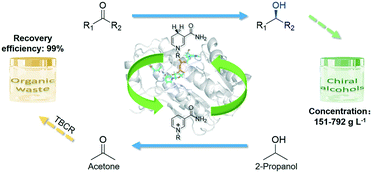
Efficient asymmetric synthesis of chiral alcohols using high 2-propanol tolerance alcohol dehydrogenase SmADH2 via an environmentally friendly TBCR system†
Zeyu
Yang
 ,
Hengwei
Fu
,
Wenjie
Ye
,
Youyu
Xie
,
Qinghai
Liu
,
Hualei
Wang
,
Hengwei
Fu
,
Wenjie
Ye
,
Youyu
Xie
,
Qinghai
Liu
,
Hualei
Wang
 * and
Dongzhi
Wei
*
* and
Dongzhi
Wei
*
State Key Laboratory of Bioreactor Engineering, New World Institute of Biotechnology, East China University of Science and Technology, Shanghai 200237, PR China. E-mail: hlwang@ecust.edu.cn; dzhwei@ecust.edu.cn
First published on 5th November 2019
Abstract
Alcohol dehydrogenases (ADHs) together with the economical substrate-coupled cofactor regeneration system play a pivotal role in the asymmetric synthesis of chiral alcohols; however, severe challenges concerning the poor tolerance of enzymes to 2-propanol and the adverse effects of the by-product, acetone, limit its applications, causing this strategy to lapse. Herein, a novel ADH gene smadh2 was identified from Stenotrophomonas maltophilia by traditional genome mining technology. The gene was cloned into Escherichia coli cells and then expressed to yield SmADH2. SmADH2 has a broad substrate spectrum and exhibits excellent tolerance and superb activity to 2-propanol even at 10.5 M (80%, v/v) concentration. Moreover, a new thermostatic bubble column reactor (TBCR) system is successfully designed to alleviate the inhibition of the by-product acetone by gas flow and continuously supplement 2-propanol. The organic waste can be simultaneously recovered for the purpose of green synthesis. In the sustainable system, structurally diverse chiral alcohols are synthesised at a high substrate loading (>150 g L−1) without adding external coenzymes. Among these, about 780 g L−1 (6 M) ethyl acetoacetate is completely converted into ethyl (R)-3-hydroxybutyrate in only 2.5 h with 99.9% ee and 7488 g L−1 d−1 space–time yield. Molecular dynamics simulation results shed light on the high catalytic activity toward the substrate. Therefore, the high 2-propanol tolerance SmADH2 with the TBCR system proves to be a potent biocatalytic strategy for the synthesis of chiral alcohols on an industrial scale.
Introduction
Chiral alcohols are important building blocks for chiral pharmaceuticals, fine chemicals and agrochemicals. In particular, in the pharmaceutical industry, chiral alcohols can serve as intermediates of many chiral drugs such as atorvastatin, enalapril, and crizotinib.1–3 Biocatalysis is proved to be a practical approach to synthesise chiral alcohols, which has attracted considerable attention due to its mild reaction conditions, high stereoselectivity and absence of heavy metals.4 Chiral resolution and asymmetric synthesis are two common ways of the biocatalytic method. Due to the limitation of a 50% theoretical yield for chiral resolution, asymmetrical synthesis becomes an ideal process to produce chiral alcohols as it can reach a theoretical yield of 100%.5It is necessary to regenerate expensive coenzymes such as NAD(P)H in ADH-mediated bioreductions, which can be achieved by coupling a procedure of oxidizing co-substrates. There are two practical ways to reach the purpose. One is to add glucose dehydrogenase (GDH) or formate dehydrogenase (FDH) to form an enzyme-coupled system;6 the other is to add co-substrates such as 2-propanol, which can be oxidised simultaneously by some ADHs to form a self-regenerating system of cofactors called the substrate-coupled system.7 The latter has two advantages: first, a higher catalytic efficiency on the microscopic level is achieved due to the presence of only one enzyme in the bioreduction; second, a relatively constant pH value and the cheap co-solvent 2-propanol collectively cause much convenience to the whole reaction on the macroscopic level. These make the substrate-coupled cofactor regeneration system more promising.
Nevertheless, ADHs exhibiting high oxidative activity to 2-propanol in previous reports are scarce, and moreover, most of them have poor tolerance to 2-propanol.8 For example, ADHs such as LSADH (Leifsonia sp.), YlCR (Yarrowia lipolytica) and LKADH (Lactobacillus kefiri) presented activity toward different ketones in a substrate-coupled system and were successfully applied in the synthesis of several chiral drug intermediates;7,9,10 however, this application is generally limited because the tolerance to 2-propanol is relatively poor, and the proportion of 2-propanol in the system cannot surpass 2.62 M (20%, v/v), leading to a low substrate loading.
Another difficulty in using this system was the accumulation of the by-product acetone during the whole process, usually leading to a depressed enzyme activity and an unfavourable side of the chemical equilibrium.11 To solve this problem, two strategies, an aeration system and a depressurizing method, were considered;12,13 however, excess 2-propanol had to be added at the beginning of the reaction to compensate for the loss of 2-propanol caused by these two strategies, which resulted in extra loss of the enzyme activity. Among these, the depressurizing method had a significant effect on acetone removal; however, it exhibited high energy consumption and was not conducive to the synthesis of low-boiling products. Therefore, it is urgent to establish a sound strategy to prevent acetone accumulation in order to achieve high substrate loading.
In this study, a novel NADH-dependent alcohol dehydrogenase SmADH2 with a broad substrate spectrum was successfully expressed by recombinant Escherichia coli cells, which can tolerate about 80% (v/v) 2-propanol. In order to solve the acetone accumulation issue during bioreduction using 2-propanol as the hydrogen donor, a newly designed thermostatic bubble column reactor (TBCR) system was established. In this system, acetone can be removed by gas flow and 2-propanol was continuously supplemented by a gas mixture and formed a dynamic equilibrium in the reaction unit. The final gas mixture of acetone and 2-propanol from the reaction unit was condensed by the collection unit as a side product, which is compatible with green synthesis. Eventually, six important chiral alcohols with quite different structures (ee >99.9%) were successfully synthesised at 150–780 g L−1 substrate loading (Scheme 1). The sound TBCR system combined with the high 2-propanol tolerance SmADH2 would have a promising prospect in future applications.
