheng, Bioresour. Technol., 2017, 224, 25–33 CrossRef CAS PubMed.
- D. K. Chaudhary and J. Kim, Int. Biodeterior. Biodegrad., 2019, 142, 58–72 CrossRef CAS.
- A. Banach-Wiśniewska, M. Tomaszewski, M. S. Hellal and A. Ziembińska-Buczyńska, Sci. Rep., 2021, 11, 840 CrossRef PubMed.
- M. M. Ureta, G. N. Martins, O. Figueira, P. F. Pires, P. C. Castilho and A. Gomez-Zavaglia, Crit. Rev. Food Sci. Nutr., 2021, 61, 2659–2690 CrossRef CAS PubMed.
- M. B. Cassidy, H. Lee and J. T. Trevors, J. Ind. Microbiol. Biotechnol., 1996, 16, 79–101 CrossRef CAS.
- X. Fu, T. Wu, H. Li, J. Xue, J. Sun, L. Li, Y. Qiao and C. Li, Environ. Technol., 2021, 1–7 CAS.
- X. Fu, Q. Zhang, Y. Gao, Y. Wu, X. Xiao, L. Li, J. Xue and B. Liu, Pet. Sci. Technol., 2019, 37, 1417–1424 CrossRef CAS.
- Y. Huang, J. Sun, D. Wu and X. Feng, Sep. Purif. Technol., 2018, 207, 142–150 CrossRef CAS.
- C. Xu, X. Zeng, Z. Yang and H. Ji, Molecules, 2022, 27, 1148 CrossRef CAS PubMed.
- Z. He, W. Wang, J. Fan, B. Bao, X. Qin and D. Yu, React. Funct. Polym., 2020, 157, 104762 CrossRef CAS.
- C. F. Li, Y. J. Wen, N. Cao, D. Sun, X. M. Song and Y. S. Yang, CIESC J., 2021, 280, 233–240 Search PubMed.
- Y. J. Wen, Y. S. Yang, H. J. Ren, X. Q. Du, X. Y. Yang, L. Y. Zhang and X. S. Wang, Chem. Eng. J., 2015, 280, 233–240 CrossRef CAS.
- Y. Wang, Y. Hou, Q. Wang and Y. Wang, J. Hazard. Mater., 2021, 413, 125377 CrossRef CAS PubMed.
- Y. Wang, Y. Hou, Y. Wang, L. Zheng and Q. Wang, Enzyme Microb. Technol., 2019, 131, 109434 CrossRef CAS PubMed.
- P. Bergauer, P. A. Fonteyne, N. Nolard, F. Schinner and R. Margesin, Chemosphere, 2005, 59, 909–918 CrossRef CAS PubMed.
- S. Sepehr, B. Shahnavaz, A. Asoodeh and M. Karrabi, J. Environ. Sci. Health, Part A: Environ. Sci. Eng., Part A, 2019, 54, 367–379 CrossRef CAS PubMed.
- S. Verma and S. Chatterjee, Int. Biodeterior. Biodegrad., 2021, 158, 105169 CrossRef CAS.
- A. Fareed, S. Riaz, I. Nawaz, M. Iqbal, R. Ahmed, J. Hussain, A. Hussain, A. Rashid and T. A. Naqvi, Heliyon, 2019, 5, e02740 CrossRef PubMed.
- N. Totubaeva, Z. Tokpaeva, J. Izakov and M. Moldobaev, Plant, Soil Environ., 2023, 69, 188–193 CrossRef CAS.
- S. Miri, T. Robert, S. M. Davoodi, S. Kaur Brar, R. Martel, T. Rouissi and J.-M. Lauzon, J. Hazard. Mater., 2023, 450, 131078 CrossRef CAS PubMed.
- J. Kim, A. H. Lee and W. Chang, Sci. Total Environ., 2018, 612, 903–913 CrossRef CAS PubMed.
- M. Abdulrasheed, N. N. Zakaria, A. F. Ahmad Roslee, M. Y. Shukor, A. Zulkharnain, S. Napis, P. Convey, S. A. Alias, G. Gonzalez-Rocha and S. A. Ahmad, Antarct. Sci., 2020, 32, 341–353 CrossRef.
- S. Bajaj and D. K. Singh, Int. Biodeterior. Biodegrad., 2015, 100, 98–105 CrossRef CAS.
