Open Access Article
Open Access ArticleBiomimetic enzyme-linked immunoassay based on a molecularly imprinted 96-well plate for the determination of triazophos residues in real samples
Sihui Honga,
Yongxin She *a,
Xiaolin Caoa,
Miao Wang
*a,
Xiaolin Caoa,
Miao Wang a,
Chao Zhanga,
Lufei Zhenga,
Shanshan Wanga,
Xingbin Mab,
Hua Shaoa,
Maojun Jina,
Fen Jina and
Jing Wang*a
a,
Chao Zhanga,
Lufei Zhenga,
Shanshan Wanga,
Xingbin Mab,
Hua Shaoa,
Maojun Jina,
Fen Jina and
Jing Wang*a
aInstitute of Quality Standard and Testing Technology for Agro-Products, Chinese Academy of Agricultural Science, Key Laboratory of Agro-Products Quality and Safety of Chinese Ministry of Agriculture, Beijing 100081, P. R. China. E-mail: 0891syx@163.com; Fax: +86 010 82106567; Tel: +86 10 82106513
bInstitute of Veterinary and Animal Husbandry, Tibet Academy of Agricultural and Animal Husbandry Sciences, Lhasa 850006, P. R. China
First published on 5th June 2018
Abstract
In this study, a direct competitive biomimetic enzyme-linked immune-sorbent assay (BELISA) based on a molecularly imprinted nanomembrane as an artificial antibody was developed for the determination of triazophos in real samples. The imprinted film was directly synthesized on the well surface of a 96-well plate using a dummy molecular template under the conditions of thermal polymerization. The developed BELISA using a hapten of triazophos as an enzyme-labeled probe is much more sensitive, simple, quick, steady and inexpensive than the other instrumental and immuno assay methods. Under optimal conditions, the linear range of the method was 0.001–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg L−1 with a good regression coefficient of 0.977. The sensitivity (IC50) and the limit of detection (LOD) of BELISA were 428 μg L−1 and 0.001 μg L−1, respectively. This method was performed to detect triazophos in cabbage and apple samples, and showed excellent recovery and relative standard deviations (RSDs) ranging from 70.5 to 119.8% and from 5.2 to 19.7%, respectively. The results correlated well with those obtained using high performance liquid chromatography-tandem mass spectrometry.
000 μg L−1 with a good regression coefficient of 0.977. The sensitivity (IC50) and the limit of detection (LOD) of BELISA were 428 μg L−1 and 0.001 μg L−1, respectively. This method was performed to detect triazophos in cabbage and apple samples, and showed excellent recovery and relative standard deviations (RSDs) ranging from 70.5 to 119.8% and from 5.2 to 19.7%, respectively. The results correlated well with those obtained using high performance liquid chromatography-tandem mass spectrometry.
1. Introduction
Triazophos is a moderately toxic, broad-spectrum organophosphate pesticide that has been widely used in agriculture to control insect pests.1 Due to its relative stability, widespread use and long half-life, residues of triazophos in agricultural products and the natural environment may result in potential hazards to human health and animals.2 Since the end of 2016, triazophos has been banned for use on vegetables by the Chinese Ministry of Agriculture, but the illegal use of triazophos for vegetables and fruits still happens now and then. Therefore, it is important to develop a highly sensitive and selective method for the detection of triazophos residues in agricultural and environmental samples.3So far, various analytical methods for the effective monitoring of triazophos have been developed, such as gas chromatography (GC),4 gas chromatography with mass spectrometry (GC/MS),5 liquid chromatography with mass spectrometry (LC/MS),6 and enzyme/antibody-based immunoassays.7 The LC, GC method, or that coupled with MS, are common methods with high sensitivity and accuracy and good reproducibility, but the instrument size, the high costs, the need for skilled technicians and the lengthy analysis time limit their wide utilization and direct field detection.8 Immunoassays, especially enzyme-linked immunosorbent assays (ELISA), are also sensitive and selective and can run many analyses simultaneously without the need for skilled technicians.9 However, they have many drawbacks, including the high commercial cost, reagent stability, difficulties associated with antibody production and the use of laboratory animals.9 Thus, it is necessary to develop a sensitive, simple, quick, stable and inexpensive method to monitor the residual quantity of triazophos in real samples.
Molecular imprinting is a method used to create selective recognition sites in synthetic polymers.10 The molecularly imprinted polymers (MIPs) have properties such as predictable specific recognition, easy preparation, low cost, high mechanical/chemical stability and good rigidity.11,12 Due to the above advantages, MIPs are described as artificial antibodies that can replace antibodies and have been applied in some biomimetic enzyme-linked immunosorbent assay (BELISA) methods.13–16
Usually, it is difficult to fully eliminate the molecular template from MIPs, so leakage of the molecular template during the use of MIPs resulted in inaccuracy of the analytical results. Triadimefon, whose structure is similar to triazophos but has a lower toxicity, would be available as a dummy template molecule to further reduce the number of false positive results. In this study, we chose triadimefon as a molecular template to synthesize MIPs as a recognition unit on a 96-well plate and also constructed a labelling probe using a hapten of triazophos labelled as horseradish peroxidase. Subsequently, a sensitive, simple, quick, and directly competitive BELISA method for the detection of triazophos was developed, using the imprinted film as an artificial antibody. The parameters affecting the performance of the BELISA were optimized in detail. This developed method was applied to the analysis of triazophos in cabbage and apple samples, and the results were validated by HPLC-MS/MS.
2. Materials and methods
2.1 Reagents and chemicals
Triazophos was obtained from Aladdin Chemistry Co. Ltd. (Shanghai, China). Parathion, chlorpyrifos, triadimefon, and methomyl were purchased from Dr Ehrenstorfer Gmbh (Augsburg, Germany). Methacrylic acid (MAA) and trimethylolpropane trimethacrylate (TRIM) were purchased from Alfa Aesar (Massachusetts, USA). The free radical initiator 2,2-azobisisobutyronitrile (AIBN) was purchased from the no. 4 reagent and H. V. Chemical Co., Ltd. (Shanghai, China). The triazophos hapten was gifted by the Institute of Pesticide and Environmental Toxicology, Zhejiang University, China. Horseradish peroxidase (HRP) was bought from Solarbio (Beijing, China). N-Dicyclohexylcarbodiimide (DCC), and N-hydroxysuccinate (NHS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The 3,3′,5,5′-tetramethylbenzidine (TMB) ELISA substrate was purchased from TransGen Biotech Co., Ltd (Beijing, China). All the other chemicals and organic solvents used in this study were of the highest available purity and at least of analytical grade. Maxisorp polystyrene 96-well plates were obtained from Corning, Inc. (Corning, NY, USA). Ultrapure water (≥18.2 MΩ cm) produced by a Milli-Q water purification system (Millipore, Bedford, MA, USA) was used in all experimental works.An assay buffer consisting of 10 mM PBS (pH 7.4), a washing buffer (PBST) (consisting of 10 mM PBS (pH 7.4) and 0.05% Tween 20) and a stopping solution (2 M H2SO4) were used in this study.
2.2 Preparation of the molecularly imprinted film on the surface of the 96-well plate
The imprinted film was directly polymerized in the wells of the 96-well plate according to the following procedure: 588 mg (2 mmol) of triadimefon was dissolved in 20 mL of acetonitrile in a 150 mL flask, and then mixed with 1020 μL (12 mmol) of MAA. After magnetically stirring for 30 min at room temperature, 1916 μL (6 mmol) of TRIM and 50 mg of AIBN were added in turn. The mixture was continuously stirred for 60 min. Then, 100 μL of the mixture was placed in each well of the 96-well plate and the reaction was carried out under vacuum for 24 h at 37 °C by a vacuum oven.After polymerization was complete, the plate was washed three times with DDW and methanol, respectively, to remove any unreacted solution. The imprinted film was first extracted with methanol/acetic acid (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) followed by methanol for 4 h using an ultrasonic cleaner until no triadimefon was detected in the eluent by HPLC. For comparison, a non-imprinted film was synthesized by the same procedure as above without the addition of triadimefon. The process is shown in Fig. 1(a).
1, v/v) followed by methanol for 4 h using an ultrasonic cleaner until no triadimefon was detected in the eluent by HPLC. For comparison, a non-imprinted film was synthesized by the same procedure as above without the addition of triadimefon. The process is shown in Fig. 1(a).
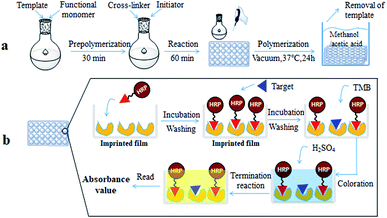 | ||
| Fig. 1 Preparation of the molecularly imprinted film on the surface of the 96-well plate (a); direct competitive BELISA procedure (b). | ||
2.3 Characterization of the imprinted film
The morphological characterization was analyzed using a field-emission scanning electron microscope (JSM-6300, Japan). The bonding effects in the polymers were evaluated by Fourier transform infrared (FT-IR) spectra (4000–400 cm−1) with a Philips PU9800 spectrometer (Philips Analytical, Cambridge, UK).2.4 Adsorption dynamic experiments
To evaluate the binding kinetics of the imprinted film, 200 μL of PBS solution containing triazophos (at 100 mg L−1) was added to the 96-well plate. After shaking for 5, 10, 20, 30, 40, 50, 60, 70, and 80 min at room temperature, the supernatants were analyzed by HPLC.To measure the adsorption capacity, the imprinted or non-imprinted film was equilibrated with 200 μL of PBS solution containing triazophos at various concentrations (10–100 mg L−1). After shaking for 50 min at room temperature, the supernatants were analyzed by HPLC. The polymer adsorption capacity (Q, μg per well) was calculated using eqn (1).17
| Q = (C0–C1)V × 1000 | (1) |
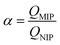 | (2) |
2.5 Synthesis of the enzyme-labeled probe of triazophos
The hapten of triazophos was supplied by Zhejiang University. The synthesis of the enzyme-labeled probe of triazophos was based on the literature19 with certain modifications. First, 18.85 mg (0.05 mmol) of the hapten of triazophos was dissolved in 0.5 mL of DMF as solution (a). Next, 17.25 mg (0.15 mmol) of NHS was added to solution (a) and stirred for 15 min as solution (b). Then, 15.46 mg (0.075 mmol) of DCC was dissolved in 0.5 mL of DMF, and added to solution (b) drop by drop. The mixture was magnetically stirred at room temperature over-night. After centrifugation (4000 rpm, 10 min), the supernatant was preserved as solution (c). Solution (d) was prepared by adding 40 mg of HRP to 4 mL of 10 mmol L−1 CBS solution. After that, 400 μL of solution (c) was added dropwise to solution (d), and the mixture was stirred for 4 h at room temperature. The enzyme conjugated solution was then dialyzed against PBS (10 mmol L−1). Finally, the volume of the conjugated enzyme was accurately calculated, and the same volume of glycerine was added to the solution, and stored at −20 °C before use.2.6 Direct competitive BELISA procedure
The direct competitive BELISA procedure was carried out using the imprinted film on the 96-well plate as a biomimetic antibody. First, the plate was washed three times with PBST (PBS with 0.05% (v/v) Tween 20). Then, 100 μL of PBS was added to the blank, 100 μL of the enzyme labelled probe was added to all wells except for the blank, and the 96-well plate was shaken for 30 min at room temperature. After that, the plate was washed with PBST ten times. Next, 5% methanol PBS (v/v) was added to the blank (100 μL) and the control (100 μL) wells, 100 μL of the standard solution or sample extracts were added to the allocated wells, and the 96-well plate was shaken for 1 h at room temperature. Following washing with PBST ten times, 100 μL of the TMB ELISA substrate solution was added to each well. After coloration for 15 min at 37 °C, the reaction was terminated by adding 50 μL of H2SO4 (2 mol L−1) per well. The whole mixture in each well was transferred to another 96-well plate, and the UV absorbance was recorded using a Labsystems 96-well plate reader (TECAN, Switzerland) in dual-wavelength mode (450–650 nm). The process is shown in Fig. 1(b). The inhibition rate (IC%) of triazophos at various concentrations was calculated using eqn (3).
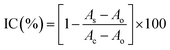 | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), followed by methanol, for the next BELISA procedure.
1), followed by methanol, for the next BELISA procedure.
2.7 Selectivity of BELISA
Cross-reactivity (CR) studies were carried out by measuring the competitive curves and selectivity properties of the structural analogs and several chemically related compounds. In this paper, parathion, chlorpyrifos, triadimefon, and methomyl were selected to evaluate the selectivity of BELISA. The CR was calculated using eqn (4).| CR% = {IC50(triazophos)/IC50(cross-reacting compound)} × 100 | (4) |
2.8 Sample preparation
To evaluate the performance of BELISA, some cabbage and apple samples were purchased from a local market to carry out the spiked recovery studies. Before spiking, the samples were determined to be free of triazophos by HPLC-MS/MS. The pretreatment of the cabbage and apple samples was based on the literature20 with slight modifications. Briefly, 10 g samples were weighed into 50 mL centrifuge tubes, which were spiked with triazophos at three levels, 10, 50, and 500 μg kg−1. Then 10 mL acetonitrile was added and shaken vigorously by a vortex for 2 min. After that, 4 g MgSO4 and 1 g NaCl were added and shaken for 2 min, and then centrifuged for 5 min at 5000 rpm. 2 mL of supernatant was transferred to a 10 mL centrifuge tube containing 100 mg PSA, 100 mg C18 and 300 mg MgSO4. After being shaken for 2 min, the mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. Half of the supernatant was used for HPLC-MS/MS detection and the other was dried using nitrogen. Finally, the residue was dissolved in 5% methanol-PBS (diluted 20 times) and analyzed by the BELISA method.