Results and discussion
Construction and screening of ADH library
About 30 genes of SmADHs (Table S1†) were identified from Stenotrophomonas maltophilia LH15 by genome mining and expressed in a heterologous host, E. coli BL21 (DE3).14 The asymmetrical reduction from ethyl acetoacetate (EAA) to ethyl (R)-3-hydroxybutyrate [(R)-EHB] was chosen as a model reaction. In this reaction, the EAA is a substrate with a simple structure and the optically pure (R)-EHB is an important intermediate for the synthesis of β-lactamase inhibitors, β-lactam antibiotics and (+)-decarestrictine L.15–17 The coenzyme dependence of ADHs was tested. Thereafter, the catalytic activity and enantioselectivity of SmADHs to EAA was detected using the corresponding coenzyme and the result revealed that 21 ADHs presented activities toward EAA. In order to build a substrate-coupled cofactor regeneration system, these enzymes were then screened for oxidation activities toward 2-propanol (Table S2†). The results revealed that only 5 ADHs had activities for 2-propanol. SmADH2 exhibited the best oxidative activity for 2-propanol and reductive activity for EAA; hence, it was chosen as the biocatalyst for further research.Characterisation of SmADH2
Sequence alignment of SmADH2 was performed with different short-chain dehydrogenases (SDRs), which exhibited high catalytic activity toward EAA, including CmCR from Candida magnoliae (GenBank: BAB21578.1), BgADH from Burkholderia gladioli BSR3 (GenBank: YP_004348055.1), LbADH from Lactobacillus brevis (GenBank: CAD66648.1) and LkADH from Lactobacillus kefiri (GenBank: WP_054768785.1).18–21 Results revealed that SmADH2 belongs to the SDR superfamily due to a conserved coenzyme binding domain (GXXXGXG) and a catalytic triad, S146-Y159-K163 (Fig. S1†). Based on the sequence alignment, the amino acid sequence of SmADH2 had merely 30–35% identity compared to those enzymes, which indicated that SmADH2 would exhibit distinct properties when catalysing the same substrate.In addition, SmADH2 showed distinct cofactor dependence with LkADH and LbADH, which may contribute partly to the low similarity. There are two key regions that determine coenzyme dependence. One is at the β2α2 domain, while the other is at the coenzyme binding motif. At the coenzyme binding motif shown as green squares in Fig. S1,† the residues of LkADH and LbADH are Thr, which can form hydrogen bonds with the 2′-phosphate of NADPH, while the residue of SmADH2 is Val, which cannot form hydrogen bonds with the 2′-phosphate of NADPH. At the β2α2 domain shown as purple squares, the residues of LkADH and LbADH are Arg and Gly residues, which can form hydrogen bonds with the 2′-phosphate of NADPH, while the residue of SmADH2 is Asp, which can only form hydrogen bonds with the adenine ribose of NADH (Fig. S1†). These differences at the key residues give rise to the distinct coenzyme dependence of the three ADHs.22,23
The purified enzyme presented a single band on SDS-PAGE at 27 kDa (Fig. S2†), and this is in accordance with the calculated result of amino acid sequences. The molecular weight of native SmADH2 was estimated to be about 110 kDa by using a gel filtration column, which indicated that SmADH2 is preserved in a tetrameric state. The specific activity of SmADH2 to EAA was 320 U mg−1.
The temperature–activity curve of the purified SmADH2 revealed that the optimum temperature is set at 30 °C (Fig. S3A†). Compared to 40 °C and 50 °C, SmADH2 was most stable under 30 °C with a half-life (t1/2) of 60 h (Fig. S3B†). At pH 7.0, the maximum activity of the enzyme was obtained (Fig. S3C†). Coincidently, SmADH2 remained most stable at pH 7.0 and presented 94% residual activity according to the pH–activity curve (Fig. S3D†). Therefore, the optimum reaction conditions were established, that is, in phosphate buffer saline (PBS; 100 mM, pH 7.0) at 30 °C.
The experiment measuring kinetic parameters was carried out in PBS (100 mM, pH 7.0) at 30 °C using NADH as a cofactor. SmADH2 presented a low Km value (0.39 mM) and a high kcat value (210 S−1), indicating that SmADH2 had a strong affinity for and reduction ability to EAA (Table 1). In addition, the Km value toward NADH was extremely low, which indicated that SmADH2 had a strong affinity for NADH. According to the kcat/Km value of SmADH2 to 2-propanol, the oxidation of 2-propanol by SmADH2 can be well used in the substrate-coupled coenzyme regeneration system.9,24
| Substrate | K m (mM) | k cat (s−1) | k cat/Km (mM−1 s−1) |
|---|---|---|---|
| Reaction conditions: EAA (0.1–5 mM, methanol, 5% v/v), purified SmADH2 (0.1 mg mL−1), NADH (0.5 mM), pH 7.0, and 30 °C toward EAA. EAA (5 mM, methanol, 5% v/v), purified SmADH2 (0.1 mg mL−1), NADH (0.005–0.05 mM), pH 7.0, and 30 °C toward NADH. 2-Propanol (0.01–0.5 M), purified SmADH2 (0.1 mg mL−1), NAD+ (0.5 mM), pH 7.0, and 30 °C toward 2-propanol. 2-Propanol (0.5 M), purified SmADH2 (0.1 mg mL−1), NAD+ (0.1–5 mM), pH 7.0, and 30 °C toward NAD+. | |||
| EAA | 0.39 | 210 | 5.4 × 102 |
| NADH | 0.013 | 188 | 1.4 × 104 |
| 2-Propanol | 66 | 192 | 2.9 |
| NAD+ | 0.15 | 120 | 8.0 × 102 |
Substrate specificity
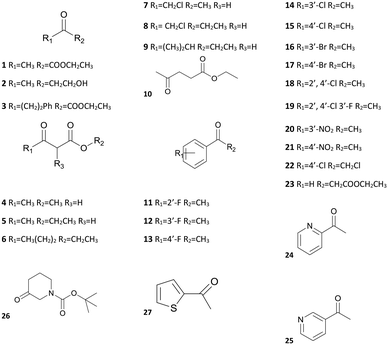
The substrate-specificity assay revealed that SmADH2 had a broad substrate spectrum. It had good to excellent catalytic activity toward EAA and 23 other ketone substrates, including α/β-ketoesters, aliphatic ketones, aryl ketones and heterocyclic ketones (Table 2). In addition, SmADH2 exhibits excellent stereoselectivity towards 14 products (ee >99.9%). Results indicate that the activity of SmADH2 to β-ketoesters is higher than that to other ketones. Among these β-ketoesters, the corresponding products 4, 5, 7 and 8 exhibited 99.9% ee values. These make it possible to synthesise optically pure β-hydroxy esters on an industrial scale.| S | Relative activitya (%) | eeb (%) | S | Relative activity (%) | ee (%) | S | Relative activity (%) | ee (%) |
|---|---|---|---|---|---|---|---|---|
| a Relative activities measured using purified SmADH2 under the standard assay protocol and expressed as percentages referred to EAA. b ee values determined by GC or HPLC analysis (Table S5†). | ||||||||
| 1 | 169 | R(95.6) | 10 | 64 | R(98.7) | 19 | — | — |
| 2 | 34 | R(99.9) | 11 | 25 | R(58.2) | 20 | 48 | R(99.9) |
| 3 | 26 | S(99.9) | 12 | 24 | R(98.5) | 21 | 14 | R(99.9) |
| 4 | 87 | R(99.9) | 13 | 56 | R(99.9) | 22 | 59 | S(96.7) |
| 5 | 100 | R(99.9) | 14 | 14 | R(68.5) | 23 | 18 | R(99.9) |
| 6 | 79 | R(87.4) | 15 | 23 | R(99.9) | 24 | 6 | R(96.7) |
| 7 | 68 | S(99.9) | 16 | — | — | 25 | 37 | R(99.9) |
| 8 | 63 | S(99.9) | 17 | 57 | R(99.9) | 26 | 27 | R(99.9) |
| 9 | 76 | R(93.9) | 18 | — | — | 27 | 12 | R(87.4) |
From the substrate spectrum, we observed that all the products follow the anti-Prelog rule and most of them are (R)-form alcohols.25 The substrates 3, 7 and 8 yield the (S)-form alcohols due to higher priority atoms (–CH2Cl > –CF3 > –phenyl > –CH3) at their chiral centres according to the Cahn–Ingold–Prelog rule.25 It is well known that relatively few enzymes follow the anti-Prelog rule,26 but anti-Prelog alcohols act as key intermediates in the synthesis of chiral drugs.27 Therefore, SmADH2 has great application potential.
2-Propanol tolerance of SmADH2 and other ADHs
Polar organic solvents such as 2-propanol usually have adverse effects on the enzyme activity. To investigate the potential of SmADH2 in the substrate-coupled cofactor regeneration system, the enzyme was incubated in different concentrations of 2-propanol and the residual enzyme activity was measured. We found that there was no significant decrease in activity of SmADH2 in 20% (v/v) 2-propanol, and the residual activity was above 95% after 24 h. When the 2-propanol content increased to 80% (v/v), it still maintained 87.3% activity (Table 3). As the concentration of 2-propanol increased, the bound water of enzyme proteins was partly stripped, resulting in unfavourable conformational changes.28 In several reported substrate-coupled cofactor regeneration systems, the ratios of 2-propanol/water (v/v) were generally less than 20% (2.63 M) to ensure that the enzyme functions well. When the ratio reached 80% (v/v), the enzyme activity decreased significantly (Table 3). Thus, the theoretical substrate concentration was limited. The excellent tolerance to 2-propanol triggers SmADH2 to achieve high substrate loading.| Entry | Enzyme | Source organism | 2-Propanol | ||
|---|---|---|---|---|---|
| 20% | 80% | Ref. | |||
| a Residual activity (%) determined in 20% (v/v) and 80% (v/v). Reaction conditions: PBS buffer (100 mM, pH 7.0) and 2-propanol total 1 mL reaction volume, 0.5 mM NADH, 5 mM EAA, 1.25 μg purified SmADH2; activities were compared with those in pure PBS (100 mM, pH 7.0) as solvent. b Dec. means declined. | |||||
| 1 | SmADH2 | Stenotrophomonas maltophilia | 95.4a | 87.3a | This work |
| 2 | LSADH | Leifsonia sp. S749 | Dec.b | — | 7 |
| 3 | YlCR | Yarrowia lipolytica ACA-DC 50109 | Dec. | — | 9 |
| 4 | PAR | Rhodococcus sp. ST-10 | Dec. | — | 12 |
| 5 | LKADH | Lactobacillus kefiri DSM 20587 | 0.6 | 0 | 8 |
| 6 | Rhoc (ReADH) | Rhodococcus erythropolis | 16 | 7.5 | 8 |
| 7 | YglC | Saccharomyces cerevisiae | 45.5 | 5.5 | 8 |
| 8 | SpbC (ScCR) | Streptomyces coelicolor | 2 | 0.8 | 8 |
Enzyme- and substrate-coupled cofactor regeneration system
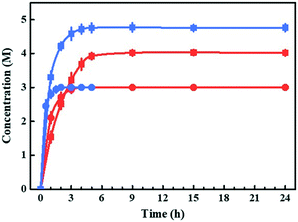
Bioreactions based on substrate- and enzyme-coupled coenzyme regeneration systems were compared in thermostatic stirred tank reactors (TSTRs) at 3 and 5 M EAA loading (Fig. 1). Considering the mass transfer efficiency of coenzymes, we co-expressed the smadh2 and gdh in E. coli cells. Both the smadh2 and gdh were successfully expressed in soluble form. All the reactions were performed under an equivalent enzyme activity of SmADH2 (512.4 U). The activity of GDH (582.3 U) is higher than that of SmADH2 and thus is sufficient for cofactor regeneration. In 3 M bioreduction, the conversions of both systems can finally reach 99.9%. However, only half the time was required for the substrate-coupled bioreduction compared with that of the enzyme-coupled system. The concentration of 5 M bioreduction based on the substrate-coupled system achieved a maximum conversion of 95% after 5 h, but that of the enzyme-coupled system could only attain 80% even after 24 h. These results indicate that the SmADH2 mediated substrate-coupled coenzyme regeneration system is more efficient. The reason may be that the additional coenzymes and substrate molecules can get into the molecules more efficiently due to the increased permeability of the cell membrane caused by high-concentration 2-propanol. Noteworthily, although SmADH2 is highly tolerant to 2-propanol, 1.25 substrate equivalents of 2-propanol are sufficient to achieve 99.9% conversion, which makes the bioreduction conform to the principle of atom economy.Construction and optimization of the TBCR system
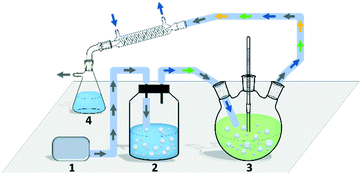
Though the conversion of 5 M EAA bioreduction carried out in the substrate-coupled system was 95% after 5 h, which indicates a good result, the accumulation of the by-product acetone resulting in adverse effects on enzyme activity and chemical equilibrium should be considered.29 In order to solve the acetone accumulation issue and then increase the substrate loading, a TBCR system was built in this study. The TBCR system comprises an air pump, a buffer unit, a reaction unit and a collection unit. All these units are connected by air paths (Fig. 2). The air pump unit provides ventilation for the entire system. The buffer unit supplies 2-propanol to the system and the air flows through this unit will become a mixture of 2-propanol and water vapour. The reaction unit is the site of bioreduction based on the substrate-coupled coenzyme regeneration system. In this unit, the gas mixture is injected through a distributor, which can remove the acetone and 2-propanol according to the gas–liquid mass transfer theory. While acetone is maintained at a relatively low concentration in the system, extra 2-propanol can be continuously supplemented via the gas mixture, forming a dynamic equilibrium. The final gas mixture of acetone and 2-propanol from the reaction unit can be recycled by the collection unit as side products.In order to enhance the performance of the TBCR, two factors containing the amount of air flow and the proportion of 2-propanol in the buffer unit were optimised (Fig. 3). At low air flow, the ability to remove acetone was limited. In this case, increasing the proportion of 2-propanol has little effect on the conversion rate. When the 2-propanol proportion in the buffer unit was low, the addition of 2-propanol was limited. With the increase of air flow, the restriction factor of the conversion rate changed from acetone accumulation to 2-propanol loss. As a result, 2.5 L min−1 air flow and 50% (v/v) 2-propanol/water ratio were selected as the best conditions for the TBCR system. The concentration of acetone in the reaction unit was monitored throughout the reaction process, and it was found that under the action of the TBCR, the concentration of acetone in the reaction system decreased significantly (Fig. S4†), contributing to the complete conversion of the bioreduction.
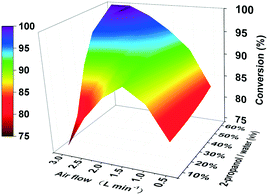 | ||
| Fig. 3 3D bivariate coordinate system optimizing the TBCR. The air flow was studied in the range of 0.5 L min−1 to 3.0 L min−1. The ratio of 2-propanol/water (v/v) was investigated from 10% to 60%. | ||
Furthermore, the amount of cell, coenzyme and substrate addition was further optimised (Table S4†). Under these conditions, a substantial amount of (R)-EHB (792 g L−1, 6 M, ee > 99.9%) was produced via asymmetric synthesis with a conversion of 99.9% in only 2.5 h without additional NADH (Table 4, entry 3). (R)-EHB was then isolated with a yield of 93%. In addition, 9.93 ml 2-propanol and 1.49 ml acetone were recovered by the collection unit and attained a 99% recovery efficiency, which solved the economic and ecological issues.
| Entry | Substrate | Reactor | Ketone | NAD+ (mM) | Cell (g L−1) | Time (h) | Conv.b (%) | eeb (%) | Yield (%) | |
|---|---|---|---|---|---|---|---|---|---|---|
| (g L−1) | (M) | |||||||||
| a Reaction conditions: 10 ml PBS (100 mM, pH 7.0), 200–500 mg lyophilised cells, 0–5 μmol NAD+, 6.5–7.8 g EAA and 4.8–5.7 ml 2-propanol (1.25 equiv.) were placed in the TBCR and reacted at 30 °C for 24 h. b Conversion and ee values determined by HPLC and GC analysis (Table S5†). | ||||||||||
| 1 | 3 | TBCR | 180 | 0.85 | 0 | 20 | 12.0 | 99.9 | S(99.9) | 89 |
| 2 | 4 | TBCR | 580 | 5 | 0 | 20 | 3.0 | 99.9 | R(99.9) | 93 |
| 3 | 5 | TBCR | 780 | 6 | 0 | 20 | 2.5 | 99.9 | R(99.9) | 93 |
| 4 | 7 | TBCR | 450 | 3 | 0 | 20 | 4.0 | 99.9 | S(99.9) | 91 |
| 5 | 8 | TBCR | 330 | 2 | 0 | 20 | 6.0 | 99.9 | S(99.9) | 89 |
| 6 | 26 | TBCR | 150 | 0.75 | 0 | 20 | 12.0 | 99.9 | R(99.9) | 88 |
From various other studies, we found that two strategies were usually adopted for the removal of acetone. One is using an aeration system,12 in which dry air is pumped into the reaction system to remove the acetone. The other includes the depressurizing method,13 which reduces the pressure of the system to remove low-boiling acetone. Compared to the aeration system and the depressurizing method, the TBCR strategy provided more advantages. Initially, water and 2-propanol vapour in the gas mixture entering the reaction unit enhanced the ability to remove acetone. Next, 2-propanol was gradually added by the gas mixture from the buffer unit to avoid a high 2-propanol content at the initial stage of the reaction. In addition, the whole reaction process was carried out at ordinary pressure with low energy consumption, which is suitable for the synthesis of products with a low boiling point. Eventually, the organic waste was collected, which realises the goal of environmental-friendliness. With the designed TBCR system, the substrate loading was increased from 5 to 6 M and the external coenzyme addition was reduced to zero.
Asymmetric synthesis of other chiral alcohols via the TBCR system
In order to extend the application scope of SmADH2, several ketones were chosen based on the substrate specificity and the application value of the products. The corresponding products 3, 4, 7, 8 and 26 can serve as important intermediates for synthesising several crucial drugs such as benidipine statins and benazepril. Thereafter, the bioreductions were carried out using the TBCR system under high substrate loading in order to asymmetrically synthesise the chiral precursors of drugs. The results revealed that 180 g L−13, 580 g L−14, 450 g L−17, 330 g L−18 and 150 g L−126 could be completely transformed into products with excellent ee of >99.9% (Table 4), which can meet the requirements of biocatalytic processes in the industry (substrate concentration >100 g L−1).30P13, P17 and P20 were also synthesised with the substrate loading of slightly less than 100 g L−1. Considering these promising results, the combination of SmADH2 and the TBCR system proves to be a potent biocatalytic strategy and will have a better prospect in drug manufacturing.Molecular dynamics simulation
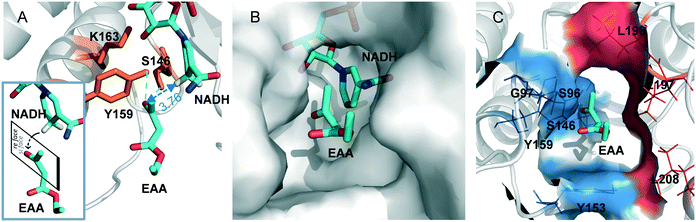
For further insight into the catalytic process, the EAA molecule was docked into the SmADH/NADH complex. Furthermore, an all-atom MD simulation of SmADH2 was performed in complex with the substrate EAA, and a trajectory of about 50 ns for the system was analysed in terms of root-mean-square deviations (RMSDs). The RMSD of Cα atoms from the starting structure reached a plateau of about 2 Å after 7 ns (Fig. S5A†), indicating the convergence of the simulation. The catalytic triad S146-Y159-K163 was an active centre of SmADH2. According to the catalytic mechanism, a proton is transferred from the hydroxyl group of Y159 to the carbonyl oxygen atom of EAA. Next, the hydrogen on the C4 of NADH attacks the carbonyl group of the substrate from the (si)-face to form an (R)-form alcohol (obeying the anti-Prelog rule; Fig. 4A).31SmADH2 possesses a semi-open substrate binding pocket with a volume of 206 Å3 (calculated using Discovery Studio 4.0 software; Fig. 4B). The volume of EAA is 112.9 Å3 and the binding energy is 18.1 kJ mol−1, indicating that EAA can easily enter the binding pocket and then interact with the active sites.
Specific polarity distribution in the substrate binding pocket of SmADH2 forms a polar cavity, which comprises hydrophobic residues (L196, L197, L208 and L217) on one side and hydrophilic residues (S96, G97 and Y159) on the other side (Fig. 4C). The strong electronegativity of oxygen atoms in EAA molecules cause the carbonyl and ester groups to be moderately polar chemical bonds. A more polar side of the substrate binding pocket is favourable for the carbonyl and ester groups of EAA. These interactions help the EAA molecules to form pre-reaction states, which could reflect early stereoselective recognition of enzymes and generate optically pure products.32
When the EAA came in a particular form into the substrate binding pocket, the carbonyl oxygen atom of EAA formed hydrogen bonds with Y159 and S146 residues separately, which anchored the EAA in the active centre. Analysis of the distances between the carbonyl group of the substrate and the catalytic centre is shown in Fig. S5B,† and the result demonstrates that the distance between the carbonyl group of EAA and the hydrogen on the C4 of NADH is only 3.76 Å, facilitating the transfer of hydrogen ions to the substrate.33 All of these factors help SmADH2 exhibit high activity and excellent stereoselectivity in reducing EAA.
Conclusions
In summary, to expand the application of the substrate-coupled coenzyme regeneration system, a novel SmADH2 which can tolerate about 80% (v/v) 2-propanol was reported and studied in detail. Then, a TBCR system was built to alleviate adverse effects on enzyme activity and chemical equilibrium caused by the by-product acetone. The newly-designed TBCR system combined with the high 2-propanol tolerance SmADH2 provided an integrated strategy for efficiently synthesising structurally diverse chiral alcohols at a high substrate loading. Furthermore, SmADH2 can be used as a practical reductase to help regenerate coenzymes in other redox reactions due to the low equivalent of 2-propanol used.Experimental
General
All the chemicals and reagents were purchased from Aladdin (Shanghai, China) or other commercial resources. Taq DNA polymerase was purchased from Yisheng (Shanghai, China). pET28a(+) vector (Novagen, Shanghai) present in E. coli DH5α was stored in Luria-Bertani (LB) medium with glycerol/water (10%, v/v) at −40 °C. The affinity chromatographic fillers, Ni–nitrilotriacetic acid (Ni–NTA) agarose, used in protein purification was obtained from SMART (Changzhou, China). Protein concentration was based on the Bradford method, and bovine serum albumin (BSA) was purchased from Tiangen (Shanghai, China). All the operations at DNA, protein or cell level were governed by standard operations or manufacturer's protocols unless additionally stated.Preparation of biocatalyst for synthesizing chiral alcohols
The SmADH2 gene was amplified by polymerase chain reaction (PCR) using the genomic DNA of S. maltophilia as a template with the following primers containing NdeI and BamHI restriction sites (forward 5′-GGAATTCCATATGATTGATTACCAGTTGACCGG-3′; reverse 5′-CGCGGATCCTCACTGCGCCAGGTAGCC-3′). Double-digested by NdeI and BamHI, the gene was then cloned into a pET28a(+) vector to form a recombinant plasmid named pET28a-SmADH2. Following the principle of translation, to-be-expressed proteins carry a His6 tag at the N-terminal site. After analysing the function of proteins by PBLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), a total of 30 SmADHs were constructed using the same strategy. Specific primers and other information are listed in Table S1.† The resulting plasmids were then transformed into E. coli BL21(DE3).The recombinant E. coli BL21(DE3) was cultivated at 37 °C in LB medium with 50 μg ml−1 kanamycin overnight. The 200 ml E. coli seed culture was then inoculated into 4 L fermentation medium (yeast extract, 24 g L−1; tryptone, 12 g L−1; glycerol, 4 g L−1; KH2PO4, 2.31 g L−1; K2HPO4, 12.54 g L−1). Ammonium hydroxide was added to maintain the pH at 6.9, whereas the agitation speed and air volume were adjusted to preserve the dissolved oxygen (DO) at less than 30%. When the OD600nm reached 20, isopropyl β-D-thiogalactopyranoside (IPTG) was added (final concentration 0.5 mM) to facilitate the expression of enzyme at 20 °C for 12 h. Once centrifuged (8000 rpm, 10 min) at 4 °C, the cells were then collected and washed twice with physiological saline solution. Some of the cells were freeze-dried for 24 h to form lyophilised cells, whereas others were resuspended using imidazole phosphate buffer (PBI; 100 mM sodium phosphate (pH 7.0), 100 mM NaCl, 10 mM imidazole) and then crushed into pieces using an ultrasonic oscillator to obtain the cell lysate.
The obtained cell lysate was centrifuged at 8000 rpm for 10 min at 4 °C to separate the cell debris and soluble components. After loading the supernatant on the Ni–NTA fast-flow (FF) column, the absorbed enzyme was eluted by imidazole in PBI with concentrations ranging from 10 to 500 mM. The cell debris, supernatant and collected fractions were analysed by SDS-PAGE. The fractions containing SmADH2 were diluted with sodium phosphate buffer (PBS; 100 mM, p 7.0) and concentrated using an ultrafiltration membrane for desalting. The purified enzyme was stored in PBS with glycerol/water (20%, v/v) at −80 °C.
Enzyme activity assay
The cofactor dependence of each SmADH was first analysed by adding NADH or NADPH separately as the coenzyme into the assay mixtures and by determining the SmADH activity. The activity of SmADHs was measured at 340 nm (ε = 6.220 M−1 cm−1; 30 °C) using a UV spectrophotometer to monitor the decrease in the absorbance of NADPH or NADH quantitatively. To test the reductive activity of the enzyme, the reaction system was designed to include 2 μmol EAA, 0.1 μmol NADPH or NADH, 97 μmol PBS (pH 7.0) and an appropriate amount of the corresponding enzyme in a total volume of 1 ml. As for the oxidative activity, the experiment was conducted, ceteris paribus, using 150 mM 2-propanol as the substrate and 0.1 μmol NADP+ or NAD+ as the cofactor instead. One unit (U) of enzyme activity is defined as the amount of enzyme that catalyses the oxidation reaction of 1 μmol NAD(P)H or the reduction reaction of 1 μmol NAD(P)+ within 1 min.Determination of optimal bioreduction conditions
The optimum temperature was studied considering various temperatures from 15 °C to 65 °C under standard conditions, whereas the optimum pH was tested in 50 mM of the following buffers: sodium citrate (pH 4.0–6.0), sodium phosphate (pH 6.0–8.0), Tris-HCl (pH 8.0–9.0), and glycine–NaOH (pH 9.0–1.0). The enzyme activity under the optimum temperature and pH was defined as 100%.Thermal stability was investigated by incubating the enzyme at different temperatures (20, 30, 40 and 50 °C) for 72 h. During this period, the enzyme activity was measured at regular intervals until the residual activity was less than 50% of the initial level. To study pH stability, SmADH2 was incubated in different buffers at pH values between 4 and 10 at 4 °C for 24 h followed by the measurement of the residual activity.
Tolerance of 2-propanol was studied under standard conditions. A certain amount of purified SmADH2 was added into the reaction mixture with various 2-propanol contents from 20% (v/v) to 80% (v/v). The residual activities were then measured at 340 nm using a UV spectrophotometer.
To investigate the influence of 2-propanol equivalent to substrate on the bioreduction, reactions containing different 2-propanol contents from 1.0 equivalent to 3.0 equivalents were performed in the mixture (20 mM EAA, 100 mM PBS pH 7.0, 0.5 mM NAD+ and an appropriate amount of SmADH2 cells in a total volume of 1 ml) for 24 h at 30 °C. The conversions were detected by GC.