- Y. Wu, Y. Liu, H. Kamyab, M. Rajasimman, N. Rajamohan, G. H. Ngo and C. Xia, Environ. Res., 2023, 232, 116363 CrossRef CAS PubMed.
- J. D. Gu, Appl. Environ. Biotechnol., 2016, 1, 92–95 CrossRef CAS.
- L. Gao and J. D. Gu, Int. Biodeterior. Biodegrad., 2021, 162, 105253 CrossRef CAS.
- A. S. Nwankwegu, L. Zhang, D. Xie, C. O. Onwosi, W. I. Muhammad, C. K. Odoh, K. Sam and J. N. Idenyi, J. Environ. Manage., 2022, 304, 114313 CrossRef CAS PubMed.
- M. Shen, J. Li, Y. Dong, Z. Zhang, Y. Zhao, Q. Li, K. Dang, J. Peng and H. Liu, Agriculture, 2021, 11, 389 CrossRef CAS.
- Y. Liu, W. Wang, S. B. Shah, G. Zanaroli, P. Xu and H. Tang, Appl. Microbiol. Biotechnol., 2020, 104, 427–437 CrossRef CAS PubMed.
- X. Q. Chi, J. J. Zhang, S. Zhao and N. Y. Zhou, Environ. Pollut., 2013, 172, 33–41 CrossRef CAS PubMed.
- L. Vergeynst, K. U. Kjeldsen, P. Lassen and S. Rysgaard, J. Hazard. Mater., 2018, 353, 127–134 CrossRef CAS PubMed.
- K. Zhang, H. Luo, Z. Zhu, W. Chen, J. Chen and Y. Mo, J. Environ. Eng., 2018, 144, 04018085 CrossRef.
- R. Simarro, N. González, L. F. Bautista and M. C. Molina, FEMS Microbiol. Ecol., 2013, 83, 438–449 CrossRef CAS PubMed.
- H. E. D. Jesus, R. S. Carreira, S. S. M. Paiva, C. Massone, A. Enrich-Prast, R. S. Peixoto, J. L. M. Rodrigues, C. K. Lee, C. Cary and A. S. Rosado, Microorganisms, 2021, 9, 609 CrossRef PubMed.
- M. Liu, F. Feng, T. Cai and S. Tang, Front. Microbiol., 2020, 11, 1164 CrossRef PubMed.
- R. Jaweria, S. A. P. S. A. Peshwe and A. G. I. A. G. Ingale, Indian J. Appl. Res., 2011, 3, 241–243 CrossRef.
- Y. Shinozaki, N. Kimura and T. Nakahara, J. Biosci. Bioeng., 2002, 93, 512–514 CrossRef CAS PubMed.
- X. Qiu, Q. Zhong, M. Li, W. Bai and B. Li, Int. Biodeterior. Biodegrad., 2007, 59, 297–301 CrossRef CAS.
- J. Min, L. Xu, S. Fang, W. Chen and X. Hu, Environ. Pollut., 2020, 258, 113703 CrossRef CAS PubMed.
- S. Karthikeyan and J. C. Spain, J. Hazard. Mater., 2016, 312, 37–44 CrossRef CAS PubMed.
- S. Banerjee and M. G. A. van der Heijden, Nat. Rev. Microbiol., 2022, 21, 6–20 CrossRef PubMed.
- Joint FAO, WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food, Agric. Serv. Bull. (F. A. O.), Rome, 2006 Search PubMed.
- S. Zeng, J. Ma, Y. Yang, S. Zhang, G.-J. Liu and F. Chen, Sci. Total Environ., 2019, 687, 642–653 CrossRef CAS PubMed.
- H. Zhang, X. Yuan, T. Xiong, H. Wang and L. Jiang, Chem. Eng. J., 2020, 398, 125657 CrossRef CAS.
- C. Zhang, Chem. Eng. J., 2023, 474, 145669 CrossRef CAS.
- G. Y. Fang, X. Q. Liu, Y. J. Jiang, X. J. Mu and B. W. Huang, Sci. Total Environ., 2024, 912, 168908 CrossRef CAS PubMed.
- L. Niu, W. Liu, A. Juhasz, J. Chen and L. Ma, Crit. Rev. Environ. Sci. Technol., 2022, 52, 4135–4146 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |