 Open Access Article
Open Access ArticleLNA-enhanced DNA FIT-probes for multicolour RNA imaging†
F.
Hövelmann
ab,
I.
Gaspar
b,
J.
Chamiolo
a,
M.
Kasper
a,
J.
Steffen
a,
A.
Ephrussi
b and
O.
Seitz
*a
aDepartment of Chemistry, Humboldt University Berlin, Brook-Taylor-Str. 2, D-12489 Berlin, Germany. E-mail: oliver.seitz@chemie.hu-berlin.de; Fax: +49 30 2093 7266
bEuropean Molecular Biology Laboratory (EMBL) Heidelberg, Meyerhofstr. 1, 69117 Heidelberg, Germany
First published on 2nd November 2015
Abstract
The simultaneous imaging of different RNA molecules in homogeneous solution is a challenge and requires optimisation to enable unambiguous staining of intracellular RNA targets. Our approach relies on single dye forced intercalation (FIT) probes, in which a visco-sensitive reporter of the thiazole orange (TO) family serves as a surrogate nucleobase and provides enhancements of fluorescence upon hybridisation. Previous FIT probes spanned the cyan and green emission range. Herein, we report for the first time chromophores for FIT probes that emit in the red range (above 600 nm). Such probes are valuable to overcome cellular auto-fluorescent background and enable multiplexed detection. In order to find suitable chromophores, we developed a submonomer approach that facilitated the rapid analysis of different TO family dyes in varied sequence positions. A carboxymethylated 4,4′-methine linked cyanine, which we named quinoline blue (QB), provided exceptional response characteristics at the 605 nm emission maximum. Exceeding previously reported base surrogates, the emission of the QB nucleotide intensified by up to 195-fold upon binding of complementary RNA. Owing to large extinction coefficients and quantum yields (up to ε = 129.000 L mol−1 cm−1 and Φ = 0.47, respectively) QB-FIT probes enable imaging of intracellular mRNA. A mixture of BO-, TO- and QB-containing FIT probes allowed the simultaneous detection of three different RNA targets in homogenous solution. TO- and QB-FIT probes were used to localize oskar mRNA and other polyadenylated mRNA molecules in developing oocytes from Drosphila melanogaster by means of wash-free fluorescent in situ hybridisation and super resolution microscopy (STED).
Introduction
Fluorophore-labelled oligonucleotide probes that increase fluorescence emission upon hybridisation with a complementary target are indispensable tools for the detection and imaging of RNA sequences in biological specimens. One of the goals is to enable the simultaneous detection of different targets, which requires a careful optimisation of label and probe design in order to achieve strong signalling without crosstalk between different readout channels. Conventional probes such as molecular beacons contain two labels, a chromophore and a quencher, and furnish up to 102-fold increases of fluorescence upon hybridisation-induced separation of the fluorophore–quencher complex.1,2 Probe designs that rely on quenching by aggregation of two chromophores have shown up to 103-fold increase in fluorescence signalling.3–11 However, substantial optimisation of the distance and the position of two reporters is required. With the exception of nanoparticle based systems,12–15 energy transfer-based probes inevitably cover a larger part of the emission spectrum which limits applications in multicolour RNA imaging. Furthermore, background might be significantly increased in complex media such as cells and lysates due to unspecific binding and reduced dye–dye interaction.5 We inferred that a set of singly labelled probes with chromophores in distinct readout channels would be ideal for RNA multiplexing.Our work relies on quencher-free forced intercalation (FIT)-probes (I, Fig. 1A), in which a single asymmetric cyanine dye of the thiazole orange family serves as an environmentally sensitive nucleobase surrogate.16–19 Hybridisation with a complementary target (I → II) forces the dye to intercalate between predetermined base pairs. The resulting viscosity increase restricts torsions around the methine bridge and prolongs the lifetime of the excited state.20 Previous studies revealed the utility of thiazole orange in real-time PCR,21 wash-free RNA-FISH22 and RNA imaging in living cells.23–29 The combined use of TO and the cyan-emitting BO-reporter allowed the simultaneous detection of two highly expressed viral RNA targets in living cells.23 Yet, the cellular auto fluorescence, caused, amongst others, by riboflavins, pyridoxine and NAD calls for new, red emissive chromophores.30,31
Herein, we present a method for rapid screening of new chromophores in various FIT probe sequences. By using this method, we discovered quinoline blue (QB) as a notably responsive chromophore in FIT-DNA, affording up to 152-fold enhancement of fluorescence upon hybridization with RNA. While studying the influence of next neighbours, we noticed that the incorporation of a “Locked Nucleic Acid” (LNA) adjacent to the dye nucleotide further improved the fluorescence properties. The combined use of BO, TO and QB nucleotides allowed the simultaneous detection of three different RNA strands without crosstalk. Moreover, the newly developed QB-reporter proved useful in dual colour imaging of total mRNA and oskar mRNA in oocytes of Drosophila melanogaster.
Results
The design of dye nucleotides that provide enhancements of fluorescence upon hybridisation and emit in the red spectral range is challenging.32–34 Red emitters typically are larger than blue or green emitters and it has to be considered that large dye nucleotides can perturb duplex formation.35 Furthermore, in order to be useful in wash-free nucleic acid imaging fluorescence turn-on probes should combine a high fold increase of emission with high brightness, and this property should prevail in different sequence contexts.In our previous work on DNA FIT probes we relied on preformed dye nucleotides comprised of TO or JO (oxazolo pyridine) dyes and an open-chain backbone analogue based on serinol.19,22 To facilitate the screening of different dyes, we developed a universal, N-trifluoroacetyl-protected serinol phosphoramidite building block (1) that simplifies the access of FIT probes by post-DNA-synthetic incorporation of chromophores by activated ester coupling (Fig. 1 and 2A). In the search for red emitting FIT probes we explored TR (thiazole red, far red emission)36 as an extended version of TO. Furthermore, we included the 2,4′- and 4,4′-methine-linked quinolinium-quinolines QV (quinoline violet) and QB (quinoline blue, names according to appearance in solution) owing to previous reports about viscosity-dependent radiation-less excited state depletion.37–40 For comparison, we also investigated the known BO (cyan emission) and TO (green emission) nucleotides.19
Synthesis
The chromophores QV, QB and TR were synthesized according to published procedures with slight modification to obtain carboxymethyl linkages, which are crucial for the use in DNA FIT-probes (see Scheme S1†).41 The universal Ser(TfA) phosphoramidite 1 is shelf-stable and was accessed in a 5-step synthesis starting from commercially available L-serine methylester (Scheme S1†). Each chromophore was placed at 15 different positions within a 27mer sequence directed against influenza H1N1-neuraminidase-mRNA (Table 1).42 TO- and BO-labelling was performed by means of the previously reported Ser(BO) and Ser(TO) phosphoramidites.19 For the introduction of the new chromophores, oligonucleotides carrying Ser(Tfa) as a universal precursor were assembled by automated DNA-synthesis (Fig. 2B) and purified by reversed phase (RP)-HPLC. The Tfa group was removed under standard cleavage conditions. Dye-NHS-esters were prepared in DMF by using the chromophore, pyridinium p-toluene sulfonate (PPTS, for enhanced solubility of the chromophores), diisopropylcarbodiimide (DIC) and N-hydroxysuccinimide (NHS) and reacted with the Ser(NH2) oligonucleotide (2) for 1 h at 30 °C (Fig. 2C). Precipitation by isopropanol and subsequent purification by RP-HPLC provided pure FIT-probes 3 in an overall 30–60% yield (Fig. 2D).43| neu- | Sequence | X = Ser(BO) | X = Ser(TO) | X = Ser(QV) | X = Ser(QB) | X = Ser(TR) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I 0 | I | I/I0 | I 0 | I | I/I0 | I 0 | I | I/I0 | I 0 | I | I/I0 | I 0 | I | I/I0 | ||
| a | GGTTTCXGTTATTATGCCGTTGTATTT | 18 | 39 | 2.2 | 4.3 | 29 | 6.7 | 0.5 | 9 | 17.7 | 0.7 | 20 | 28.3 | 94 | 86 | 0.9 |
| b | GGTTTCAXTTATTATGCCGTTGTATTT | 19 | 66 | 3.4 | 8.6 | 48 | 5.6 | 1.1 | 11 | 9.9 | 1.1 | 28 | 25.5 | 101 | 98 | 1.0 |
| c | GGTTTCAGXTATTATGCCGTTGTATTT | 32 | 89 | 2.8 | 5.6 | 68 | 12.2 | 0.9 | 11 | 12.3 | 1.6 | 24 | 15.4 | 105 | 74 | 0.7 |
| d | GGTTTCAGTXATTATGCCGTTGTATTT | 20 | 22 | 1.1 | 4.9 | 18 | 3.7 | 0.5 | 4 | 6.9 | 0.9 | 27 | 30.3 | 98 | 82 | 0.8 |
| e | GGTTTCAGTTXTTATGCCGTTGTATTT | 15 | 46 | 3.0 | 6.2 | 66 | 10.6 | 0.5 | 20 | 39.8 | 0.9 | 58 | 63.6 | 92 | 111 | 1.2 |
| f | GGTTTCAGTTAXTATGCCGTTGTATTT | 36 | 41 | 1.1 | 3.8 | 19 | 5.0 | 0.9 | 4 | 4.6 | 1.1 | 24 | 22.6 | 114 | 91 | 0.8 |
| g | GGTTTCAGTTATXATGCCGTTGTATTT | 37 | 20 | 0.6 | 9.7 | 16 | 1.6 | 1.0 | 4 | 3.7 | 1.4 | 19 | 13.7 | 125 | 86 | 0.7 |
| h | GGTTTCAGTTATTXTGCCGTTGTATTT | 21 | 31 | 1.5 | 8.6 | 54 | 6.2 | 1.0 | 20 | 20.9 | 0.9 | 38 | 42.3 | 117 | 128 | 1.1 |
| i | GGTTTCAGTTATTAXGCCGTTGTATTT | 33 | 18 | 0.6 | 7.6 | 11 | 1.4 | 1.3 | 2 | 1.4 | 1.4 | 9 | 6.3 | 102 | 72 | 0.7 |
| j | GGTTTCAGTTATTATXCCGTTGTATTT | 32 | 45 | 1.4 | 13.5 | 42 | 3.2 | 1.6 | 18 | 11.1 | 3.0 | 30 | 10.1 | 110 | 99 | 0.9 |
| k | GGTTTCAGTTATTATGXCGTTGTATTT | 17 | 99 | 5.6 | 8.7 | 42 | 4.8 | 1.3 | 10 | 8.0 | 1.6 | 21 | 13.6 | 98 | 56 | 0.6 |
| l | GGTTTCAGTTATTATGCXGTTGTATTT | 23 | 79 | 3.7 | 3.5 | 23 | 6.6 | 0.4 | 4 | 10.7 | 0.7 | 29 | 43.7 | 83 | 54 | 0.7 |
| m | GGTTTCAGTTATTATGCCXTTGTATTT | 36 | 50 | 1.4 | 4.2 | 41 | 9.7 | 0.4 | 20 | 49.8 | 0.5 | 74 | 152 | 81 | 142 | 1.8 |
| n | GGTTTCAGTTATTATGCCGXTGTATTT | 17 | 66 | 3.8 | 4.4 | 52 | 12.0 | 0.5 | 7 | 15.3 | 0.8 | 25 | 31.5 | 66 | 104 | 1.0 |
| o | GGTTTCAGTTATTATGCCGTXGTATTT | 25 | 22 | 0.9 | 3.5 | 11 | 3.3 | 0.5 | 2 | 4.7 | 1.2 | 43 | 35.4 | 117 | 79 | 0.7 |
| Mean values | 25 | 49 | 2.2 | 6 | 36 | 6.2 | 1 | 10 | 14.5 | 1 | 31 | 35.6 | 100 | 91 | 0.9 | |
Fluorescence properties
The fluorescence emission of the colour expanded FIT probes was measured before (I0) and after hybridisation (I) with complementary RNA target. The probe concentration was assessed by UV-absorbance measurements and fluorescence emission was corrected accordingly (Table 1 and Fig. 3). Previous experiments had revealed that the introduction of a locked nucleic acid (LNA) monomer adjacent to the dye nucleotide afforded particularly bright DNA FIT probes.28 Yet for simplicity we excluded LNA in the initial screen. The known Ser(TO) nucleotide was walked through the 15 positions of the neuraminidase mRNA probe neu. As shown previously,19 the fluorescence enhancement for TO ranges from 1.6 to 12-fold with 4 probes that show I/I0 ≥ 10. In contrast, the BO-nucleotide provided lower response factors (0.6–5.6-fold), but high overall intensity. Of note, QV-labelled probes furnished up to 50-fold fluorescence enhancement (ranging from 1.4 to 50-fold), but the absolute intensity of QV emission was rather low (∼1/3 of intensity of TO emission, see Table 1 and Fig. 3C). Though low brightness is not an issue for in vitro measurements, we expect limitations in cellular RNA imaging. A remarkably strong fluorescence intensification upon hybridisation (152-fold, neu-QB-m, Fig. 3D) was obtained with the QB-labelled FIT probes. The minimal fluorescence enhancement obtained in this study was still 6.3-fold. In contrast to QV, the intensity of QB was comparable to TO, suggesting a usefulness in RNA imaging studies (vide infra). The TR-containing oligonucleotides showed very bright fluorescence emission. However, the emission was strong in both single stranded and target-bound states and so the TR probes remained virtually non-responsive towards hybridisation to complementary RNA. | ||
| Fig. 3 Absorbance and emission spectra of the most responsive probes carrying (A) BO, (B) TO, (C) QV, (D) QB and (E) TR. Spectra are corrected for probe concentration. Conditions: 0.5 μM probe and 5 eq. RNA target, when added, in PBS at 37 °C. BO: λex = 455 nm; TO: λex = 485 nm; QV: λex = 530 nm; QB: λex = 560 nm; λex = 610 nm; slitex = 5 nm, slitem = 5 nm. Analytical data, absorbance and emission spectra of all probes are provided in the ESI (Table S1–S6 and Fig. S1–S5†). | ||
The study suggested QB as a particularly promising candidate for a red-emitting FIT probe. In the single stranded state, the absorption spectra of the QB-FIT probes showed a rather broad band with a peak at 589–596 nm that is blue-shifted by up to 10 nm and in many instances sharpened upon duplex formation (Fig. 3D, see also Table S5 and Fig. S4†). This and the accompanying increase of fluorescence emission reflects the transition from a low viscosity environment to a more confined high viscosity environment in the duplex form.
Locked nucleic acid-enhanced QB-probes
For thiazole orange it has been demonstrated that the excited state lifetime is reduced by rotations around the central methine bridge.28,44 Recently, we have shown that the introduction of LNA adjacent to the Ser(TO) nucleotide led to increases of the brightness of TO emission,28 presumably because LNA rigidifies the structure of the probe-RNA duplex in a compact, A-type conformation.45 The combined use of 2′-O-Me and LNA in so-called “gapmers” (fully 2′-O-Me-modified probes with a formal serinol-LNA dinucleotide, flanked by a desoxyribonucleotide) conferred nuclease resistance, brightness and responsiveness to the TO-containing FIT probes. We envisioned that this strategy will extend beyond the use of TO and therefore we tested the influence of LNA on QB-labelled probes. We explored the QB base surrogate in the context of sequences directed against vasa mRNA in zebrafish. The experiments shown in Table 1 suggested that the best fluorescence properties were observed when a pyrimidine was 3′-adjacent to the reporter (GxC in neu-BO-k, GxT in neu-TO-c and CxT in neu-QV-m, neu-QB-m and neu-TR-m), while a purine 3′ to the reporter usually gave low fluorescence enhancement (e.g. TxA in neu-BO-g, TxG in neu-TO-o and AxG in neu-QB-i). Therefore, the “QB base” was placed between pyrimidine nucleotides of each of the 4 vasa sequences evaluated. The probes were synthesized as 2′-O-Me-gapmers carrying QB with and without a 3′-adjacent LNA nucleotide (Table 2). Even though the fluorescence properties depend on a multitude of parameters, such as length of the probe, the position of the reporter and the larger sequence context, the rather simplistic sequence guide (avoid purines as 3'-neighbours of QB) proved successful by yielding 14–52-fold enhancement of fluorescence upon hybridisation.| Name | Sequencea, X = Ser(QB) | Φ 0 | Φ | Φ/Φ0 | I/I0 |
|---|---|---|---|---|---|
| a LNA nucleotides are printed in boldface and indicated by subscript “L”, 2′-O-Me nucleotides are underlined. | |||||
| vasa-QB-1 |

|
0.007 | 0.18 | 25.7 | 65.0 |
| vasa-QB-1LNA |

|
0.004 | 0.45 | 112.5 | 195.1 |
| vasa-1-RNA |

|
||||
| vasa-QB-2 |

|
0.004 | 0.31 | 77.5 | 52.2 |
| vasa-QB-2LNA |

|
0.009 | 0.47 | 52.2 | 69.6 |
| vasa-2-RNA |

|
||||
| vasa-QB-3 |

|
0.006 | 0.27 | 45.0 | 42.8 |
| vasa-QB-3LNA |

|
0.007 | 0.47 | 67.1 | 125.2 |
| vasa-3-RNA |

|
||||
| vasa-QB-4 |
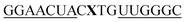
|
0.020 | 0.24 | 12.0 | 14.0 |
| vasa-QB-4LNA |

|
0.012 | 0.45 | 37.5 | 68.9 |
| vasa-4-RNA |

|
||||
As previously reported for TO-labelled probes28 the fluorescence intensity and quantum yield (I and Φ, respectively) of QB-containing probe-target duplexes increased when an LNA rather than a DNA building block was placed adjacent to the Ser(QB) nucleotide (Table 2 and Fig. 4). Again, the response factor I/I0 proved to be extremely high (69–195-fold, see also Table S8 and Fig. S6†). Such strong enhancements of fluorescence upon hybridization of single fluorophore labelled oligonucleotides with RNA are, to the best of our knowledge, without precedence. The high quantum yield (up to 0.47) of the probe-target complex combines with the sharpening of the absorption (Fig. 4) which leads to a hybridization induced increase of the extinction coefficient (ε(max) = 129.000 L mol−1 cm−1). As a result, the achievable brightness (upon excitation at the absorption maximum) amounted to 55–58 L mmol−1 cm−1 for the LNA-flanked QB-FIT probes, while QB-FIT probes without LNA afforded brightness values in a range of 13–16 L mmol−1 cm−1 (see Table S8†). This compares with commonly used fluorescence labels in oligonucleotides46 such as fluorescein (48 L mmol−1 cm−1) or TAMRA (69 L mmol−1 cm−1), which provide bright fluorescence, yet are virtually nonresponsive.47
Multi-colour readout
The ability to simultaneously detect multiple RNA targets is envisioned to facilitate RNA imaging experiments, particularly when different RNAs of interest or splicing variants are to be detected. The fluorescence spectra of BO, TO and QB can be resolved and cover the cyan (BO), green (TO) and red (QB) range of the visible light spectrum. Thus, multi-colour readout by FIT probes can be carried out by standard fluorescence microscopy equipment. The Ser(BO)-, Ser(TO)- and Ser(QB)-nucleotides were included in three gapmer FIT probes targeting three different RNA sequences. The second LNA building block in the BO-probe was included to improve the target affinity of the rather short oligonucleotide. The three readout channels were simultaneously detected in a kinetic experiment (Fig. 5). The probes were sequentially added to buffer (BO: 3 min, TO: 7 min, QB: 10 min) followed by sequential addition of each complementary RNA (BO-RNA: 13 min, TO-RNA: 17 min, QB-RNA: 20 min). The ascending order from blue to red emission range was chosen to assess potential cross-excitation. In spite of the combination of probes and the excess of targets, neither cross-excitation nor energy transfer of the different readout channels was observed. The addition of probe resulted immediately in a stable signal, indicating the absence of folding or aggregation. Furthermore, the probes responded rapidly to the addition of target RNA and within seconds afforded 20–69-fold enhancements of fluorescence, while non-complementary RNA produced only a marginal increase in fluorescence, if at all. The near instantaneous response points to one of the advantages provided by non-structured probes not shared by molecular beacon type probes.48 Owing to the LNA enhancement, the probes exhibited fluorescence of comparable intensities, which will facilitate multiple colour readouts in cells. The low background signal and the high brightness of FIT probes in their target-bound state allowed the detection of complementary target at low μM concentrations by the naked eye upon excitation with a handheld UV-lamp (see Fig. S7†).Wash-free FISH
To assess the usefulness of QB-labelled probes in multicolour RNA imaging, we set out to analyse the localisation of oskar mRNA and total mRNA in fixed Drosophila egg-chambers by wash-free FISH.22 The egg-chamber consists of an oocyte connected to its 15 sibling nurse cells and is encapsulated by a single layer of follicular epithelial cells. To demonstrate that multiplexing works in situ, we used an 18mer LNA-modified oligo-dT-probe (TTTTTTTT-Ser(QB)-TLTTTTTTTT) dT(18)-QB targeted against the poly-A tail of mRNA, which should be distributed all over the specimen, and a mixture of five LNA-modified TO-labeled probes targeted against oskar mRNA (osk-TO-LNA1-5), which should localize at the posterior pole of the oocyte.28 In cuvette experiments, the oligo-dT-probe provided 89-fold enhancement of QB emission (605 nm) upon hybridisation with complementary RNA (Fig. S8†). Previously, we have shown that the oskar mRNA specific TO-FIT probes afforded response factors I/I0 = 6–13 (Table S9†).28The oskar mRNA is produced in the nurse cells and transported to the posterior pole of the oocytes. The fluorescence microscopy analysis (Fig. 6A and A′) confirmed that the TO signal was mainly detected at the posterior, where oskar localizes.19,28 In contrast, imaging QB showed a more uniform distribution of the poly-dT probe, as expected. Interestingly, however, we observed a mild accumulation of QB signals at the posterior pole (Fig. 6A′′, yellow arrow), where not only oskar but many other mRNAs were shown to enrich.49 Accordingly, when we performed super-resolution imaging on the posterior pole of stage 10 oocytes using stimulated emission depletion (STED) microscopy, we found that many but not all dT18-QB positive particles are positive for oskar (Fig. 6D, see also Fig. S10A and B†). As a control, staining was attempted with the vasa-QB-2LNA probe (Fig. S9†). This probe has no target in Drosophila melanogaster and therefore only showed dim background signals.
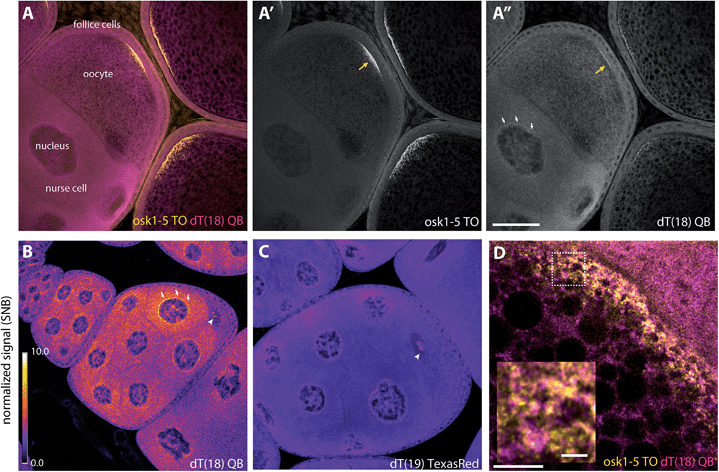 | ||
| Fig. 6 Wash-free FISH of fixed Drosophila egg chambers. (A–A′′) Egg chambers comprising the translationally active nurse cells interconnected to the meiotic oocyte, surrounded by a layer of somatic follicular epithelium were co-labelled with a mixture of (A′) five TO-labelled oskar probes (osk-TO-LNA1-5, 0.1 μM each) and (A′′) dT(18) (0.5 μM) QB. (B and C) Normalized images of egg chambers labelled with (B) dT18-QB, (C) TexasRed-labelled dT19 (0.5 μM). Scale bars represent 50 microns (A–C). Normalization was done against the background signal measured within the oocyte nuclei (Fig. S10B,† cyan dashed circle). White arrows indicate accumulated RNA around the nurse cell nuclei; yellow arrows indicate accumulation of oskar mRNA at the posterior pole; white arrowheads indicate speckles in the oocyte nucleus. (D) Super-resolution image (STED) of the oocyte posterior region of a stage 10 egg chamber, where oskar mRNA is enriched. The inset is an enlarged version of the image area within the dashed box. Scale bar represents five microns; scale bar in the inset represents one micron. Wash-free FISH of fixed Drosophila egg chambers. | ||
We also found accumulation of the oligo-dT-QB probe around the nurse cell nuclei (Fig. 6A′′, white arrows). These cells – unlike the oocyte – are transcriptionally highly active, yet a large fraction of the synthesized mRNA species enrich within the oocyte.49 We think that this accumulation of poly-A(+) mRNA around the nurse cell nuclei could represent molecules awaiting to be actively transported into the oocyte. One of such molecules is oskar mRNA itself,50 and similar to what we observed at the oocyte posterior pole, a fraction of dT18-QB particles co-localized with oskar (Fig. S9C and D†). Interestingly, the dT18-QB-stained particles within the perinuclear region are relatively large and appear to be regularly spaced (Fig. S10C and D†). Another previously undetected enrichment of dT probe signal was observed within the meiotic and thus translationally silent nucleus of the oocyte (Fig. 6B, arrowhead), where we observed speckles of unknown nature (Fig. S9, ESI movie†). Of note, these speckles also appeared when staining was performed with Texas Red labelled dT19-probes, though contrast after normalization was significantly lower than for the dT18-QB probe (Fig. 6C).
Discussion
The aim of this work was to expand the colour repertoire of DNA-based FIT-probes in order to enable multicolour RNA imaging. To streamline the screening for suitable dyes we established a method for the synthesis of DNA FIT probes based on a universal N-Tfa-protected serinol phosphoramidite, which allowed the post-synthetic coupling of dye-NHS-esters onto oligonucleotides. Similar methods based on NHS-ester coupling are routinely used for terminal labelling of e.g. molecular beacons or for modification of nucleobases in ECHO-probes.3,51 The coupling succeeded on small scale and as a result the method greatly facilitated the study of different chromophores in equal sequence contexts.We identified a 4,4′-linked cyanine as a particularly responsive chromophore in FIT-probes which provided 102-fold fluorescence enhancement upon hybridization with RNA target. The dye has been the subject of intense photophysical studies in which the barrier-less isomerization was scrutinized in fine detail.38,40,52–56 The viscosity-dependence of excited state depletion by photoisomerisation is a property common to many monomethine chromophores of the thiazole orange family of DNA intercalators. To illustrate this relation we coined the term quinoline blue (describes structural detail and colour in solution). Like many TO family members QB has low fluorescence in free form,56 yet experiences dramatic enhancements of fluorescence upon binding to DNA. This has been noticed previously when QB was attached to terminal and internal positions of oligonucleotides by means of 8–12 atom-long linkers.39 However, the fluorescence of the previously explored DNA-conjugates was high regardless of the hybridization state. This illustrates a key feature of FIT probes. The use of a very short carboxymethyl linker presumably provides steric hindrance and perturbs dye–nucleotide interactions in cis and in trans. As a result, FIT probes remain dark unless hybridization with a target molecule enforces the intercalation between predetermined base pairs. The accompanying viscosity increase hinders torsional motions around the methine bridge so that fluorescence can occur. This effect is augmented when LNA is placed adjacent to FIT-reporters. As previously reported for TO, a single LNA adjacent to the QB reporter results in significantly increased quantum yield (DNA(mean) Φ = 0.25; LNA(mean) Φ = 0.46) and brightness (see also Table S8†).28,57 Similar improvements of the photophysical properties of dyes on introduction of flanking LNA monomers have recently been reported by Hrdlicka.57 Moreover, the emission in the red range (575–700 nm) is orthogonal to BO and TO and allows RNA multiplexing without any false positive signalling by the presence of other probes or non-complementary RNA target.
The fluorescence microscopic analysis of developing oocytes from Drosophila melanogaster revealed the localization of two different mRNA targets. The poly-A-tail specific QB-FIT probes revealed a quite uniform distribution of total mRNA in the specimen with enrichment at the perinuclear region of the transcriptionally active nuclei of the nurse cells. The discovery of speckles in the transcriptionally silent nucleus of the oocyte observed with the QB-FIT probes and, at lower overall contrast, with Texas Red-dT19 probes is noteworthy. The combined use of the two spectrally resolvable probes confirmed the enrichment of mRNA at the posterior pole from mid-oogenesis onwards, including oskar mRNA and other poly-A tail-containing mRNA molecules.
Conclusions
We developed a method for the simplified screening of DNA FIT probes that respond to hybridisation by showing enhancement of fluorescence in the red spectral range. The synthesis method relies upon an N-trifluoroacetyl-protected serinol phosphoramidite, which is introduced during automated DNA synthesis and allows the solution-phase coupling of thiazole orange family cyanine dyes. According to the design criteria for forced intercalation probes, the obtained oligonucleotide conjugates contain a carboxymethyl-linked cyanine dye rather than a nucleobase at a sequence-internal position. The comparison of 5 different dyes in 15 different sequence positions revealed a quinoline based 4,4′-cyanine (QB) as a particularly suitable dye. Among the single-labelled hybridisation probes reported in the literature, QB-FIT probes provide the highest fluorescence enhancement (up to 195-fold) upon hybridisation with complementary RNA. The red emission (λem(max) ≈ 605 nm) is orthogonal to the previously reported BO- and TO-FIT probes and allows their combined use in multiplexing experiments. The brightness of QB can be significantly improved by introducing an adjacent LNA-nucleotide. The usefulness of the QB dye in FIT probes was demonstrated in dual colour RNA imaging in developing oocytes from Drosophila melanogaster. The combined use of five different TO-FIT probes targeted against oskar mRNA and a QB-FIT probe designed for recognition of the mRNA poly-A tail revealed the different localization of the two targets when used in wash-free fluorescent in situ hybridisation, and exposed hitherto unknown signals from fluorescence-labelled oligo-dT probes in the meiotic nucleus of the oocyte. In this investigation we also showed for the first time super-resolution microscopy with FIT probes by stimulated emission depletion (STED). Based on the robustness and brightness of fluorescence signalling by QB-FIT probes and the documented usefulness of TO-FIT probes we are convinced that FIT probes will probe useful in many other multiplexed RNA imaging endeavours.Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft, the Einstein-Stiftung Berlin and the EMBL.References
- S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS PubMed.
- R. Häner, S. M. Biner, S. M. Langenegger, T. Meng and V. L. Malinovskii, Angew. Chem., Int. Ed., 2010, 49, 1227–1230 CrossRef PubMed.
- S. Ikeda and A. Okamoto, Chem.–Asian J., 2008, 3, 958–968 CrossRef CAS PubMed.
- E. Socher, A. Knoll and O. Seitz, Org. Biomol. Chem., 2012, 10, 7363–7371 CAS.
- H. Asanuma, M. Akahane, R. Niwa, H. Kashida and Y. Kamiya, Angew. Chem., Int. Ed., 2015, 54, 4315–4319 CrossRef CAS PubMed.
- C. Holzhauser and H. A. Wagenknecht, Angew. Chem., Int. Ed., 2011, 50, 7268–7272 CrossRef CAS PubMed.
- S. Berndl and H. A. Wagenknecht, Angew. Chem., Int. Ed., 2009, 48, 2418–2421 CrossRef CAS PubMed.
- H. Asanuma, T. Osawa, H. Kashida, T. Fujii, X. G. Liang, K. Niwa, Y. Yoshida, N. Shimadad and A. Maruyama, Chem. Commun., 2012, 48, 1760–1762 RSC.
- D. O. Wang and A. Okamoto, J. Photochem. Photobiol., C, 2012, 13, 112–123 CrossRef CAS.
- A. Okamoto, Chem. Soc. Rev., 2011, 40, 5815–5828 RSC.
- T. Kubota, S. Ikeda, H. Yanagisawa, M. Yuki and A. Okamoto, PLoS One, 2010, 5, e13003 Search PubMed.
- N. Li, C. Chang, W. Pan and B. Tang, Angew. Chem., Int. Ed., 2012, 51, 7426–7430 CrossRef CAS PubMed.
- X. H. Peng, Z. H. Cao, J. T. Xia, G. W. Carlson, M. M. Lewis, W. C. Wood and L. Yang, Cancer Res., 2005, 65, 1909–1917 CrossRef CAS PubMed.
- A. E. Prigodich, P. S. Randeria, W. E. Briley, N. J. Kim, W. L. Daniel, D. A. Giljohann and C. A. Mirkin, Anal. Chem., 2012, 84, 2062–2066 CrossRef CAS PubMed.
- G. Qiao, Y. Gao, N. Li, Z. Yu, L. Zhuo and B. Tang, Chem.–Eur. J., 2011, 17, 11210–11215 CrossRef CAS PubMed.
- O. Seitz, F. Bergmann and D. Heindl, Angew. Chem., Int. Ed., 1999, 38, 2203–2206 CrossRef CAS.
- O. Köhler and O. Seitz, Chem. Commun., 2003, 2938–2939, 10.1039/b308299g.
- O. Kohler, D. Venkatrao, D. V. Jarikote and O. Seitz, ChemBioChem, 2005, 6, 69–77 CrossRef PubMed.
- F. Hövelmann, L. Bethge and O. Seitz, ChemBioChem, 2012, 13, 2072–2081 CrossRef PubMed.
- V. Karunakaran, J. L. Perez Lustres, L. Zhao, N. P. Ernsting and O. Seitz, J. Am. Chem. Soc., 2006, 128, 2954–2962 CrossRef CAS PubMed.
- E. Socher, D. V. Jarikote, A. Knoll, L. Röglin, J. Burmeister and O. Seitz, Anal. Biochem., 2008, 375, 318–330 CrossRef CAS PubMed.
- F. Hövelmann, I. Gaspar, A. Ephrussi and O. Seitz, J. Am. Chem. Soc., 2013, 135, 19025–19032 CrossRef PubMed.
- S. Kummer, A. Knoll, E. Socher, L. Bethge, A. Herrmann and O. Seitz, Bioconjugate Chem., 2012, 23, 2051–2060 CrossRef CAS PubMed.
- S. Kummer, A. Knoll, E. Socher, L. Bethge, A. Herrmann and O. Seitz, Angew. Chem., Int. Ed., 2011, 50, 1931–1934 CrossRef CAS PubMed.
- Y. Kam, A. Rubinstein, A. Nissan, D. Halle and E. Yavin, Mol. Pharm., 2012, 9, 685–693 CrossRef CAS PubMed.
- M. V. Sonar, M. E. Wampole, Y. Y. Jin, C. P. Chen, M. L. Thakur and E. Wickstrom, Bioconjugate Chem., 2014, 25, 1697–1708 CrossRef CAS PubMed.
- Y. Kam, A. Rubinstein, S. Naik, I. Djavsarov, D. Halle, I. Ariel, A. O. Gure, A. Stojadinovic, H. Pan, V. Tsivin, A. Nissan and E. Yavin, Cancer Lett., 2014, 352, 90–96 CrossRef CAS PubMed.
- F. Hövelmann, I. Gaspar, S. Loibl, E. A. Ermilov, B. Röder, J. Wengel, A. Ephrussi and O. Seitz, Angew. Chem., Int. Ed., 2014, 53, 11370–11375 CrossRef PubMed.
- A. G. Torres, M. M. Fabani, E. Vigorito, D. Williams, N. Al-Obaidi, F. Wojciechowski, R. H. Hudson, O. Seitz and M. J. Gait, Nucleic Acids Res., 2012, 40, 2152–2167 CrossRef CAS PubMed.
- I. Georgakoudi, B. C. Jacobson, M. G. Muller, E. E. Sheets, K. Badizadegan, D. L. Carr-Locke, C. P. Crum, C. W. Boone, R. R. Dasari, J. van Dam and M. S. Feld, Cancer Res., 2002, 62, 682–687 CAS.
- W. R. Zipfel, R. M. Williams, R. Christie, A. Y. Nikitin, B. T. Hyman and W. W. Webb, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 7075–7080 CrossRef CAS PubMed.
- L. Bethge, D. V. Jarikote and O. Seitz, Bioorg. Med. Chem., 2008, 16, 114–125 CrossRef CAS PubMed.
- S. Ikeda, T. Kubota, M. Yuki and A. Okamoto, Angew. Chem., Int. Ed., 2009, 48, 6480–6484 CrossRef CAS PubMed.
- C. Holzhauser, S. Berndl, F. Menacher, M. Breunig, A. Göpferich and H. A. Wagenknecht, Eur. J. Org. Chem., 2010, 1239–1248, DOI:10.1002/ejoc.200901423.
- H. Asanuma, M. Akahane, N. Kondo, T. Osawa, T. Kato and H. Kashida, Chem. Sci., 2012, 3, 3165–3169 RSC.
- S. Ikeda, H. Yanagisawa, A. Nakamura, D. O. Wang, M. Yuki and A. Okamoto, Org. Biomol. Chem., 2011, 9, 4199–4204 CAS.
- A. Miethe and G. Book, Ber. Dtsch. Chem. Ges., 1904, 37, 2821–2824 CrossRef CAS.
- V. Sundström and T. Gillbro, J. Phys. Chem., 1982, 86, 1788–1794 CrossRef.
- R. Lartia and U. Asseline, Chem.–Eur. J., 2006, 12, 2270–2281 CrossRef CAS PubMed.
- V. Sundström and T. Gillbro, Chem. Phys., 1981, 61, 257–269 CrossRef.
- L. Bethge, I. Singh and O. Seitz, Org. Biomol. Chem., 2010, 8, 2439–2448 CAS.
- W. Wang, Z. Q. Cui, H. Han, Z. P. Zhang, H. P. Wei, Y. F. Zhou, Z. Chen and X. E. Zhang, Nucleic Acids Res., 2008, 36, 4913–4928 CrossRef CAS PubMed.
- A detailed labelling protocol is provided in the ESI†.
- D. V. Jarikote, N. Krebs, S. Tannert, B. Röder and O. Seitz, Chem.–Eur. J., 2007, 13, 300–310 CrossRef CAS PubMed.
- K. E. Nielsen, J. Rasmussen, R. Kumar, J. Wengel, J. P. Jacobsen and M. Petersen, Bioconjugate Chem., 2004, 15, 449–457 CrossRef CAS PubMed.
- L. Wang, A. K. Gaigalas, J. Blasic and M. J. Holden, Spectrochim. Acta, Part A, 2004, 60, 2741–2750 CrossRef CAS PubMed.
- Brightness values are calculated based on the published data for extinction coefficients (93000 L mol−1 cm−1 for Fluorescein and 97000 L mol−1 cm−1 for TAMRA) and qunatum yields (see ref. 46).
- T. N. Grossmann, L. Röglin and O. Seitz, Angew. Chem., Int. Ed., 2007, 46, 5223–5225 CrossRef CAS PubMed.
- H. Jambor, V. Surendranath, A. T. Kalinka, P. Mejstrik, S. Saalfeld and P. Tomancak, eLife, 2015, 4, e05003 Search PubMed.
- S. C. Little, K. S. Sinsimer, J. J. Lee, E. F. Wieschaus and E. R. Gavis, Nat. Cell Biol., 2015, 17, 558–568 CrossRef CAS PubMed.
- A. Okamoto, S. Ikeda and T. Kubota, EP2130835B1, 2008.
- U. Åberg, E. Åkesson and V. Sundström, Chem. Phys. Lett., 1993, 215, 388–394 CrossRef.
- U. Åberg, E. Åkesson, J. L. Alvarez, I. Fedchenia and V. Sundström, Chem. Phys., 1994, 183, 269–288 CrossRef.
- J.-L. Alvarez, A. Yartsev, U. Åberg, E. Åkesson and V. Sundström, J. Phys. Chem. B, 1998, 102, 7651–7658 CrossRef CAS.
- Q.-H. Xu and G. R. Fleming, J. Phys. Chem. A, 2001, 105, 10187–10195 CrossRef CAS.
- C. A. Guarin, J. P. Villabona-Monsalve, R. López-Arteaga and J. Peon, J. Phys. Chem. B, 2013, 117, 7352–7362 CrossRef CAS PubMed.
- M. Kaura and P. J. Hrdlicka, Org. Biomol. Chem., 2015, 13, 7236–7247 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis, NMR spectra, fluorescence and absorbance spectra, analytical data of probes and super resolution images. See DOI: 10.1039/c5sc03053f |
| This journal is © The Royal Society of Chemistry 2016 |