000 rpm for 5 min. Half of the supernatant was used for HPLC-MS/MS detection and the other was dried using nitrogen. Finally, the residue was dissolved in 5% methanol-PBS (diluted 20 times) and analyzed by the BELISA method.
2.9 HPLC analysis and HPLC-MS/MS validation
The HPLC analysis was carried out using a Waters 2695 Alliance HPLC system (Waters Corporation, Milford, MA, USA) equipped with a DAD detector at 246 nm for triazophos and 224 nm for triadimefon. Separation was carried out using a Waters XBridge C18 column (4.6 × 250 mm, particle size 5 μm). The column temperature was 25 °C. The mobile phase was water–acetonitrile (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80, v/v) for triazophos and triadimefon in isocratic mode at a flow rate of 0.8 mL min−1, and the injection volume was 10 μL.
80, v/v) for triazophos and triadimefon in isocratic mode at a flow rate of 0.8 mL min−1, and the injection volume was 10 μL.
HPLC-MS/MS analysis was carried out using an Agilent Series 1200 liquid chromatograph (Agilent Technologies, Palo Alto, CA, USA), with an API2000 triple quadrupole mass spectrometer equipped with a turbo-ion spray ionization source (Applied Biosystem/MDS Sciex, Foster City, CA, USA). The MS was operated in the multiple reaction monitoring (MRM) mode using positive ionization. Triazophos was identified in the positive scan mode at m/z 314.1, and the quantifier ion (m/z) and qualifier ion (m/z) were 162.2 and 286.0, whose declustering potential (DP, v/v) values were both 52.60, while the collision energy (CE, v/v) values were 24.96 and 28.36, respectively. The ion source temperature was 450 °C and the gas pressure of the collision activated dissociation (CAD), the curtain gas (CUR), ion source gas 1 (GS1), and ion source gas 2 (GS2) were 10, 50, 35, and 35 psi, respectively. The mobile phase consisted of 0.1% formic acid and 5 mM ammonium acetate in ultra-pure water (A)-acetonitrile (B) (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 v/v), using an XBridge C18 column (150 × 2.1 mm, 5 μm, Waters, Ireland) at a flow rate of 250 μL min−1. The injection volume was set to 5 μL.
60 v/v), using an XBridge C18 column (150 × 2.1 mm, 5 μm, Waters, Ireland) at a flow rate of 250 μL min−1. The injection volume was set to 5 μL.
3. Results and discussion
3.1 Optimizing the preparation of the MIP films
The synthesis conditions, including the mass ratio of template to functional monomer and the mass ratio of template to cross-linker, were optimized based on the adsorption capacity of the MIP/NIP films for triazophos. The MIP films were synthesized at different mass ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 1
4, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, and 1
5, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) of template to functional monomer and different mass ratios (1
6) of template to functional monomer and different mass ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and 1
4, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) of template to cross-linker. The results in Fig. 2(a) and (b) show that the imprinting factor reaches a maximal value of 1.17 and 1.51 at the mass ratio (1
5) of template to cross-linker. The results in Fig. 2(a) and (b) show that the imprinting factor reaches a maximal value of 1.17 and 1.51 at the mass ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) of template to functional monomer and the mass ratio (1
6) of template to functional monomer and the mass ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) of template to cross-linker, respectively.
3) of template to cross-linker, respectively.
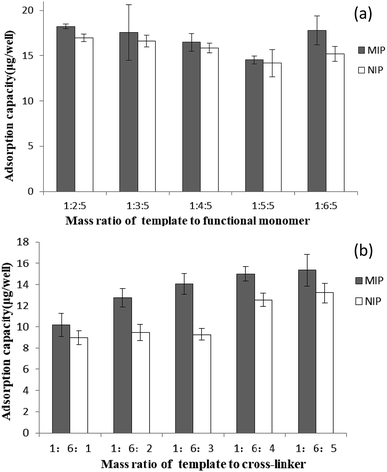 | ||
| Fig. 2 Adsorption capacities of the MIP and NIP films synthesized at different mass ratios of template to functional monomer (a); and different mass ratios of template to cross-linker (b). | ||
3.2 FT-IR spectra and SEM of the imprinted film
The FT-IR spectra of triadimefon (a), the imprinted film before extraction of the template (b), the imprinted film after extraction of the template (c) and the non-imprinted film (d) are shown in Fig. 3. In both spectra (a) and (b), there are peaks 670 cm−1 and 649 cm−1, which are characteristic of the benzene ring and the chlorine on the benzene ring in triadimefon, which are not found in spectra (c) and (d). Spectrum (c) is similar to spectrum (d). These results indicate that the template molecule was anchored on the imprinted polymer framework successfully and could be completely removed after extraction. Meanwhile, after the extraction of the template, the peak in the MIP spectrum near 830 cm−1 disappeared and the peak near 1726 cm−1 was weakened, as shown in Fig. 3(c), and the same peaks are present for the NIP.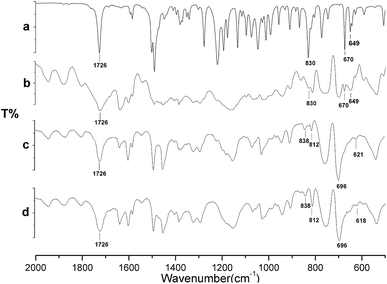 | ||
| Fig. 3 FT-IR spectra of triadimefon (a), the imprinted film before extraction (b), the imprinted film after extraction (c) and the non-imprinted film (d). | ||
In order to investigate the structure, the imprinted film before extraction (a), the imprinted film after extraction (b) and the non-imprinted film (c) were visualized using SEM (Fig. 4). The SEM images revealed that the surface of the imprinted film was markedly different from that after the removal of the template from the MIP. The surface of the imprinted film was much rougher than the non-imprinted film, which manifested that the specific binding sites could be formed in the imprinted film.
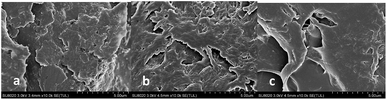 | ||
| Fig. 4 SEM images of the imprinted film before extraction (a), the imprinted film after extraction (b) and the non-imprinted film (c). | ||
3.3 Adsorption properties of the imprinted film
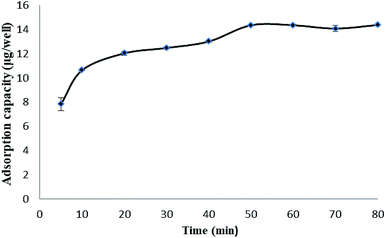
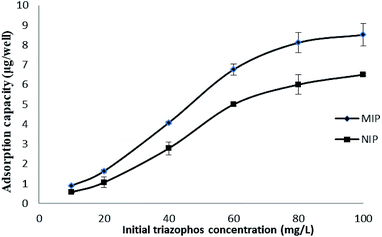
The adsorption dynamics of triazophos on the imprinted film was investigated in 100 mg L−1 of triazophos solution. As shown in Fig. 5, the imprinted film had a fast adsorption capacity, 74.1% of binding was achieved within a short period of 10 min, and an equilibrium value was reached at 50 min.Besides this, the isothermal adsorption curves of the imprinted and the non-imprinted films were obtained using a series of triazophos-PBS solutions (10–100 mg L−1), and are plotted in Fig. 6. The results indicate that the adsorption capacities of the imprinted and the non-imprinted films increased with the initial concentration, and the imprinted films exhibited higher adsorption capacities than the non-imprinted films at the same triazophos level.
3.4 Optimization of the BELISA conditions
In order to achieve optimal sensitivity and precision for the BELISA method, the parameters, including the order of adding the enzyme labelled probe and the standard solution, the number of times the plate was washed, the enzyme labelled probe concentration, the preparation of the solvent and the thickness of the imprinted film, were optimized in detail.The order of adding the enzyme labelled probe and the standard solution could affect the BELISA. The experimental results show that the competitive effect of the separate addition of the enzyme labelled probe and the standard solution was much better than the simultaneous addition of both of them. The number of times the plate was washed could affect the sensitivity of the method. In order to investigate the influence on the performance of the assay, the number of times the plate was washed (5, 10, and 15) was tested by comparing the inhibition. Washing ten times had a higher inhibition. Washing more times may reduce nonspecific adsorption to increase the competitive effect, but washing too much may wash away specific adsorption at the same time.
The sensitivity of the BELISA method can be affected by the concentration of the enzyme labelled probe. Different dilution ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1
100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, 1
200, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, 1
400, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600, 1
600, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, and 1
800, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
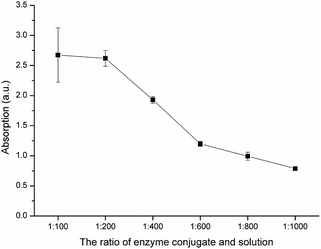
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) of the enzyme labelled probe were used in the BELISA procedure. Fig. 7 shows that the absorbance value decreased with decreasing concentration of the enzyme labelled probe. The concentration of the enzyme labelled probe is usually determined by the titre of enzyme tracers to yield absorbance values ranging from 1 to 1.2. In this study, the enzyme labelled probe was diluted to 1
1000) of the enzyme labelled probe were used in the BELISA procedure. Fig. 7 shows that the absorbance value decreased with decreasing concentration of the enzyme labelled probe. The concentration of the enzyme labelled probe is usually determined by the titre of enzyme tracers to yield absorbance values ranging from 1 to 1.2. In this study, the enzyme labelled probe was diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 before use.
600 before use.
The solvent used for the preparation of the standard solution and the samples could affect the selectivity and the recognition ability of the imprinted films, and the sensitivity of BELISA. In order to investigate the influence on the performance of the assay, methanol solutions at 0, 5, 10, and 15% in PBS were tested by comparing the inhibition and the IC50 value obtained from the standard curves. The results (Fig. 8) indicate that the inhibition ratio and the sensitivity were the highest in the 5% methanol PBS system. Therefore, the 5% methanol PBS solution was chosen as the preparation solvent for the routine analysis of triazophos in real samples.
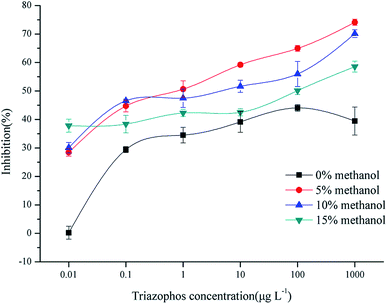 | ||
| Fig. 8 BELISA standard curves of triazophos using the imprinted film as an artificial antibody in PBS containing different concentrations of methanol. | ||
The thickness of the imprinted film may have a direct effect on the assay sensitivity. In order to investigate the influence, the coating amount of the imprinted film containing different volumes of pre-polymer solution (50, 100, and 200 μL) were tested. The results show that the imprinted film could not fully coat on the well surface of the 96-well plate using 50 μL of pre-polymer solution, and it was too thick and had a lower sensitivity using 200 μL. Thus, 100 μL of pre-polymer solution was added into each well of the plate to form a thin and uniform imprinted film.
3.5 Cross-reactivity of the BELISA method
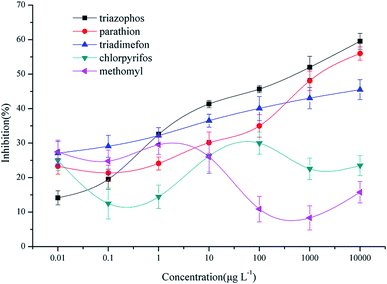
The cross-reactivity of the BELISA method was evaluated using competitive assays with related compounds. Parathion and chlorpyrifos were selected as organophosphorus pesticides with similar molecular structures to triazophos; triadimefon was selected as a dummy molecular template; methomyl was selected as a non-benzene ring compound. The results are shown in Fig. 9 and Table 1.Relative to triazophos, the CR of parathion and triadimefon are 7.47% and 0.25%, respectively. This result could indicate that the imprinted films have specific recognition capability toward compounds with a triazole ring, a benzene ring or a phosphoester bond when selecting triadimefon as the dummy template and the hapten of triazophos as the enzyme labelled probe. However, the CR values for parathion and triadimefon were less than 10%, which indicates that the imprinted films still have high selectivity toward triazophos. Besides this, the CR of chlorpyrifos and methomyl were less than 0.04%, which shows that the imprinted films have low sensitivity toward non phenyl pesticides.
3.6 Parameters of the BELISA method
The normalized competition curves obtained using the novel imprinted film and the non-imprinted film as the artificial antibody for triazophos at concentrations ranging from 0.001 μg L−1 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg L−1 in 5% methanol PBS solution are shown in Fig. 10. At the same triazophos concentration, the inhibition rate of the MIP was higher than that of the NIP. When the concentration of triazophos was 10
000 μg L−1 in 5% methanol PBS solution are shown in Fig. 10. At the same triazophos concentration, the inhibition rate of the MIP was higher than that of the NIP. When the concentration of triazophos was 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg L−1, the inhibition rate of the MIP was 59.5% and that of the NIP was only 19.9%. Thus, the MIP had better selectivity and adsorption capacity for triazophos than the NIP.
000 μg L−1, the inhibition rate of the MIP was 59.5% and that of the NIP was only 19.9%. Thus, the MIP had better selectivity and adsorption capacity for triazophos than the NIP.
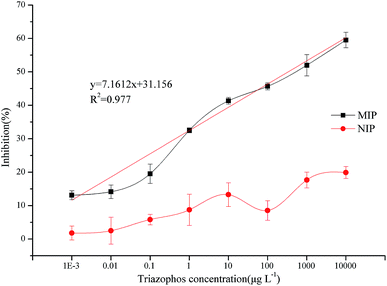 | ||
| Fig. 10 Triazophos BELISA standard curves using the novel imprinted film and the non-imprinted film as the antibody. | ||
Under optimal conditions, the limit of detection (LOD) and the sensitivity (IC50) were 0.001 μg L−1 and 428 μg L−1, respectively. The maximum residue level of triazophos in primary products in China is 0.05 mg kg−1. This method can meet the need of sensitivity in trace triazophos determination. Thus, the BELISA method is fully sensitive for detecting triazophos in real samples.
3.7 Real sample application
To evaluate the suitability and the applicability of BELISA for the measurement of real samples, cabbage and apple samples spiked at 10, 50, and 500 μg kg−1 were investigated (with three replicates for each concentration). The results are shown in Table 2. The recovery of BELISA ranged from 72.0 to 119.8% in cabbage and from 70.5 to 119.7% in apple, with the RSD of BELISA ranging from 15.6 to 17.5% in the cabbage sample and from 5.2 to 19.7% in the apple sample. The accuracy of BELISA was validated by comparative analysis of the spiked cabbage and apple samples by HPLC-MS/MS with a correlation coefficient (r2) of 0.9927. These results indicate that the BELISA method has good accuracy and precision for triazophos detection in agricultural products.| Sample | Spiking concentration (μg kg−1) | Recoveries/RSD (%) | |
|---|---|---|---|
| BELISA | HPLC-MS/MS | ||
| Cabbage | 10 | 72.0/15.6 | 85.3/15.0 |
| 50 | 115.1/17.4 | 100.9/13.6 | |
| 500 | 119.8/17.5 | 103.8/7.0 | |
| Apple | 10 | 119.7/18.9 | 78.6/2.8 |
| 50 | 70.5/5.2 | 105.2/3.5 | |
| 500 | 105.9/19.7 | 103.7/4.7 |
3.8 Comparison between various methods
Various methods for triazophos detection are compared in Table 3. We can see that BELISA has a wide linear range, a low LOD and low cost. Besides this, BELISA doesn’t need expensive instruments, skilled technicians and a hard prepared antibody. With these advantages, BELISA has potential for application in the quantitative determination of triazophos in agro-products.| Method | Scheme | Sample | Recovery (%) | RSD (%) | Linear range (μg L−1) | LOD (μg L−1) | Sensitivity (IC50, μg L−1) |
|---|---|---|---|---|---|---|---|
| MMIPs3 | Magnetic molecularly imprinted polymer, solid-phase extraction, GC analysis | Cabbage, lettuce, spinach, and amaranth | 94.4 | 3.8 | 0.1525–6260 | 0.10016 | — |
| ELISA21 | A direct competitive ELISA based on the THHe monoclonal antibody (McAb) and a heterologous enzyme tracer (THBu-HRP) | Water and soil | 92.0 | 11.4 | — | 0.02 | 0.21 |
| BBCCIA22 | Indirect competitive, AuNPs probe modified with a HRP-labeled bio-barcode, hybridization | Apple, cabbage, orange, and rice | 92.1 | 14.0 | 0.015–40 | 0.014 | 0.604 |
| CFBBCIA20 | Indirect competitive, AuNPs, probe modified with a FAM-labeled bio-barcode | Water, rice, cucumber, apple, and cabbage | 96.1 | 12.3 | 0.01–20 | 0.006 | 0.25 |
| BELISA | Direct competitive, enzyme-labeled hapten | Cabbage and apple | 100.5 | 15.7 | 0.001–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.001 | 428 |
4. Conclusions
A sensitive, simple, quick, and direct biomimetic enzyme-linked immunosorbent assay based on a dummy template for the determination of triazophos residues was developed. This method exhibited good performance in the detection of triazophos residues in agro-products after pretreatment. Under optimal conditions, the limit of detection (LOD) and the sensitivity (IC50) of the developed BELISA were 0.001 μg L−1 and 428 μg L−1, respectively. The methodological evaluation of this method showed that it has excellent recovery ranging from 70.5 to 119.8% and relative standard deviations (RSDs) ranging from 5.2 to 19.7%, when it was used to detect triazophos in cabbage and apple samples. This study also indicated that the developed novel BELISA had particular selectivity toward triazophos using the hapten of triazophos labelled as horseradish peroxidase.Conflicts of interest
All of the authors have declared no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Contact No. 31471654, 31772071,31401590) and the Key Laboratory of Agrifood safety and Quality for MOA (2016-KF-10), the Central Public-interest Scientific Institution Basal Research Fund (No. 1610072016009), and the China Agriculture Research System (CARS-05-05A-03).References
- C. Chen, Y. Wang, X. Zhao, Q. Wang and Y. Qian, Ecotoxicology, 2014, 23, 221–228 CrossRef PubMed.
- W. J. Gui, S. T. Wang, Y. R. Guo and G. N. Zhu, Anal. Biochem., 2008, 377, 202–208 CrossRef PubMed.
- H. Li, T. Xie, L. Ye, Y. Wang and C. Xie, Microchim. Acta, 2017, 184, 1011–1019 CrossRef.
- J. Xiong and B. Hu, J. Chromatogr. A, 2008, 1193, 7–18 CrossRef PubMed.
- Z. Huang, Y. Li, B. Chen and S. Yao, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 853, 154–162 CrossRef PubMed.
- R. Romero-Gonzalez, A. Garrido Frenich and J. L. Martinez Vidal, Talanta, 2008, 76, 211–225 CrossRef PubMed.
- Y. R. Guo, S. Y. Liu, W. J. Gui and G. N. Zhu, Anal. Biochem., 2009, 389, 32–39 CrossRef PubMed.
- C. Ramansuri, R. Boro, Y. Nangia, S. Gandhi, P. Sharma, N. Wangoo, K. Rajesh and G. Shekhawat, TrAC, Trends Anal. Chem., 2009, 28, 29–39 CrossRef.
- S. Wang, Z. Xu, G. Fang, Y. Zhang, B. Liu and H. Zhu, J. Agric. Food Chem., 2009, 57, 4528–4534 CrossRef PubMed.
- K. Mosbach, Trends Biochem. Sci., 1994, 19, 9 CrossRef PubMed.
- X. Kan, Y. Zhao, Z. Geng, A. Zhilin Wang and J. J. Zhu, J. Phys. Chem. C, 2008, 112, 4849–4854 Search PubMed.
- L. Li, Z. Lin, A. Peng, H. Zhong, X. Chen and Z. Huang, Food Chem., 2017, 229, 403–408 CrossRef PubMed.
- Q. Sun, L. Xu, Y. Ma, X. Qiao and Z. Xu, J. Sci. Food Agric., 2014, 94, 102–108 CrossRef PubMed.
- C. Shi, X. Liu, L. Song, X. Qiao and Z. Xu, Food Anal. Method, 2015, 8, 2496–2503 CrossRef.
- W. Lian, S. Liu, L. Wang and H. Liu, Biosens. Bioelectron., 2015, 73, 214–220 CrossRef PubMed.
- L. Cenci, C. Piotto, P. Bettotti and B. Maria, Talanta, 2018, 178, 772–779 CrossRef PubMed.
- D. Zhao, X. Qiao, Z. Xu, R. Xu and Z. Yan, J. Immunoassay Immunochem., 2013, 34, 16–29 CrossRef PubMed.
- J. Pan, X. Zou, X. Wang, W. Guan, Y. Yan and J. Han, Chem. Eng. J., 2010, 162, 910–918 CrossRef.
- W. J. Gui, R. Y. Jin, Z. L. Chen, J. L. Cheng and G. N. Zhu, Anal. Biochem., 2006, 357, 9–14 CrossRef PubMed.
- C. Zhang, P. Du, Z. Jiang, M. Jin, G. Chen, X. Cao, X. Cui, Y. Zhang, R. Li, A. M. Abd El-Aty and J. Wang, Anal. Chim. Acta, 2018, 999, 123–131 CrossRef PubMed.
- C. Liang, R. Jin, W. Gui and G. Zhu, Environ. Sci. Technol., 2007, 41, 6783 CrossRef PubMed.
- P. Du, M. Jin, G. Chen, C. Zhang, X. Cui, Y. Zhang, Y. Zhang, P. Zou, Z. Jiang, X. Cao, Y. She, F. Jin and J. Wang, Microchim. Acta, 2017, 184, 3705–3712 CrossRef.
| This journal is © The Royal Society of Chemistry 2018 |