Kinetic parameters
Kinetic parameters for the reduction of EAA and the oxidation of 2-propanol were tested using the purified enzyme. Initially, kinetic parameters for EAA were determined at different EAA concentrations (0.1–5 mM, methanol, 5% v/v) with the concentration of NADH kept at 0.5 mM. Similarly, the apparent Km value for NADH was measured at various NADH concentrations from 0.005 mM to 0.05 mM. As for 2-propanol, the concentrations were set in the range of 0.01–0.5 M with a fixed NAD+ concentration at 0.5 mM. The apparent Km value for NAD+ was measured at different NAD+ concentrations (0.1–5 mM) in the presence of 0.5 M 2-propanol. All data were fitted to the Michaelis–Menten equation and the corresponding Km and Vmax values were calculated using Graphpad Prism v5.0 (GraphPad Software, San Diego, CA, USA).Substrate specificity
The substrate specificity assay was performed to measure the reduction activity of SmADH2. The substrate spectrum contains the data of enzyme activity and stereoselectivity with regard to different substrates. The enzyme activity was tested using 5 mM substrates under standard conditions, determined as a percentage relative to 100% activity (the enzyme activity toward EAA was fixed at 100%). To determine the ee value of corresponding products, the reaction system containing different substrates (final concentration 20 mM) and other substances (100 mM PBS pH 7.0, 0.5 mM NAD+, 25 mM glucose 1 mg lyophilised GDH cells and 1 mg lyophilised SmADH2 cells in a total volume of 1 ml) was shaken for 24 h at 30 °C. Thereafter, the mixture was extracted with ethyl acetate and dried using anhydrous sodium sulphate. The ee value was then analysed by GC and HPLC with the corresponding chiral chromatographic columns (Table S5†).Optimum strategy for asymmetric synthesis of chiral alcohols using SmADH2
Two primers containing EcoRI and HindIII restriction sites (forward 5′-CCGGAATTCAAGGAGATATACATATGTATCCGGATTTAAAAGGAAA-3′; reverse 5′-CCCAAGCTTTTAACCGCGGCCTGCC-3′) were designed using the gdh gene as a template; the Shine–Dalgarno (SD) sequence (AAGGAG) and the aligned spacing (AS) sequence (ATATACAT) were inserted into the primer sd-as-gdhF. The PCR product was double digested and ligated to the plasmid pET28a-SmADH2. The resulting plasmid (pET28a-SmADH2–GDH), which contained both enzyme genes, was transformed into E. coli BL21 (DE3) cells. Co-expression of SmADH2 and GDH was performed as described above. The recombinant E. coli cells were freeze-dried for 24 h to form lyophilised cells. After the enzymatic assay, the activity of SmADH2 expressed in lyophilised co-expression cells was 77% of that in lyophilised SmADH2 cells.The reaction mixture of the enzyme-coupled cofactor regeneration system using co-expressed cells contained 10 ml PBS (100 mM, pH 7.0), 650 mg lyophilised E. coli cells co-expressing SmADH2 and GDH, 5 μmol NAD+, 3.9–6.5 g EAA and 6.8–11.3 g glucose. The aforementioned substances were then added into the TSTR system, and the pH was controlled at 7.0. The bioreaction was performed at 30 °C for 24 h. The reaction mixture of the substrate-coupled cofactor regeneration system contained 10 ml PBS (100 mM, pH 7.0), 500 mg lyophilised SmADH2 cells, 5 μmol NAD+, 3.9–6.5 g EAA and 2.9–4.8 ml 2-propanol. The aforementioned substances were also added into the TSTR system. The bioreaction was performed at 30 °C for 24 h.
To select the optimal strategy for the substrate-coupled regeneration system, two identical portions of reaction mixture containing 10 ml PBS (100 mM, pH 7.0), 500 mg lyophilised cells, 5 μmol NAD+, 6.5 g EAA and 4.8 ml 2-propanol (1.25 equiv.) were placed in the TSTR and TBCR separately and reacted at 30 °C for 24 h. In the TBCR, the gas flow varies between 0.5 and 3 L min−1, whereas the ratio of 2-propanol to water in the buffer unit of the TBCR ranges from 10% to 60% (v/v). A mixture of 10 ml PBS (100 mM, pH 7.0), 500 mg lyophilised cells, 0–5 μmol NAD+, 6.5–7.8 g EAA and 4.8–5.7 ml 2-propanol (1.25 equiv.) was poured into the TBCR (2.5 L min−1 gas flow, 50% 2-propanol in the buffer unit) and maintained at 30 °C for 4 h to decide the optimal amount of lyophilised cells added. To determine the optimum amount of cofactor in the system, 10 ml PBS (100 mM, pH 7.0), 100–500 mg lyophilised cells, 6.5–7.8 g EAA and 4.8–5.7 ml 2-propanol (1.25 equiv.) were reacted in the TBCR (2.5 L min−1 gas flow, 50% 2-propanol in the buffer unit) at 30 °C for 4 h.
Preparation of (R)-EHB and other chiral alcohols in the TBCR system
Based on a substrate-coupled cofactor regeneration system, the bioreductions for six ketones 3, 4, 5, 7, 8 and 26 were performed in a 50 ml TBCR (2.5 L min−1 gas flow, 50% 2-propanol in the buffer unit). Considering EAA as an example, the reaction mixture composed of 10 ml PBS (100 mM, pH 7.0) and 200 mg lyophilised cells were pre-heated under 30 °C for 10 min followed by the addition of 7.8 g EAA and 5.7 ml 2-propanol (1.25 equiv.). After 4 h, the mixture was extracted twice with an equivalent volume of ethyl acetate and centrifuged at 8000 rpm for 10 min. The resulting organic phase was dried using anhydrous sodium sulphate and evaporated to obtain the crude (R)-EHB as a light yellow oily liquid. The molar conversions and ee values for these six ketoesters were detected by HPLC and GC (Table S5, Fig. S6–S11†) and the final products were identified by NMR (Fig. S12–S23†).Modelling, docking and MD simulation
The structural model of SmADH2 was built using the SWISS-MODEL web server (http://www.swissmodel.expasy.org/) based on the crystal structure of an alcohol dehydrogenase (ADH) from Aromatoleum aromaticum EbN1 (PDB code: 4URE) with 49% sequence identity.34 Thereafter, the geometrically optimised NADH was superimposed into the 3D structure of SmADH2 to form the SmADH2/NADH complex.Docking was carried out using AutoDock Vina software (http://vina.scripps.edu/) under default processes using EAA as the ligand and the SmADH2/NADH complex as the receptor molecule. The centre of the grid box was located at the catalytic triad region and each dimension of the grid box was set at 20 Å. Once the docking was finished, the optimal conformation was selected from the 12 results according to the binding energies and the attacking direction of NADH to the carbonyl group.
MD simulation was carried out using the Antechamber package and the BCC method to generate force field parameters. The LEaP module in Amber16 was used to add missing hydrogen atoms. Amber FF14SB force field parameters were used for SmADH2. The final simulation system contained about 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms. Gromacs 4.6.5 was used for running the MD simulation process.
000 atoms. Gromacs 4.6.5 was used for running the MD simulation process.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21776084/B060804) and the Fundamental Research Funds for the Central Universities (22221818014).Notes and references
- S. K. Ma, J. Gruber, C. Davis, L. Newman, D. Gray, A. Wang, J. Grate, G. W. Huisman and R. A. Sheldon, Green Chem., 2010, 12, 81–86 RSC.
- G. Iwasaki, R. Kimura, N. Numao and K. Kondo, Chem. Pharm. Bull., 1989, 37, 280–283 CrossRef CAS.
- F. Y. Qin, B. Qin, T. Mori, Y. Wang, L. X. Meng, X. Zhang, X. Jia, I. Abe and S. You, ACS Catal., 2016, 6, 6135–6140 CrossRef CAS.
- K. Goldberg, K. Schroer, S. Lutz and A. Liese, Appl. Microbiol. Biotechnol., 2007, 76, 249–255 CrossRef CAS.
- P. F. Mugford, U. G. Wagner, Y. Jiang, K. Faber and R. J. Kazlauskas, Angew. Chem., Int. Ed., 2008, 47, 8782–8793 CrossRef CAS.
- S. Ludeke, M. Richter and M. Muller, Adv. Synth. Catal., 2009, 351, 253–259 CrossRef.
- K. Inoue, Y. Makino and N. Itoh, Appl. Environ. Microbiol., 2005, 71, 3633–3641 CrossRef CAS.
- M. J. Sorgedrager, F. van Rantwijk, G. W. Huisman and R. A. Sheldon, Adv. Synth. Catal., 2008, 350, 2322–2328 CrossRef CAS.
- Q. Xu, X. Xu, H. Huang and S. Li, Biochem. Eng. J., 2015, 103, 277–285 CrossRef CAS.
- E. Pekala, A. Godawska-Matysik and D. Zelaszczyk, Biotechnol. J., 2007, 2, 492–496 CrossRef CAS.
- S. Kara, J. H. Schrittwieser, F. Hollmann and M. B. Ansorge-Schumacher, Appl. Microbiol. Biotechnol., 2014, 98, 1517–1529 CrossRef CAS.
- N. Itoh, K. Isotani, M. Nakamura, K. Inoue, Y. Isogai and Y. Makino, Appl. Microbiol. Biotechnol., 2012, 93, 1075–1085 CrossRef CAS.
- K. Inoue, Y. Makino and N. Itoh, Tetrahedron: Asymmetry, 2005, 16, 2539–2549 CrossRef CAS.
- H. Dong, X. M. Li, C. H. Xue and X. Z. Mao, Biotechnol. Prog., 2016, 32, 649–656 CrossRef CAS.
- D. M. Tschaen, L. M. Fuentes, J. E. Lynch, W. L. Laswell, R. P. Volante and I. Shinkai, Tetrahedron Lett., 1988, 29, 2779–2782 CrossRef CAS.
- M. T. Reetz, J. Am. Chem. Soc., 2013, 135, 12480–12496 CrossRef CAS.
- J. S. Clark, T. C. Fessard and G. A. Whitlock, Tetrahedron, 2006, 62, 73–78 CrossRef CAS.
- Y. Yasohara, N. Kizaki, J. Hasegawa, M. Wada, M. Kataoka and S. Shimizu, Biosci., Biotechnol., Biochem., 2000, 64, 1430–1436 CrossRef CAS.
- Y. S. Seo, J. Lim, B. S. Choi, H. Kim, E. Goo, B. Lee, J. S. Lim, I. Y. Choi, J. S. Moon, J. Kim and I. Hwang, J. Bacteriol., 2011, 193, 3149–3149 CrossRef CAS.
- M. Wolberg, M. Villela Filho, S. Bode, P. Geilenkirchen, R. Feldmann, A. Liese, W. Hummel and M. Mueller, Bioprocess Biosyst. Eng., 2008, 31, 183–191 CrossRef CAS.
- M. Amidjojo, E. Franco-Lara, A. Nowak, H. Link and D. Weuster-Botz, Appl. Microbiol. Biotechnol., 2005, 69, 9–15 CrossRef CAS PubMed.
- R. Zhang, Y. Xu and R. Xiao, Biotechnol. Adv., 2015, 33, 1671–1684 CrossRef CAS.
- Z.-N. You, Q. Chen, S.-C. Shi, M.-M. Zheng, J. Pan, X.-L. Qian, C.-X. Li and J.-H. Xu, ACS Catal., 2018, 9, 466–473 CrossRef.
- M. Li, Z. J. Zhang, X. D. Kong, H. L. Yu, J. Zhou and J. H. Xu, Appl. Environ. Microbiol., 2017, 83, e00603-17 CrossRef PubMed.
- V. Prelog, Pure Appl. Chem., 1964, 9, 119–130 CAS.
- A. Li, L. Ye, X. Yang, C. Yang, J. Gu and H. Yu, Chem. Commun., 2016, 52, 6284–6287 RSC.
- N. Itoh, Appl. Microbiol. Biotechnol., 2014, 98, 3889–3904 CrossRef CAS.
- M. Nakasako, J. L. Finney, P. Rand, J. B. F. N. Engberts, R. McKendry and G. Zaccai, Philos. Trans. R. Soc., B, 2004, 359, 1191–1206 CrossRef CAS.
- K. Goldberg, K. Edegger, W. Kroutil and A. Liese, Biotechnol. Bioeng., 2006, 95, 192–198 CrossRef CAS.
- G. W. Huisman, J. Liang and A. Krebber, Curr. Opin. Chem. Biol., 2010, 14, 122–129 CrossRef CAS.
- N. Tanaka, T. Nonaka, K. T. Nakamura and A. Hara, Curr. Org. Chem., 2001, 5, 89–111 CrossRef CAS.
- J. Y. Zhou, Y. Wang, G. C. Xu, L. Wu, R. Z. Han, U. Schwaneberg, Y. J. Rao, Y. L. Zhao, J. H. Zhou and Y. Ni, J. Am. Chem. Soc., 2018, 140, 12645–12654 CrossRef CAS.
- P. K. Agarwal, S. P. Webb and S. Hammes-Schiffer, J. Am. Chem. Soc., 2000, 122, 4803–4812 CrossRef CAS.
- I. Busing, H. W. Hoffken, M. Breuer, L. Wohlbrand, B. Hauer and R. Rabus, J. Mol. Microbiol. Biotechnol., 2015, 25, 327–339 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cy01794a |
| This journal is © The Royal Society of Chemistry 2020 |